Abstract
Hedgehog (Hh) signaling plays a major role in multiple aspects of embryonic development. A key issue is how negative regulation of Hh signaling might contribute to generating differential responses over tens of cell diameters. In cells that respond to Hh, two proteins that are up-regulated are Patched1 (Ptch1), the Hh receptor, a general target in both invertebrate and vertebrate organisms, and Hip1, a Hh-binding protein that is vertebrate specific. To address the developmental role of Hip1 in the context of Hh signaling, we generated Hip1 mutants in the mouse. Loss of Hip1 function results in specific defects in two Hh target issues, the lung, a target of Sonic hedgehog (Shh) signaling, and the endochondral skeleton, a target of Indian hedgehog (Ihh) signaling. Hh signaling was up-regulated in Hip1 mutants, substantiating Hip1's general role in negatively regulating Hh signaling. Our studies focused on Hip1 in the lung. Here, a dynamic interaction between Hh and fibroblast growth factor (Fgf) signaling, modulated at least in part by Hip1, controls early lung branching.
Keywords: Hedgehog, Hip1, Fgf10, feedback regulation, lung, branching morphogenesis
Supplemental material is available at http://www.genesdev.org.
The Hedgehog (Hh) signaling pathway plays a central role in the regulation of invertebrate and vertebrate development (for reviews, see Ingham and McMahon 2001; McMahon et al. 2003). Several components of the pathway have been identified that are essential for transducing the Hh signal; these include membrane proteins, cytoplasmic components, and transcriptional activators. Hh signal transduction is regulated by many cellular processes, including proteolysis, phosphorylation, and transcriptional activation of negative regulators. A Hh signal is transduced on binding of Hh ligand to its receptor, Patched1 (Ptch1), a multipass transmembrane protein. Genetic and molecular studies suggest that Ptch1 inhibits the signaling activity of Smoothened (Smo), a seven-transmembrane protein that shares sequence similarity with G-protein coupled receptors. Though the precise molecular mechanism remains to be elucidated, Hh binding to Ptch1 appears to relieve a Ptch1-mediated repression of Smo, resulting in elevated levels of a phosphorylated form of Smo at the surface of the cell. In its activated form, Smo can initiate the signaling cascade, activating transcriptional targets of the Hh signaling pathway.
An important aspect of Hh signaling is induction of the genes encoding Hh-binding proteins, Ptc/Ptch1 and Hedgehog-interacting protein 1 (Hip1; for review, see Ingham and McMahon 2001). Whereas up-regulation of Ptc/Ptch1 in target cells is a highly conserved response, no Hip1 homologs have been identified in invertebrates. Increased Ptc on the cell surface of Hh-responsive cells sequesters Hh signal, limiting the range of Hh action in its target field, and may help shape the cell's response to Hh signaling (see Ingham and McMahon 2001). How Hip1 activity features in Hh signaling is less well understood.
Hip1 encodes a membrane-bound protein that directly binds all mammalian Hh proteins (Chuang and McMahon 1999). Like Ptch1, Hip1 is transcriptionally activated in response to Hh signaling, overlapping the expression domains of Ptch1 (Goodrich et al. 1996; Chuang and McMahon 1999). Further, gain-of-function experiments indicate that Hip1 binding of Hh ligands attenuates Hh signaling (Chuang and McMahon 1999). Here we demonstrate that loss-of-function mutants in Hip1 result in an up-regulation of Hh signaling in the mouse embryo, disrupting cell interactions essential for the normal morphogenesis of the lung and skeleton (see Supplemental Material).
Results and Discussion
Targeted disruption of Hip1 results in neonatal lethality with respiratory failure
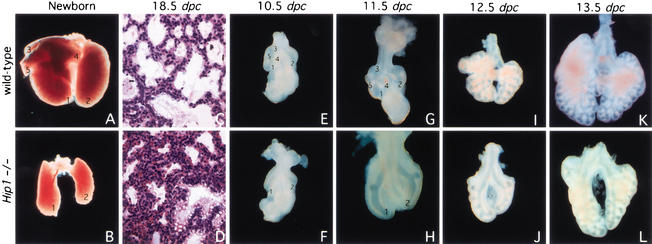
To generate a null allele of the Hip1 gene in mice, a standard positive/negative targeting vector was constructed. The details are described in the Supplemental Material and Supplementary Figure 1A. Loss of Hip1 activity leads to recessive postnatal lethality. The ratio of Hip1+/+:Hip1+/−:Hip1−/− (55:118:51) newborn pups approximates a 1:2:1 Mendelian distribution (Supplementary Fig. 1B), but all homozygous Hip1 mutant pups die a few hours after birth due to respiratory failure. Hip1 mutants are superficially identical to their wild-type littermates, indicating that Hip1 activity does not appear to be essential for normal patterning of limbs, hair, or whisker, all of which are regulated by Hh signaling (Supplementary Fig. 1C; McMahon et al. 2003). Histological analysis revealed that dorsal-ventral patterning of the neural tube, development of the somites, and the organization of most internal organs appeared grossly normal in Hip1 mutants (data not shown). In contrast, Hip1 mutants have only one right and one left lung lobe rather than the five lobes (four on the right side and one on the left side) typical of wild-type mice (Fig. 1A–B,E–L). Mutant lungs do not inflate after birth and Hip1 mutants die of respiratory failure.
Figure 1.
