Abstract
Recently a Drosophila p53 protein has been identified that mediates apoptosis via a novel pathway involving the activation of the Reaper gene and subsequent inhibition of the inhibitors of apoptosis (IAPs). The present study found that CIAP1, a major mammalian homolog of Drosophila IAPs, is irreversibly inhibited (cleaved) during p53-dependent apoptosis and this cleavage is mediated by a serine protease. Serine protease inhibitors that block CIAP1 cleavage inhibit p53-dependent apoptosis. Furthermore, activation of the p53 protein increases the transcription of the HTRA2 gene, which encodes a serine protease that interacts with CIAP1 and potentiates apoptosis. These results demonstrate that the mammalian p53 protein may activate apoptosis through a novel pathway functionally similar to that in Drosophila, which involves HTRA2 and subsequent inhibition of CIAP1 by cleavage.
Keywords: Inhibitor of apoptosis (IAP), CIAP1, serine protease HTRA2, p53-dependent apoptosis, AEBSF, z-VAD
The p53 protein is a sequence-specific DNA-binding transcription factor whose function is to integrate a variety of stress signals (i.e., DNA damage, hypoxia, or oncogene activation) in a cell (Levine 1997). After DNA damage, p53 protein levels and activities rise, and a series of downstream target genes is transcribed at a high rate (Vogelstein et al. 2000; Jin and Levine 2001). These genes initiate programs of cell cycle arrest, apoptosis, DNA repair, cellular senescence, or antiangiogenesis. Among the cellular programs triggered by p53, apoptosis has been shown to be central for its tumor-suppressor function (Schmitt and Lowe 2002) in at least some tumors. There are many known p53-regulated genes that can contribute to apoptosis, including Bax, Fas ligand, Killer/DR5, Pig3, Puma, Perp, Noxa, and p53AIP-1 (Vogelstein et al. 2000). However, studies using knockout mice of many of these genes have indicated that none of these genes is entirely essential for p53-dependent apoptosis (Knudson et al. 1995; Zhang et al. 1998). It seems that p53 induces apoptosis in mammals by activating a highly redundant set of proapoptotic genes in a tissue-specific manner, in a scheme in which many of the p53-inducible genes are dispensable for this process. It remains formally possible, however, that some of the above genes that have not been tested in knockout models, or genes yet to be found, may play a major or indispensable role.
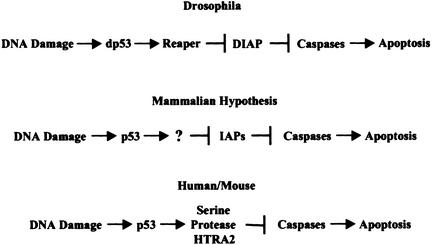
Recent studies have demonstrated that the p53 gene and its functions are conserved in evolution. Homologs of human p53 have been found in worms (Caenorhabditis elegans; Derry et al. 2001), flies (Drosophila; Brodsky et al. 2000; Jin et al. 2000; Ollmann et al. 2000), a clam (Mya arenaria; Van Beneden et al. 1997), and all vertebrates (Soussi et al. 1990). Amino acid sequence homology and biochemical analyses reveal that the p53 proteins from all the species are transcription factors containing a similar DNA-binding domain. Moreover, apoptosis is probably the most conserved cellular response induced by p53 in all of these organisms. A detailed investigation into the mechanisms by which p53 induces apoptosis in Drosophila has revealed a pathway not observed in mammalian systems to date (Fig. 1). In this pathway, p53 activates apoptosis via transcriptional activation of the Reaper gene, which encodes a protein that inhibits IAPs (inhibitors of apoptosis; Brodsky et al. 2000). IAPs are a family of proteins that contain one or multiple BIR (baculoviral IAP repeat) domains. Among other functions, IAPs are endogenous apoptosis inhibitors that interact with caspases, suppressing apoptosis by preventing procaspase activation and inhibiting the enzymatic activities of caspases (Deveraux and Reed 1999). Reaper is a protein composed of 65 amino acids, which, like other members of its family (Hid, Grim, and Sickle in Drosophila), contains an N-terminal tetrapeptide motif (AVAF) responsible for IAP interaction and relief of caspase inhibition (Wu et al. 2000). Moreover, recent studies showed that Reaper can also inhibit IAP by promoting DIAP1 ubiquitination and degradation (Holley et al. 2002; Yoo et al. 2002). Induction of the Reaper protein by p53 in Drosophila activates caspases, and consequently triggers apoptosis. Notably, this pathway seems to be the major p53-dependent apoptotic pathway in Drosophila, because loss of the Reaper locus completely blocks apoptosis induced by ionizing irradiation (Nordstrom et al. 1996).
Figure 1.
p53-dependent apoptotic pathway in Drosophila, working hypothesis in mammalian cells, and a similar novel pathway in human/mouse discovered in this study.
