Abstract
Euchromatin, which has an open structure and is frequently transcribed, tends to replicate in early S phase. Heterochromatin, which is more condensed and rarely transcribed, usually replicates in late S phase. Here, we report significant deviation from this correlation in the fission yeast, Schizosaccharomyces pombe. We found that heterochromatic centromeres and silent mating-type cassettes replicate in early S phase. Only heterochromatic telomeres replicate in late S phase. Research in other laboratories has shown that occasionally other organisms also replicate some of their heterochromatin in early S phase. Thus, late replication is not an obligatory feature of heterochromatin.
Keywords: Heterochromatin, DNA replication, timing, centromere, telomere, mating type
“Heterochromatin” was originally defined as chromatin that remains condensed during interphase, whereas “euchromatin” decondenses during interphase (Heitz 1928, 1929). Later, the discovery that genes from euchromatin can become epigenetically inactivated if they are translocated into heterochromatin (for review, see Grewal and Elgin 2002) provided an additional, functional definition for heterochromatin. Recent studies have begun to provide a molecular definition as well (for review, see Grewal and Elgin 2002). Many forms of heterochromatin are characterized by hypermethylation of Lys 9 on histone H3 (H3-K9). In these cases, a protein with a chromodomain and a chromo-shadow domain (similar to Drosophila Hp1 and fission yeast Swi6) binds to methylated H3-K9 with its chromodomain, and then recruits additional proteins to the heterochromatic region with its chromo-shadow domain.
Until now, replication in late S phase has been considered to be another distinguishing feature of heterochromatin. The paradigm of late heterochromatin replication was first articulated by Lima-de-Faria and Jaworska (1968) on the basis of studies in a wide range of eukaryotic organisms. With minor exceptions, this paradigm has withstood the test of time (for review, see Gilbert 2002). Now, however, we report significant deviation from this paradigm in fission yeast.
Results and Discussion
Previous results: telomeres
Chromatin near fission yeast telomeres is heterochromatic, because bringing normally active genes into the proximity of telomeres epigenetically inactivates the genes, and this inactivation is partially dependent on standard heterochromatin proteins including Swi6 (Nimmo et al. 1994; Allshire et al. 1995; Ekwall et al. 1995, 1996). Epigenetic inactivation of euchromatic genes introduced into telomere-proximal positions is called Telomere Position Effect (TPE) and is conserved from budding yeast (Gottschling et al. 1990; Tham and Zakian 2002) to humans (Baur et al. 2001).
Using several independent synchronization procedures, we have demonstrated previously that the terminal HindIII restriction fragments of fission yeast chromosomes I and II replicate in very late S phase (Kim and Huberman 2001), consistent with the paradigm of late heterochromatin replication. However, we were surprised when the same studies revealed that the outer portions of fission yeast centromeres replicate in early S phase.
Previous results: outer centromeres
Fission yeast centromeres consist of variable numbers of outer repeat (otr) sequences arranged, in inverted orientation, around an inner portion consisting of chromosome-specific innermost repeats (imr) inverted around a central sequence (cnt) that is fully or partially unique for each chromosome (Chikashige et al. 1989; Takahashi et al. 1992; Smith et al. 1995).
The otr region consists primarily of dg and dh repeats. These repeats are heterochromatic, because an indicator gene transplaced into otr becomes silenced (Allshire et al. 1995), and otr is associated with the heterochromatin protein, Swi6 (Partridge et al. 2000). Our previous results led to the surprising conclusion that the HindIII fragments from the dg repeats (also called K repeats) within the otr portions of the centromeres contain active chromosomal replication origins and replicate in very early S phase (Kim and Huberman 2001).
Inner centromeres
In fission yeast, the chromatin within the inner portions of centromeres is heterochromatic in the sense that euchromatic genes introduced into inner centromeres become epigenetically silenced (Allshire et al. 1994, 1995). However, in fission yeast as in all eukaryotic organisms, histone H3 within the inner centromeres is replaced by a histone H3 paralog, frequently called CENP-A (for review, see Smith 2002). Because the N-terminal portion of CENP-A differs substantially from that of histone H3, CENP-A is not subject to K9 methylation, and it does not bind Hp1/Swi6-like proteins. Instead, inner centromeres are associated with the Mis6 protein (Partridge et al. 2000; Takahashi et al. 2000; Kniola et al. 2001).