The Drosophila apoptotic machinery is by and large conserved in mammalian systems (Meier et al. 2000). Humans have at least five IAPs: CIAP1, CIAP2, XIAP, NAIP, and Survivin. Among these, CIAP1, CIAP2, and XIAP are structurally and functionally similar to the two major IAPs in Drosophila: DIAP1 and DIAP2 (Deveraux and Reed 1999). These proteins are widely distributed and involved in the regulation of apoptosis in a wide variety of tissues. Although no Reaper ortholog has been found in the human genome, two human proteins have been found to be functional homologs of Drosophila Reaper. Both of these contain the N-terminal Reaper-like motif (AVPI), which interacts with human IAPs and is required for promoting apoptosis. One protein is called Smac, a mitochondrial protein that interacts with and antagonizes IAPs after release from mitochondria (Du et al. 2000; Verhagen et al. 2000). The other protein is called HTRA2/Omi (Suzuki et al. 2001; Hegde et al. 2002; Martins et al. 2002; Verhagen et al. 2002), a unique serine protease with a PDZ domain that is conserved from bacteria to humans. HTRA2 requires an intrinsic serine protease activity for its proapoptotic function. Little is known about the processes that may use these proapoptotic proteins in humans.
The evolutionary conservation of the p53 protein and its function in inducing apoptosis, as well as the conservation of the apoptotic machinery, prompted us to hypothesize that the mammalian p53 protein may induce apoptosis via a pathway similar to that in Drosophila. In the present study we demonstrate that CIAP1, a mammalian IAP, is involved in p53-induced apoptosis. In both HeLa cells and primary mouse thymocytes, CIAP1 is cleaved by serine protease(s). Moreover, this cleavage is p53-dependent and is required for apoptosis. We also demonstrate that p53 activates transcription of the HTRA2 gene, which encodes just such a proapoptotic serine protease. Furthermore, the HTRA2 protein has been shown to interact with CIAP1 in mammalian cells via an N-terminal Reaper-like motif, which then activates apoptosis via its intrinsic serine protease activity. These data are consistent with a model wherein p53 induces apoptosis through a mammalian pathway functionally similar to that found in Drosophila (Fig. 1).
Results
CIAP1 cleaved during etoposide-induced apoptosis in HeLa cells
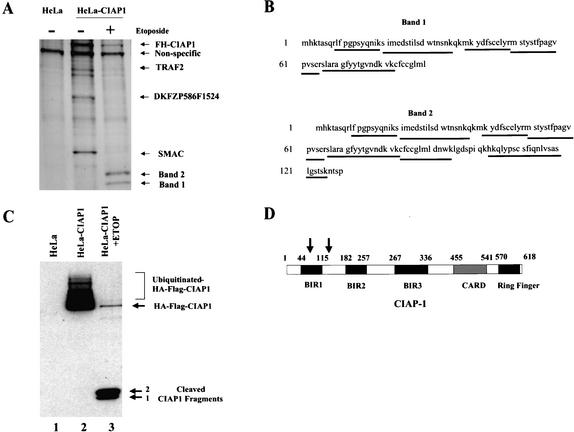
The Drosophila p53 pathway that initiates apoptosis responds to DNA damage by activating p53, which in turn initiates the transcription of the Reaper gene. The Reaper protein binds to DIAP, inactivating its ability to bind and inhibit caspases. The active caspases then initiate apoptosis (Fig. 1). The Reaper intermediate in this pathway is missing in mammalian cells, thus a study was initiated to possibly detect a protein that will bind to and/or inactivate the mammalian IAP (Fig. 1). CIAP1 was chosen as the human IAP bait to detect a Reaper-like protein because CIAP1 is an ortholog to Drosophila DIAP1 and DIAP2, and it is a major mammalian IAP with wide tissue distribution, including the thymus, where p53-dependent apoptosis occurs upon DNA damage. A mammalian expression vector was constructed that expresses an HA-Flag-tagged CIAP1 fusion protein, and was used to generate stable clonal HeLa cell lines. HeLa cells were chosen for expression because this line harbors a wild-type p53 (kept at a low level by HPV E6 protein), which can be activated by etoposide treatment. Moreover, HeLa cells are easy to expand and are therefore ideal for large-scale protein purifications. As described in Materials and Methods, the CIAP1 protein and its interacting proteins were copurified from HeLa cell lysates using M2-agrose beads that recognize and bind Flag-tagged proteins. Proteins associated with CIAP1 identified under normal growth conditions of HeLa cells include the previously reported CIAP1 interacting proteins TRAF2 (Rothe et al. 1995), SMAC (Du et al. 2000; Verhagen et al. 2000), and a protein of unknown function (DKFZP586F1524, accession no. NP_056399; Fig. 2A, lane 2). After the HeLa cells were treated with etoposide, two proteins of lower molecular weights were isolated by M2-agrose beads (Fig. 2A, lane 3). Surprisingly, these proteins were identified by mass spectrometry not as CIAP1-interacting proteins, but as N-terminal fragments of CIAP1 (Fig. 2B). The identities of these two small proteins were further confirmed by immunoblotting analysis (Fig. 2C). These results indicate that the CIAP1 protein is cleaved during etoposide-induced apoptosis in HeLa cells. Based on mass spectrometry (Fig. 2B), the approximate cleavage sites reside in the first BIR domain and at the junction between the first and second BIR domain of CIAP1 (Fig. 2D). Thus, p53 induction in HeLa cells resulted in CIAP1 cleavage.
Figure 2.
CIAP1 is cleaved in HeLa cells upon etoposide treatment. (A) Silver stain analysis of proteins immunoprecipitated from cell lysates of control untreated HeLa cells (lane 1), untreated HeLa-CIAP1 cells (lane 2) or etoposide-treated HeLa-CIAP1 cells (lane 3). The proteins were identified by MALDI-quadrupole ion trap mass spectrometric analysis using both MS and MS/MS modes. (B) MALDI-quadruple mass spectrometric analysis of the two small proteins purified from etoposide-treated HeLa-CIAP1 cell lysate (as indicated in A). Underlined sequences are tryptic peptides identified from each protein, all mapping to the N-terminal CIAP1 sequence. (C) Immunoprecipitated proteins from cell lysates of control untreated HeLa cells (lane 1), untreated HeLa-CIAP1 cells (lane 2), or etoposide-treated HeLa-CIAP1 cells (lane 3), analyzed by immunoblotting with anti-HA antibody. (D) Domain structure of CIAP1 and the approximate cleavage sites, as indicated by arrows. BIR, birculoviral IAP Repeat domain; CARD, caspase recruitment domain; Ring Finger domain.
CIAP1 cleavage, which is associated with p53 activation, is caspase-independent and is not the consequence of apoptosis
Two possible models may explain the mechanism by which CIAP1 is cleaved during the process of apoptosis. One model is that CIAP1 cleavage is a cause of apoptosis. In this model, activated p53 turns on the transcription of an unknown protease with a function similar to that of Drosophila Reaper (Fig. 1). This protease inhibits mammalian IAPs by their irreversible cleavage, resulting in caspase activation and apoptosis. An alternative model is that CIAP1 is simply cleaved as a consequence of apoptosis, probably by caspases that are activated during apoptosis. To distinguish between these two models, apoptosis was induced in HeLa cells by a different reagent, anti-Fas antibody, which is known to give rise to p53-independent apoptosis. Compared with etoposide treatment, which activates p53 and induces apoptosis (Fig. 3, lane 3), anti-Fas antibody induced apoptosis without activating p53 (Fig. 3, lane 2). The CIAP1 cleavage patterns in these two cases were strikingly different. The two characteristic cleavage products in etoposide-treated cells (two lower-molecular-weight bands as indicated as non-caspase-cleavage products in Fig. 3, lanes 3,4) were not present during Fas-antibody-induced apoptosis (Fig. 3, lane 2). These data suggest that the etoposide-induced CIAP1 cleavage is not a result of apoptosis in general, because it is not induced by Fas-antibody-induced apoptosis. To further test whether CIAP1 cleavage is not merely the consequence of apoptosis, z-VAD [z-val-ala-asp (Ome)-fluoromethylketone; Biomol], a general caspase inhibitor, was used to inhibit caspases and apoptosis induced by etoposide. As shown in Figure 3, treatment with the caspase inhibitor z-VAD inhibited apoptosis, as detected by PARP cleavage (Fig. 3, lane 4); however, it had no effect on etoposide-specific CIAP1 cleavage. This experiment directly demonstrates that the CIAP1 cleavage is not a result of apoptosis, and moreover, the CIAP1 cleavage is caspase-independent. In addition to the two major CIAP1 cleavage products that are specific to etoposide-induced apoptosis, there is a minor cleavage product with a higher molecular weight in etoposide-treated cells (Fig. 3, lane 3). This cleavage product also appears in Fas-antibody-treated cells (Fig. 3, lane 2) and is inhibited with the treatment of z-VAD (Fig. 3, lane 4), indicating that it is caused by caspases. These results suggested that CIAP1 is down-regulated differently in etoposide- and Fas-antibody-induced apoptosis. Etoposide treatment, which activates p53, leads to the down-regulation of CIAP1 mainly by a caspase-independent cleavage. In Fas-antibody-induced apoptosis, CIAP1 is cleared primarily by caspases, presumably as the result of apoptosis, and other mechanisms, probably ubiquitin-dependent protein degradation.
Figure 3.
Western blot analysis showing CIAP1 cleavage is associated with p53 induction and is caspase- and apoptosis-independent. Cell lysates from HeLa-CIAP1 with different treatments (as indicated on top: Fas-Ab, treatment of 1 μg/mL Fas antibody for 24 h; Etop, treatment of 10 μM etoposide for 24 h; z-VAD, treatment of 100 μg/mL z-VAD for 25 h; z-VAD + ETOP, treatment of 100 μg/mL z-VAD for 1 h prior to additional 10 μM etoposide treatment another 24 h) were analyzed by immunoblot assay with different antibodies (as indicated on the left side). The identities of each band are indicated to the right. The results are representative data from three independent experiments.