The surprising early replication of the outer portions of the centromeres raised the following question: Do the inner portions, which have a different heterochromatin structure, also replicate in early S phase? To answer this question, we used the cdc10 block and release procedure (Kim and Huberman 2001) to synchronize cells, and we used two-dimensional agarose gel electrophoresis (Brewer and Fangman 1987) to measure the abundance of replication intermediates (RIs). In Figure 1A (bottom panel), the course of S phase is indicated by the position of the flow cytometry peak, which shifts from a DNA content of 1N to 2N primarily between 50 and 90 min after temperature downshift. Examples of complete flow cytometry profiles for this experiment are shown in Figure 3 of Kim and Huberman (2001). We found that RIs (primarily Y arcs) from the cnt sequences were most abundant in early S phase—between 50 and 70 min after shift to 25°C. The weaker signals from RIs at earlier and later time points may be due to imperfect synchrony or may be indicative of some heterogeneity of replication times.
Figure 1.
The central portions of the centromeres are replicated in early S phase. (A) The cdc10 temperature block and release procedure (Kim and Huberman 2001) was used to synchronize passage through S phase. This experiment is identical to the one shown in Figure 4 in Kim and Huberman (2001). The Southern membrane used in that experiment was stripped and rehybridized with probes specific for the cnt sequences. (B) The hydroxyurea (HU) block and release procedure (Kim and Huberman 2001) was used to synchronize passage through S phase. This experiment is identical to the one shown in Figure 7 in Kim and Huberman (2001). The Southern membrane used in that experiment was stripped and rehybridized with probes specific for the cnt sequences. Note that the cells in the zero-minute time point had been exposed to HU for 5 h at 25°C.
We confirmed the early replication of cnt sequences by using the independent hydroxyurea (HU) block and release procedure. RIs corresponding to cnt sequences accumulate in the presence of HU (Fig. 1B; zero-minute time point). After HU is removed, cnt RIs disappear as replication forks run off. This behavior is typical of sequences that replicate in early S phase (Kim and Huberman 2001). Examples of flow cytometry profiles for this experiment are in Figure 6 of Kim and Huberman (2001).
Comparison of the cdc10 block and release results for cnt sequences in Figure 1A with our previously published cdc10 results for outer centromere (dg) sequences (Fig. 4B in Kim and Huberman 2001) suggests that the dg sequences replicate in very early S phase, whereas the cnt sequences replicate about 10 min later, but still in early S phase. The later replication of the cnt sequences is consistent with their having few, if any, active replication origins (note the near absence of bubble arc signals in Fig. 1). Our results suggest that the cnt sequences are primarily replicated by replication forks coming from active origins in the dg or dh sequences, all within early S phase.
Silent mating-type cassettes
The remaining well-characterized heterochromatic region in fission yeast is the region containing the silent mating-type cassettes. There are two mating types in fission yeast, plus and minus. Wild-type fission yeast cells have three sets of genes (cassettes) encoding mating-type information, Plus or Minus. The three cassettes—mat1, mat2, and mat3—are located within 30 kb of each other on chromosome II. This wild-type configuration, h90, is diagrammed in Figure 2 (top panel). The region between mat1 and mat2 is called the “L region,” and that between mat2 and mat3 is called the “K region.” The genes (Plus or Minus) in mat1 are expressed (mat1-P or mat1-M). Those in mat2 (usually Plus; mat2-P) and mat3 (usually Minus; mat3-M) are silenced by heterochromatinization that is partially dependent on Swi6. The silenced state is indicated in the diagram by a gray (rather than black) plus sign or minus sign. The silenced, heterochromatic region is located between two inverted repeats that form boundaries between heterochromatin and euchromatin (IR-L and IR-R; Noma et al. 2001; Singh and Klar 2002; Thon et al. 2002). Halfway between these inverted repeats is a stretch with strong sequence similarity to a portion of the otr of CEN2. This “CEN homology” domain contributes significantly to formation of regional heterochromatin (Hall et al. 2002).
Figure 2.
Structures of the major fission yeast mating-type locus configurations. These diagrams are based on nucleotide sequence information (GenBank accession nos. AL035065, AL356712, AL353012, and U57841) and on previous studies describing the derivation of h−S and h+N by recombination from h90 (Beach and Klar 1984). All three configurations are shown at the same scale. Restriction fragments studied for replication timing are indicated by horizontal boxes in the upper portions of the diagrams. The position of the hybridization probe used to identify each restriction fragment is shown as a thick black line immediately below the restriction fragment. The horizontally striped arrow in the CEN homology region represents dg sequences. The vertically striped arrow represents dh sequences, and the black arrowhead indicates an otr repeat that is frequently located between dh and dg sequences. See the text for additional description.