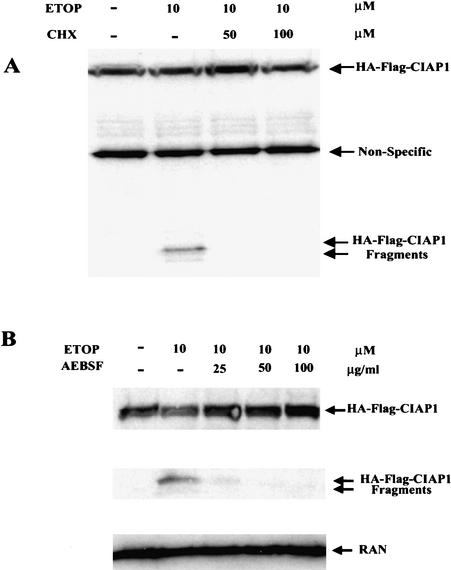
CIAP1 cleavage requires de novo protein synthesis and is dependent on serine proteases
To test whether CIAP1 cleavage requires de novo protein synthesis, which is one of the characteristics of p53-dependent apoptosis, cells were treated with cycloheximide. As shown in Figure 4A, cycloheximide treatment blocked CIAP1 cleavage, indicating the requirement of de novo protein synthesis for CIAP1 cleavage. Because the majority of CIAP1 cleavage upon etoposide treatment is not caused by caspases (as shown in Fig. 3), we tested the involvement of other types of proteases. In addition to cysteine proteases, which include caspases, certain serine proteases such as granzyme B have been reported to be involved in activating apoptosis (Russell and Ley 2002). To test whether serine proteases are involved in the CIAP1 cleavage process, we used a general serine protease inhibitor, AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride; Roche], which effectively inhibits serine proteases but not other types of proteases. As shown in Figure 4B, CIAP1 cleavage induced by etoposide is inhibited by AEBSF. Experiments with another serine protease inhibitor with distinct structure, TCPK (N-tosyl-L-phenylalanine chloromethylketone; Sigma), gave essentially the same results (data not shown). Also, ubiquitin-dependent protein degradation plays no role in cleaving CIAP1 because treatment with the proteosome inhibitor lactacystin (Calbiochem) had little effect on the CIAP1 cleavage (data not shown). The inhibitory effect of AEBSF on CIAP1 cleavage seems to be direct, because it also blocks CIAP1 cleavage in an in vitro assay that is comprised of cell lysate from etoposide-treated HeLa cells and affinity-purified Flag-HA-tagged CIAP1 (data not shown). These data indicate that CIAP1 is cleaved by a serine protease induced by etoposide treatment.
Figure 4.
(A) CIAP1 cleavage requires de novo protein synthesis. (B) CIAP1 cleavage induced by etoposide is inhibited by the serine protease inhibitor AEBSF. Cell lysates from HeLa-CIAP1 cells without or with different treatments (as indicated) were immunoblotted with anti-HA antibody, or reprobed with anti-RAN antibody as indicated. The results are representative data from three independent experiments.
The serine protease inhibitor AEBSF, which inhibits CIAP1 cleavage, blocks apoptosis induced by etoposide
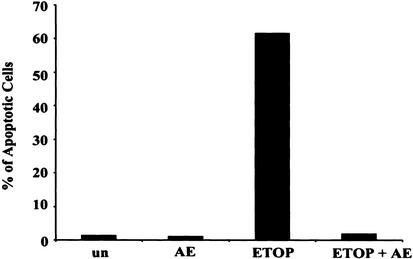
The results presented above suggest a direct involvement of CIAP1 in p53-dependent apoptosis. To explore whether the serine protease-dependent CIAP1 cleavage is a causal event leading to apoptosis in etoposide-treated HeLa cells, these cells were treated with this DNA-damaging agent and AEBSF. As shown in Figure 5, pretreatment with AEBSF blocks etoposide-dependent CIAP1 cleavage and totally inhibits apoptosis, suggesting that serine protease-dependent CIAP1 cleavage is one of the causal events that is essential for apoptosis induced by etoposide treatment in HeLa cells.
Figure 5.
Serine protease AEBSF inhibits apoptosis induced by etoposide treatment in HeLa cells. HeLa-CIAP1 cells were treated as indicated (un, untreated; ETOP, 10 μM etoposide treatment for 24 h; AE, 100 μg/mL AEBSF treatment for 25 h; AE + ETOP, 100 μg/mL AEBSF was added 1 h prior to additional 10 μM etoposide for another 24 h), followed by TUNEL analysis to determine the apoptotic profile. The results are representative data from two independent experiments.
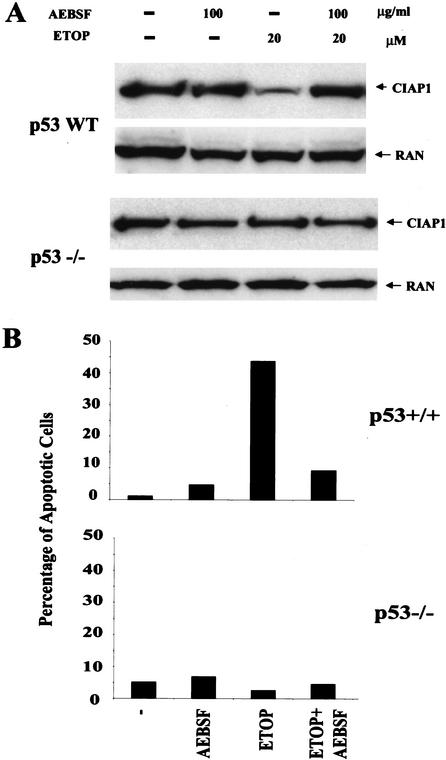
CIAP1 cleavage in primary mouse thymocytes is serine protease-dependent and is essential for p53-dependent apoptosis
It has been well established that DNA damage by etoposide treatment induces p53-dependent apoptosis in primary mouse thymocytes (Lowe et al. 1993; Soengas et al. 1999). The involvement of a serine protease-dependent CIAP1 cleavage in apoptosis in mouse primary thymocytes was therefore analyzed. As shown in Figure 6A, etoposide treatment led to the reduction of endogenous CIAP1 in primary thymocytes derived from wild-type 129sv mice but not in primary thymocytes from isogenic 129sv p53 null mice. In addition, this reduction in murine thymocytes was inhibited by treatment with the serine protease inhibitor AEBSF, indicating it was caused by a serine protease cleavage. Furthermore, AEBSF treatment of mouse thymocytes, which inhibits CIAP1 cleavage (Fig. 6A), totally inhibits apoptosis induced by etoposide (Fig. 6B). These data strongly suggest that a serine protease-dependent CIAP1 cleavage plays an important role in p53-dependent apoptosis in mouse primary thymocytes.