To measure replication timing in the mating-type region, we used fission yeast strains bearing the cdc10-M17 mutation, which facilitates cell cycle synchronization (Kim and Huberman 2001). These are haploid strains with alterations of their mating-type regions that inhibit mating-type switching. The alterations are called h−S and h+N, and they are diagrammed in Figure 2 (bottom panel). h−S strains arise from mat1-M h90 cells by recombination between homologous sequences at the borders of mat2-P and mat3-M, leading to deletion of the K region (Beach and Klar 1984). In h−S strains, the retained silent cassette contains Minus information. In Figure 2 (middle panel, h−S), the silent cassette is designated mat2:3-M, to indicate that the sequences to its left are identical to those to the left of mat2 in h90, and the sequences to its right are identical to those to the right of mat3 in h90. Similarly, h+N strains arise from mat1-P h90 (Beach and Klar 1984), but in this case, an extra K region is inserted by recombination into mat1. The leftmost cassette (mat1:2-P) in the h+N configuration remains transcriptionally active and expresses Plus information. The second cassette (mat3:1) is inactive for transcription, but it retains the cis-acting sequences necessary for mating-type switching, and it is capable of switching. The switching has no phenotypic consequences, as the second cassette is silenced.
The results of our analyses of mating-type region replication timing with cells synchronized by cdc10 block and release (Kim and Huberman 2001) are shown in Figure 3, and the locations of the tested fragments are shown as horizontal boxes in Figure 2 (middle and bottom panels). The probes used to detect the restriction fragments in Southern blotting are indicated by thick black lines immediately below these boxes. It is evident in Figure 3 that all tested fragments replicated in early S phase. The maximum abundance of RIs occurred at 40–60 min after temperature downshift. In contrast, the maximum abundance of RIs from telomeres occurred at 90–100 min after downshift in similar experiments (Kim and Huberman 2001).
Figure 3.
The mating-type region is replicated in early S phase. Locations of the studied restriction fragments are shown in Figure 2. (A) The cdc10 block and release procedure was used to synchronize the h−S strain. This experiment is identical to the one shown in Kim and Huberman (2001). The membrane was stripped and rehybridized with probes for the indicated restriction fragments. (B) The same procedure was used to synchronize the h+N strain. The experiment is the same as in Figure 5B in Kim and Huberman (2001). The membrane was stripped and rehybridized with probes for the indicated restriction fragments.
Two features of the results in Figure 3 merit comment. First, the probe used to detect restriction fragment 3b (EcoRI, 4.0 kb) also detects restriction fragment 3a (EcoRI, 3.9 kb; Figs. 2, bottom panel, 3B, top panel). These two fragments are too close in size to be resolved. Therefore, we cannot determine whether the bubble arc detected by this probe arises from fragment 3a, fragment 3b, or both. It is interesting that this probe also detects a spot near the apex of the Y arc (Fig. 3B, top panel). This spot is probably due to replication fork pausing in one or both of the detected restriction fragments.
Another result that deserves comment is the behavior of the two h+N EcoRI fragments (5a and 5b) detected by the probe used in the bottom two-dimensional gel panel of Figure 3B. Generation of fragment 5a requires an EcoRI site that is located within Minus—but not within Plus—sequences. It can be calculated from the information in Figure 2 (bottom panel) that the overall signal strength from fragment 5a should be three times greater than from fragment 5b. This ratio is consistent with our observations.
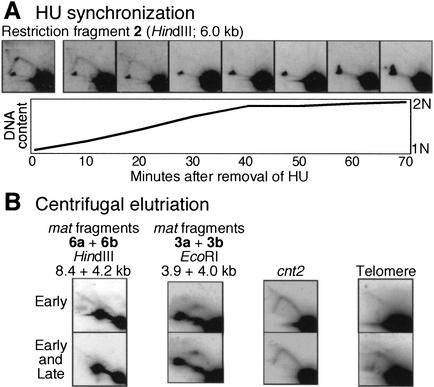
We have used two additional synchronization procedures to confirm the early replication timing of some of the restriction fragments from the mating-type region. Use of HU block and release showed that restriction fragment 2 (Fig. 2) accumulated RIs in the presence of HU, and the abundance of RIs decreased after HU was removed (Fig. 4A). Thus, this restriction fragment is also replicated in early S phase (Kim and Huberman 2001).
Figure 4.