Figure 6.
Serine protease inhibitor AEBSF blocks p53-dependent CIAP1 cleavage and apoptosis in primary mouse thymocytes. (A) Immunoblot analysis of cell lysates from primary mouse thymocytes [p53 wild type (WT) or p53−/− as indicated] without or with different treatments (ETOP, 20 μM etoposide treatment for 8 h; AEBSF, 100 μg/mL AEBSF was added to the medium 1 h prior to additional 20 μM etoposide treatment for another 8 h) using CIAP1 antibody. (B) Apoptotic cell population in primary mouse thymocytes [p53 wild type (WT) or p53−/−, respectively] upon similar treatment in A, as determined by PI staining and Facs analysis. The results are representative data from two independent experiments.
HTRA2, a serine protease previously demonstrated to interact with CIAP1, and also previously implicated in apoptosis, is induced by p53
Several groups have independently discovered that HTRA2/OMI, a serine protease containing a PDZ domain, interacts with IAPs in mammalian cells using a Reaper IAP-interacting motif (AVPS) and promotes apoptosis. Moreover, the catalytic activity of HTRA2 is essential for this activation of apoptosis. To test whether this HTRA2 serine protease is regulated by p53, a Northern blot analysis was carried out to examine HTRA2 mRNA levels in HeLa cells upon etoposide treatment. As shown in Figure 7A, HTRA2 mRNA increased by sevenfold upon etoposide treatment, as normalized against GAPDH mRNA. To further test whether this increase is caused by p53 activation, HeLa and H1299 cells (a human lung epithelial cell line that lacks endogenous p53) were transfected with a p53 expression vector. The levels of HTRA2 mRNA were then examined with or without exogenous p53 expression. As shown in Figure 7B, ectopic expression of p53 increased HTRA2 mRNA by approximately sevenfold, suggesting that HTRA2 is a p53 downstream target gene.
Figure 7.
mRNA of HTRA2 in cultured cells increases upon etoposide treatment or p53 expression. (A) Northern blot analysis of HTRA2 mRNA induction upon etoposide treatment. Total RNA was harvested from HeLa cells without or with etoposide treatment for the indicated amount of time. mRNA of HTRA2 or GAPDH genes was detected using 32P-labeled cDNA probes, quantified by phosphorimager. The fold of induction was normalized against the GAPDH level. (B) RT–PCR analysis of HTRA2 and β-actin mRNA from HeLa cells or H1299 cells transfected with p53 expression vector or their respective control (mock-transfected) cells. Intensity of the radioactive species was quantified by PhosphorImager. The fold of induction was normalized against β-actin. These data are representative results from two independent experiments.
Discussion
This investigation was prompted by the discovery and characterization of a Drosophila p53 protein (Brodsky et al. 2000; Jin et al. 2000; Ollmann et al. 2000) and a novel p53-dependent apoptotic pathway in Drosophila. In addition to sequence homology and structural similarity, Drosophila p53 is biochemically and functionally similar to mammalian p53 in DNA binding, transcriptional activation, and apoptosis activation. Interestingly, Drosophila p53 activates apoptosis in a pathway that had not been observed in mammals. Drosophila p53 activates the transcription of the Reaper gene, which encodes a small protein with an N-terminal tetrapeptide motif that interacts with DIAPs. The Reaper protein thus relieves caspase inhibition by DIAP and activates apoptosis. In both HeLa and primary mouse thymocytes, the experiments described here demonstrate that CIAP1 is inactivated by protease cleavage in a p53-dependent manner. This cleavage is caused by serine proteases and requires de novo protein synthesis. Moreover, inhibition of CIAP1 cleavage by the serine protease inhibitor AEBSF totally blocks p53-dependent apoptosis. Finally, the activation of p53 has been shown to increase the transcription of the HTRA2 gene, which encodes a serine protease that has previously been shown to interact with CIAP1 and potentiate apoptosis. These data define a sequential pathway in mammalian cells in which DNA damage activates p53, which, in turn, increases the transcription of the serine protease gene HTRA2. The HTRA2 protein then interacts with and cleaves CIAP1, relieving caspase inhibition and activating apoptosis. This is a pathway that is functionally similar to that found in Drosophila, because in both pathways, p53 activates transcription of a protein that antagonizes and down-regulates an IAP, leading to caspase activation and apoptosis. In Drosophila, the mechanism for IAP down-regulation is through ubiquitin-dependent degradation, whereas in mammalian cells, CIAP1 inhibition is achieved through protein cleavage.
The IAPs are a family of evolutionally conserved proteins containing one or multiple BIR (baculoviral IAP repeat) domains. In mammalian cells, CIAP1, CIAP2, and XIAP have been shown to play important roles in regulating apoptosis, although the different functions of these IAPs are not clear. The deregulation of IAPs has been reported in cancers, for example; IAPs are overexpressed in cell lines derived from a variety of leukemias, lymphomas, and other cancers (Tamm et al. 2000); and the CIAP1 gene is amplified in esophageal squamous cell carcinomas (Imoto et al. 2001). Also, a chromosome translocation, t(11,8)(q21,q21), fuses a human IAP gene CIAP2 to the MLT1/MALT1 locus, leading to the mucosa-associated lymphoid tissue (MALT) lymphoma in humans (Dierlamm et al. 1999; Stoffel and Le Beau 2001). This study demonstrates that IAPs are directly involved in p53-dependent apoptosis, and it will now be useful to explore the relationship of p53 mutations and IAP amplifications in human cancers. Consistent with this observation is that CIAP1 overexpression is associated with greater resistance to DNA-damaging chemotherapeutic drugs (Tamm et al. 2000) in many cancer cells.
HTRA2 is a member of the HTRA serine protease protein family, which contains a C-terminal PDZ domain and a central serine protease domain. There are three members in humans, HTRA1, HTRA2, and HTRA3 (Li et al. 2002). Proteins in this family share homology to the bacterial HTRA (high-temperature requirement A) protein, which is essential for bacteria to survive at high temperature. Interestingly, this bacterial protein has molecular chaperon activity at lower temperatures and serine protease activity at higher temperatures. Its function seems to be acting on denatured proteins under stressful conditions, either by refolding (chaperon) or destroying (protease) the denatured proteins. The mammalian HTRA2 has been reported to localize in different cellular compartments, including mitochondria (Suzuki et al. 2001; Hegde et al. 2002; Martins et al. 2002; Verhagen et al. 2002), nuclei (Gray et al. 2000), and the endoplasmic reticulum (Faccio et al. 2000b). Transcription of HTRA2 has been shown to be up-regulated in kidney ischemia in humans (Faccio et al. 2000a), and it is also induced by a variety of cellular stresses, including high temperature and tunicamycin treatment (Gray et al. 2000). The identification of HTRA2 in mammalian cells as a downstream target gene of p53 suggests that p53 could also be involved in modifying other stress responses, including the cellular responses to increased temperature, protein unfolding, or protein aggregation.
Material and methods
Plasmids, cell lines, and cell culture
Flag-HA-tagged CIAP1 expression vector pFH-CIAP1 was constructed by inserting the CIAP1 cDNA into the pFH-IRESneo vector (Teichmann et al. 2000). The CIAP1 cDNA was PCR-amplified using oligonucleotides Nhe-CIAP1: 5′-ACGATGC TAGCATGCACAAAACTGCCTCCCAAAG-3′ and EcoRI-CI AP1: 5′-ACCGGAATTCGTTAAGAGAGAAATGTACGAAC AG-3′ with pCDNA3-6-myc CIAP1 as template. The PCR product was gel-purified and digested with NheI/EcoRI restriction enzymes, and ligated into NheI/EcoRI-digested pFH-IRESneo vector. The final construct was sequence verified by the sequencing facility at Rockefeller University.
The HeLa-S cell line (Teichmann et al. 2000) was maintained in DMEM medium supplemented with 10% fetal calf serum, 10 units/mL penicillin, and 10 μg/mL streptomycin. The HeLa-CIAP1 cell line was generated by transfecting HeLa-S cells with pFH-CIAP1 vector using calcium phosphate precipitation methods and selected with 500 μg/mL of G418 essentially as described (Kindston 1997). Individual colonies were picked and amplified. The chosen clone expresses a moderate level (compared with endogenous level ) of Flag-HA-tagged CIAP1, as detected by Western blot assay using M2, anti-HA, and CIAP1 antibodies, and was established for a large scale of protein purification. The HeLa-CIAP1 cells were maintained with medium as described above supplemented with 250 μg/mL G418.
Primary thymocytes from 129sv wild-type mice or p53−/− mice were immediately cultured in RPMI medium supplemented with 10% fetal calf serum, 10 units/mL penicillin, 10 μg/mL streptomycin, and 50 μM 2-mercaptomethanol. For experimental treatments, cells were treated with 100 μg/mL AEBSF (Sigma) alone for 9 h, or 20 μM etoposide (Sigma) for 8 h, or 100 μg/ml AEBSF was added to the medium 1 h before 20 μM etoposide (Sigma) was added, and the cells were further grown for another 8 h.