Early replication of the mating-type region. (A) The HU synchronization procedure was used with the h−S strain. This experiment is identical to the one shown in Figure 7 in Kim and Huberman (2001). The membrane was stripped and rehybridized with a probe for restriction fragment 2 (Fig. 2). (B) The centrifugal elutriation procedure was used with the h+N strain. This experiment is the same as in Figures 1 and 2 in Kim and Huberman (2001). The membrane was stripped and rehybridized with probes for the indicated regions. See the text and Materials and Methods for further details on the probes used.
We found previously that sufficient synchronization of passage through S phase could be obtained by centrifugal elutriation to allow distinction between early- and late-replicating restriction fragments (Kim and Huberman 2001). In this procedure, the smallest cells in an unsynchronized population growing in rich medium are separated from the other cells by centrifugation and permitted to continue growing in rich medium. The small cells, which are initially in early G2 phase, continue through G2 and mitosis and then through G1 and S phase of the following cell cycle. Although the quality of the S-phase synchrony is not as good as with the other procedures described here, and only low numbers of cells are obtained, this procedure has the advantage that no cell cycle blocks are required. Consequently, this procedure should be free of potential artifacts that might be produced by temporarily blocking the cell cycle.
For the experiment in Figure 4B, two samples were collected from the synchronized culture for analysis of RIs. One sample, called “Early” in Figure 4B, contained mostly cells in early S phase with very few cells in late S phase. The other sample, called “Early and Late,” contained cells in both early and late S phase (Kim and Huberman 2001). The proportions of cells in early and late S phase in these two samples can be estimated from the quantitative fluorescence microscopy data in Figure 1 of Kim and Huberman (2001). The RIs detected by the 6a+6b and 3a+3b probes appear stronger in the “Early” sample, but the spot detected by the 3a+3b probe that is probably due to replication fork pausing appears stronger in the “Early and Late” sample, consistent with the relative apparent strengths of the Y arc and probable pause spot at different time points in the top panel of Figure 3B. Thus, both of the mating-type probes used in this experiment detect restriction fragments that are replicated predominantly in early S phase. For comparison, Figure 4B also shows results obtained with a probe specific for cnt2 (roughly equal signal strengths in the “Early” and “Early and Late” samples, consistent with cnt2 replication in mid-early S phase) and with a telomere probe (much stronger Y arc signals in the “Early and Late” sample, consistent with late telomere replication).
Occasional early heterochromatin replication in other organisms
Heterochromatin sometimes replicates early in other organisms, but examples in other organisms have been sufficiently rare that they have not called into question the paradigm of late heterochromatin replication. These examples include (1) early initiation of replication within bovine satellite I, which comprises 5% of bovine genomic DNA and is located in the centromeric heterochromatin of most chromosomes (Matsumoto and Gerbi 1982); (2) early initiation of replication in the pericentric heterochromatin and centromeres of mouse chromosomes (Holló et al. 1996); (3) early replication of some human telomeres (Wright et al. 1999), despite the fact that human telomeres exhibit TPE (Baur et al. 2001); (4) occasional early replication of autosomal genes inactivated by spreading of heterochromatin from a translocated human X chromosome (Sharp et al. 2001); (5) early replication of inner centromeres in Drosophila (Ahmad and Henikoff 2001, 2002; Sullivan and Karpen 2001); and (6) early replication of Drosophila β-heterochromatin (the form of heterochromatin containing occasional expressed genes and located immediately distal to pericentric α-heterochromatin; Schübeler et al. 2002).
No essential connection between heterochromatin and late replication
These examples from other organisms suggest that formation and maintenance of heterochromatin do not necessarily require late replication, but the surprising early replication of most heterochromatin in fission yeast forces us to recognize that the current paradigm of late heterochromatin replication requires revision. We do not make this statement lightly, because the association between heterochromatin and late replication is a strong one in most eukaryotic organisms. Yet the early-replicating heterochromatin in fission yeast appears to resemble heterochromatin in other organisms in all essential respects except replication timing. The best-studied fission yeast heterochromatin (in the outer centromeres and in the mating-type region) is physically associated with the Hp1-like Swi6 protein and with hypoacetylated histones. Its histone H3 is undermethylated on Lys 4 and overmethylated on K9. Normally active genes introduced into this heterochromatin become epigenetically inactivated (for review, see Grewal and Elgin 2002). The RNA interference machinery (RNAi) contributes to establishment and (in the case of centromeres) maintenance of this heterochromatin (Hall et al. 2002; Volpe et al. 2002). During interphase, the centromeres are condensed and associated with the spindle pole body at the nuclear periphery (Funabiki et al. 1993; Ekwall et al. 1995; Kniola et al. 2001). The mating-type region is also condensed and frequently associated with centromeres and/or telomeres at the nuclear periphery (Ekwall et al. 1995). Nevertheless, despite possessing all of these properties of classic heterochromatin, the fission yeast centromeres and mating-type region replicate in very early S phase. Thus, if there is a link between late-replication timing and heterochromatin, that link must involve a nonessential feature that is commonly, but not universally, associated with heterochromatin.