Affinity purification of proteins interacting with Flag-HA-tagged CIAP1 and mass spectrometry
HeLa-CIAP1 cells (or control HeLa-S cells) were grown on 150-mm-diameter plates to 80% confluency, followed by either treatment with 10 μM etoposide for 24 h or growing for another 24 h without treatment. The cells were then scraped in ice-cold 1× PBS, washed twice with PBS by centrifugation at 300g, resuspended in cold BC solution (20 mM Tris-HCl at pH 7.9, 4°C, 20% glycerol, 0.2 mM EDTA, 0.5 mM PMSF, 1 mM dithiothreitol; 4 mL/109 cells) with 100 mM KCl, 0.2% NP-40, and protease inhibitors (Complete, 1 tablet/10 mL, Roche Applied Sciences), and lysed by 10 strokes of a dounce homogenizer (B-type pestle). The homogenate was checked microscopically for cell lysis and centrifuged for 10 min at 10,000g to pellet nuclei. The supernatant was collected and centrifuged for 60 min at 100,000g (Sovall). The supernatant was then filtered through a 0.45-μm syringe filter (Nalgene). Next 25 μL of M2 agarose beads was added into 20 mL of lysate and tumbled overnight at 4°C. The beads were then washed five times in BC solution with 300 mM KCl and 0.1% NP-40, and eluted at 4°C with 25 μL of Flag peptide solution (300 μg/mL, dissolved in BC solution with 100 mM KCl). Then 2 μL of eluate was analyzed by 4%–20% gradient polyacrylamide gel electrophoresis (Novex) and silver stain (Invitrogen) as recommended by the vendor. The rest of the eluate was separated by polyacrylamide gel electrophoresis (Novex precast mini gel), and Zn-stained (Castellanos-Serra et al. 1999). The corresponding bands were excised out and digested with trypsin. The tryptic peptides were analyzed and identified by using a novel MALDI-quadrupole mass spectrometer in single stage mass spectrometry (MS) and tandem mass spectrometry (MS/MS) modes (Krutchinsky et al. 2001).
Immunoblot (Western) blot analysis, Northern blot analysis, and semiquantitative PCR
Cell lysates or purified proteins were separated by 4%–20% gradient polyacrylamide gel electrophoresis (Novex) and transferred to Immobilon-P membrane (Millipore). Immunoblot and ECL (Enhanced Chemiluminescence) were performed as recommended by the vendor (Amersham). The antibodies and dilutions used are the following: HA (Covance Mono HA.11 MMS-101P), 1:1,000; CIAP1 (Pharmingen, 66791A), 1:1000; PARP (Santa Cruz, SC-7150), 1:500; p53 (Santa Cruz, SC-126), 1:500; and RAN (Santa Cruz, SC-1156), 1:2000.
RNA was purified from HeLa cells or mouse thymocytes using Trizol Reagent (Life Technology) as recommended by the vendor. A total of 10 μg of RNA was separated by agarose gel electrophoresis (Kindston 1997), and Northern blotting was performed as described (Kindston 1997). The cDNA fragment of HTRA2 to be used as a Northern blot probe was excised from pCMV-Sport6 HTRA2 EST clone (accession no. AI571444, sequence verified) using EcoRI enzyme. The cDNA fragment of the GAPDH gene for the Northern blot probe was an EcoRI–SacI fragment from pSGX (Jin and Scotto 1998). Semiquantitative PCR was performed as described (Hoh et al. 2002) using either HTRA2 or β-actin-specific oligonucleotide pairs, HTRA2: 5′-ACTTATGGGACCCCCAGTCT-3′ and 5′-CGATATAGAC CACGGCAGGT-3′; β-actin, 5′-TCACCCTGAAGTACCCCA TC-3′ and 5′-CTCCTTAATGTCACGCACGA-3′.
Flow cytometry and TUNEL assay
For flow cytometric analysis of apoptosis by propidium iodide staining, floating and attached cells were collected at the indicated times after etoposide or AEBSF treatment, combined, and washed three times in PBS, and fixed overnight or longer in 80% ethanol at −20°C. Fixed cells were washed twice in PBS/1% BSA/0.1% Tween, and stained in 1 mL of PBS containing 50 μg of propidium iodide and 10 μg of boiled RNase, at 4 °C for 1 h or longer. A total of 5 × 105 cells were analyzed on a flow cytometer (FACScalibur, BD). For flow cytometric analysis of apoptosis by TUNEL staining, floating and attached cells were combined and washed three times in PBS, and then fixed in 2% formaldehyde at room temperature for 1 h. Fixed cells were washed three times in PBS, and transferred to 80% ethanol at −20°C for overnight or longer. TUNEL staining was performed using the In Situ Cell Death Detection Kit (Roche), as per the manufacturer's protocol. A total of 5 × 105 cells were analyzed on a flow cytometer (FACScalibur, BD).
Acknowledgments
We thank Angelika Teresky for excellent technical assistance, Kenan Onel for critical reading of the manuscript, Eric Lacasse for the CIAP1 cDNA, and all members of the Arnold J. Levine lab for helpful discussion. S.J. is supported by NIH grant P01CA87497. M.K. and B.T.C. are supported by NIH grant 1R33CA89810-01.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL alevine@ias.edu; FAX (609) 924-7592.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1047003.