In many cell types, heterochromatin is located predominantly at the nuclear periphery, where it replicates in late S phase. For these reasons, the hypothesis that late replication is determined by peripheral nuclear positioning is currently popular (for review, see Gilbert 2002). As indicated above, however, peripheral location is not a guarantee of late replication timing in fission yeast. It is particularly interesting that in some fission yeast cells early-replicating mating-type regions appear to colocalize with late-replicating telomeres in regions that contain abundant Swi6 protein (Ekwall et al. 1995). This observation argues that cis-acting sequences (which may be involved in specifying type of heterochromatin) are likely to be more important than nuclear position as primary determinants of replication timing in fission yeast. Results from other laboratories suggest that the same may be true in budding yeast (Zappulla et al. 2002) and in mammals (Simon et al. 2001).
Materials and methods
Strains
Easily synchronized cdc10-M17 ade6-704 leu1-32 ura4-D18 strains [kindly sent to us by Tony Carr (Sussex University, Falmer, Brighton, UK)] were used for all experiments. The mating-type configuration was h−S unless indicated otherwise in the text and figure legends.
Cell cycle synchronization and flow cytometry
Synchronization by centrifugal elutriation, cdc10 block and release, and HU block and release was carried out as described previously (Kim and Huberman 2001). Evaluations of cell cycle position by flow cytometry or by quantitative fluorescence microscopy were as described (Kim and Huberman 2001).
Two-dimensional agarose gel analyses
Our procedures for purification of DNA from synchronized cell populations and for detection of RIs by two-dimensional gel electrophoresis have been described previously (Kim and Huberman 2001).
Hybridization probes
To detect telomeres, we used a previously described (Nimmo et al. 1994; Kim and Huberman 2001) 750-bp TAS probe. Cnt1 and cnt3 were detected using a 3.4-kb EcoRI–ClaI fragment from cnt1, whereas cnt2 was detected using a 2.15-kb EcoRI–PvuII fragment. The cnt probes were kindly sent to us by Mary Baum (University of California at Santa Barbara) and Louise Clarke (University of California at Santa Barbara). Probes for the mating-type region were obtained by appropriate restriction digestion of plasmids kindly sent to us by Amar Klar (Frederick Cancer Research and Development Center, Frederick, MD). In h−S cells, the 2.5-kb EcoRI and 6.0-kb HindIII fragments were detected by a 700-bp EcoRV–HindIII probe. In h+N cells, the 3.9- and 4.0-kb EcoRI fragments were detected by an 890-bp XbaI–HindIII probe. The 3.7-kb EcoRI fragment was detected by a 2.1-kb HindIII–EcoRI probe. The 5.7- and 8.1-kb EcoRI fragments, and the 4.2- and 8.4-kb HindIII fragments were detected with a 1.4-kb HindIII–NsiI probe.
Acknowledgments
We thank Elaine Nimmo, Janet Partridge, Robin Allshire, Tony Carr, Mary Baum, Louise Clarke, and Amar Klar for their generous gifts of strains, plasmids and advice. We thank Carl Schildkraut, Paolo Norio, Steve Henikoff, Kami Ahmad, Dave Gilbert, Mark Groudine, and Karuna Sharma for their constructive criticisms of the manuscript. This research was supported by grants to J.A.H. from the National Institutes of Health (GM49294, CA84302, and CA095908). We are grateful for support of the Roswell Park Cancer Institute Flow Cytometry Facility by the Roswell Park Cancer Center Support Grant (P30 CA16056-26).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL joel.huberman@roswellpark.org; FAX (716) 845-8126.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1046203.
References
- Ahmad K, Henikoff S. Centromeres are specialized replication domains in heterochromatin. J Cell Biol. 2001;153:101–109. doi: 10.1083/jcb.153.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci. 2002;99 (Suppl. 4):16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Javerzat J-P, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science. 2001;292:2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- Beach DH, Klar AJS. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J. 1984;3:603–610. doi: 10.1002/j.1460-2075.1984.tb01855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Composite motifs and repeat symmetry in S. pombe centromeres: Direct analysis by integration of NotI restriction sites. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R. The chromodomain protein Swi6: A key component of fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63:751–62. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Elgin SCR. Heterochromatin: New possibilities for the inheritance of structure. Curr Opin Genet Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K-I, Ayoub N, Cohen A, Grewal SIS. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Heitz E. Das Heterochromatin der Moose. Jahrb Wiss Botanik. 1928;69:762–818. [Google Scholar]
- ————— Heterochromatin, chromozentren, chromomeren. Ber Deutsch Bot Ges. 1929;47:274–284. [Google Scholar]
- Holló G, Keresõ J, Praznovszky T, Cserpán I, Fodor K, Katona R, Csonka E, Fatyol K, Szeles A, Szalay AA, et al. Evidence for a megareplicon covering megabases of centromeric chromosome segments. Chromosome Res. 1996;4:240–247. doi: 10.1007/BF02254965. [DOI] [PubMed] [Google Scholar]
- Kim S-M, Huberman JA. Regulation of replication timing in fission yeast. EMBO J. 2001;20:6115–6126. doi: 10.1093/emboj/20.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniola B, O'Toole E, McIntosh JR, Mellone B, Allshire R, Mengarelli S, Hultenby K, Ekwall K. The domain structure of centromeres is conserved from fission yeast to humans. Mol Biol Cell. 2001;12:2767–2775. doi: 10.1091/mbc.12.9.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-de-Faria A, Jaworska H. Late DNA synthesis in heterochromatin. Nature. 1968;217:138–142. doi: 10.1038/217138a0. [DOI] [PubMed] [Google Scholar]
- Matsumoto LH, Gerbi SA. Early initiation of bovine satellite I DNA replication. Exp Cell Biol. 1982;140:47–54. doi: 10.1016/0014-4827(82)90154-9. [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Cranston G, Allshire RC. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J. 1994;13:3801–3811. doi: 10.1002/j.1460-2075.1994.tb06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K-I, Allis CD, Grewal SIS. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Borgstrøm B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes & Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: A link between transcription and replication timing. Nature Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- Sharp A, Robinson DO, Jacobs P. Absence of correlation between late-replication and spreading of X inactivation in an X;autosome translocation. Hum Genet. 2001;109:295–302. doi: 10.1007/s004390100578. [DOI] [PubMed] [Google Scholar]
- Simon I, Tenzen T, Mostoslavsky R, Fibach E, Lande L, Milot E, Gribnau J, Grosveld F, Fraser P, Cedar H. Developmental regulation of DNA replication timing at the human β globin locus. EMBO J. 2001;20:6150–6157. doi: 10.1093/emboj/20.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Klar AJS. The 2.1-kb inverted repeat DNA sequences flank the mat2,3 silent region in two species of Schizosaccharomyces and are involved in epigenetic silencing in Schizosaccharomyces pombe. Genetics. 2002;162:591–602. doi: 10.1093/genetics/162.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM. Centromeres and variant histones: What, where, when and why? Curr Opin Cell Biol. 2002;14:279–285. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- Smith JG, Caddle MS, Bulboaca GH, Wohlgemuth JG, Baum M, Clarke L, Calos MP. Replication of centromere II of Schizosaccharomyces pombe. Mol Cell Biol. 1995;15:5165–5172. doi: 10.1128/mcb.15.9.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B, Karpen G. Centromere identity in Drosophila is not determined in vivo by replication timing. J Cell Biol. 2001;144:683–690. doi: 10.1083/jcb.200103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Tham WH, Zakian VA. Transcriptional silencing at Saccharomyces telomeres: Implications for other organisms. Oncogene. 2002;21:512–21. doi: 10.1038/sj.onc.1205078. [DOI] [PubMed] [Google Scholar]
- Thon G, Bjerling P, Bünner CM, Verhein-Hansen J. Expression-state boundaries in the mating-type region of fission yeast. Genetics. 2002;161:611–622. doi: 10.1093/genetics/161.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Liao ML, Shay JW. Normal human telomeres are not late replicating. Exp Cell Res. 1999;251:492–499. doi: 10.1006/excr.1999.4602. [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Sternglanz R, Leatherwood J. Control of replication timing by a transcriptional silencer. Curr Biol. 2002;12:869–875. doi: 10.1016/s0960-9822(02)00871-0. [DOI] [PubMed] [Google Scholar]