References
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the Reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- Castellanos-Serra L, Proenza W, Huerta V, Moritz RL, Simpson RJ. Proteome analysis of polyacrylamide gel-separated proteins visualized by reversible negative staining using imidazole-zinc salts. Electrophoresis. 1999;20:732–737. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<732::AID-ELPS732>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: Role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Reed JC. IAP family proteins—Suppressors of apoptosis. Genes & Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21)p6 associated with mucosa-associated lymphoid tissue lymphomas. Blood. 1999;93:3601–3609. [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Faccio L, Fusco C, Chen A, Martinotti S, Bonventre JV, Zervos AS. Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J Biol Chem. 2000a;275:2581–2588. doi: 10.1074/jbc.275.4.2581. [DOI] [PubMed] [Google Scholar]
- Faccio L, Fusco C, Viel A, Zervos AS. Tissue-specific splicing of Omi stress-regulated endoprotease leads to an inactive protease with a modified PDZ motif. Genomics. 2000b;68:343–347. doi: 10.1006/geno.2000.6263. [DOI] [PubMed] [Google Scholar]
- Gray CW, Ward RV, Karran E, Turconi S, Rowles A, Viglienghi D, Southan C, Barton A, Fantom KG, West A, et al. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur J Biochem. 2000;267:5699–5710. doi: 10.1046/j.1432-1327.2000.01589.x. [DOI] [PubMed] [Google Scholar]
- Hegde R, Srinivasula SM, Zhang Z, Wassell R, Mukattash R, Cilenti L, DuBois G, Lazebnik Y, Zervos AS, Fernandes-Alnemri T, et al. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein–caspase interaction. J Biol Chem. 2002;277:432–438. doi: 10.1074/jbc.M109721200. [DOI] [PubMed] [Google Scholar]
- Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol. 2002;4:439–444. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto I, Yang ZQ, Pimkhaokham A, Tsuda H, Shimada Y, Imamura M, Ohki M, Inazawa J. Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res. 2001;61:6629–6634. [PubMed] [Google Scholar]
- Jin S, Levine AJ. The p53 functional circuit. J Cell Sci. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- Jin S, Scotto KW. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Martinek S, Joo WS, Wortman JR, Mirkovic N, Sali A, Yandell MD, Pavletich NP, Young MW, Levine AJ. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc Natl Acad Sci. 2000;97:7301–7306. doi: 10.1073/pnas.97.13.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindston RE. Preparation and analysis of RNA, pp. 4.9.1–4.9.13 and Introduction of DNA into mammalian cells, pp. 9.1.5–9.1.7. In: Ausubel FM, et al., editors. Current protocols in molecular biology. New York: John Wiley & Sons; 1997. [Google Scholar]
- Knudson CM, Tung KS, Tourtellotte WG, Brown GA, Korsmeyer SJ. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- Krutchinsky AN, Kalkum M, Chait BT. Automatic identification of proteins with a MALDI-quadrupole ion trap mass spectrometer. Anal Chem. 2001;73:5066–5077. doi: 10.1021/ac010682o. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Li W, Srinivasula SM, Chai J, Li P, Wu JW, Zhang Z, Alnemri ES, Shi Y. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat Struct Biol. 2002;9:436–441. doi: 10.1038/nsb795. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- Martins LM, Iaccarino I, Tenev T, Gschmeissner S, Totty NF, Lemoine NR, Savopoulos J, Gray CW, Creasy CL, Dingwall C, et al. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J Biol Chem. 2002;277:439–444. doi: 10.1074/jbc.M109784200. [DOI] [PubMed] [Google Scholar]
- Meier P, Finch A, Evan G. Apoptosis in development. Nature. 2000;407:796–801. doi: 10.1038/35037734. [DOI] [PubMed] [Google Scholar]
- Nordstrom W, Chen P, Steller H, Abrams JM. Activation of the reaper gene during ectopic cell killing in Drosophila. Dev Biol. 1996;180:213–226. doi: 10.1006/dbio.1996.0296. [DOI] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2–TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu Rev Immunol. 2002;20:323–370. doi: 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed] [Google Scholar]
- Schmitt CA, Lowe SW. Apoptosis and chemoresistance in transgenic cancer models. J Mol Med. 2002;80:137–146. doi: 10.1007/s00109-001-0293-3. [DOI] [PubMed] [Google Scholar]
- Soengas MS, Alarcon RM, Yoshida H, Giaccia AJ, Hakem R, Mak TW, Lowe SW. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284:156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- Soussi T, Caron de Fromentel C, May P. Structural aspects of the p53 protein in relation to gene evolution. Oncogene. 1990;5:945–952. [PubMed] [Google Scholar]
- Stoffel A, Le Beau MM. The API2/MALT1 fusion product may lead to germinal center B cell lymphomas by suppression of apoptosis. Hum Hered. 2001;51:1–7. doi: 10.1159/000022952. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- Tamm I, Kornblau SM, Segall H, Krajewski S, Welsh K, Kitada S, Scudiero DA, Tudor G, Qui YH, Monks A, et al. Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin Cancer Res. 2000;6:1796–1803. [PubMed] [Google Scholar]
- Teichmann M, Wang Z, Roeder RG. A stable complex of a novel transcription factor IIB-related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc Natl Acad Sci. 2000;97:14200–14205. doi: 10.1073/pnas.97.26.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beneden RJ, Walker CW, Laughner ES. Characterization of gene expression of a p53 homologue in the soft-shell clam (Mya arenaria) Mol Mar Biol Biotechnol. 1997;6:116–122. [PubMed] [Google Scholar]
- Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Silke J, Ekert PG, Pakusch M, Kaufmann H, Connolly LM, Day CL, Tikoo A, Burke R, Wrobel C, et al. HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J Biol Chem. 2002;277:445–454. doi: 10.1074/jbc.M109891200. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RM, Clem RJ, Muller HA, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cado D, Chen A, Kabra NH, Winoto A. Fas-mediated apoptosis and activation-induced T-cell proliferation are defective in mice lacking FADD/Mort1. Nature. 1998;392:296–300. doi: 10.1038/32681. [DOI] [PubMed] [Google Scholar]