Defective branching morphogenesis in Hip1 mutant lungs. (A–B,E–L) Ventral view of lungs dissected at indicated stages from wild-type (A,E,G,I,K) and Hip1 mutant (B,F,H,J,L) embryos or pups. (C,D) Hematoxylin/eosin-stained sections of wild-type (C) and Hip1 mutant (D) lungs at 18.5 dpc. (D) A decrease in airway space and a relative increase in the number of mesenchymal cells was observed in Hip1 mutant lungs. (E) The primary buds (labeled 1 and 2) in the wild-type lung can be seen at 10.5 dpc as well as the initial swellings (labeled 3–5) associated with the secondary branches of the right bud that establish the additional lobes of the right lung. (G) These elongate, generating three clear secondary branches from the right bud by 11.5 dpc. (F,H) The primary buds (labeled 1, 2) form in Hip1 mutants, but initial secondary branching fails to occur during specification of lobulation. The lung bud of Hip1 mutants in H was taken at a higher magnification in order to clearly visualize the branching pattern in Hip1 mutant lungs. (B) As a consequence, only one lobe is generated from the right lung bud. (I,K) Extensive dichotomous branching ensues, giving rise to the respiratory tree. Dichotomous branching occurs at a slower rate in Hip1 mutant lungs, resulting in a smaller sized left lung lobe and single right lobe and a generally stunted respiratory tree (cf. B and A).
To investigate the nature of the lung defects in Hip1−/− embryos, lungs were collected from embryos between 9.5 and 18.5 days postcoitus (dpc) (see Supplementary Fig. 1A for genotyping data). Lung development initiates at 9.5 dpc with outgrowth of paired buds of ventral foregut endoderm into surrounding splanchnic mesenchyme (for review, see Hogan 1999). At 10.5 dpc, two primary lung buds elongate, the right more rapidly than the left, and undergo further branching in an invariant pattern that is species specific, giving rise to five buds, four on the right and one on the left of the mouse embryo (Fig. 1E). In contrast, although both the left and right buds grew out in Hip1 mutants, and the left-right asymmetry in their growth was conserved, the initial stereotyped branching from the two primary buds was absent in Hip1−/− lungs (Fig. 1F). These results suggest that, in the absence of Hip1 activity, there is a failure to specify the early, invariant lateral branches that determine the lobulation pattern. At 11.5 dpc, the Hip1−/− lung exhibits two prominent lateral bulges, one on each lobe, indicating that partial secondary branching has begun (Fig. 1H). But the characteristic branching pattern (Fig. 1G) was never generated in Hip1 mutant lungs (Fig. 1H). Thus, the failure to generate a complete respiratory tree explains the much smaller lung of mutants that are only 1/4 to 1/3 the size of wild-type lungs at birth (Fig. 1, cf. A and B). However, the single left lobe is also significantly reduced, indicating that Hip1 is also likely to play a later role in the branching process. Interestingly, histological analysis of the Hip1−/− lungs at these later stages revealed reduced airway space and an increased number of mesenchymal cells (Fig. 1, cf. C and D; data not shown for 16.5 and 17.5 dpc), but proximodistal epithelial differentiation appeared normal, as judged by histological and marker analysis with a number of regional or cell type-specific markers, including CC10, SP-A, SP-B, SP-C, and CFTR (data not shown). Thus, Hip1 is required for normal branching morphogenesis of the airways, but not for proximodistal patterning.
Hedgehog signaling is up-regulated in Hip1 mutants
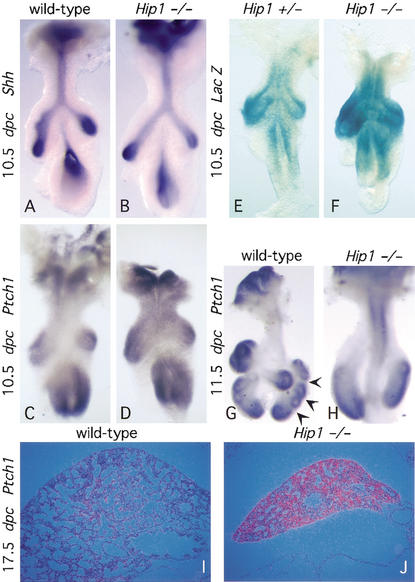
As discussed earlier, initial studies suggested that Hip1 may directly antagonize Hh signaling (Chuang and McMahon 1999). If correct, Hh targets, which include Ptch1 and Hip1, would be expected to be transcriptionally up-regulated in the absence of Hip1 function. In the wild-type lung at 10.5 dpc, Ptch1 (Fig. 2C) is expressed in the mesenchyme surrounding the developing airway epithelium where Sonic hedgehog (Shh) is expressed (Fig. 2A), but expression is elevated at the distal tips of the lung buds in response to an up-regulation of Shh expression in the adjacent epithelium (Litingtung et al. 1998; Pepicelli et al. 1998). In Hip1−/− lungs, Shh expression was unaltered (Fig. 2B) but Ptch1 expression in the mesenchyme was moderately increased and expanded (Fig. 2D). In wild-type lungs at 11.5 dpc, higher levels of Ptch1 expression are found at the tips of the newly formed secondary buds as compared with the regions between buds (arrows in Fig. 2G). In contrast, Ptch1 expression remained quite uniform in the mesenchyme of the two primary lung buds of Hip1 mutants (Fig. 2H). As development proceeds, Ptch1 expression is further down-regulated in the wild-type lungs (Fig. 2I), yet Ptch1 expression remained high in Hip1−/− lungs throughout later embryonic development (Fig. 2J; data not shown for 16.5 and 18.5 dpc). These results suggest that Shh signaling is up-regulated in the absence of Hip1. Consistent with this model, β-galactosidase activity at the targeted Hip1 locus indicated that Hip1 expression was also up-regulated in Hip1 mutant lungs from 10.5 dpc, even when taking into account differences in lacZ gene dosage (Fig. 2E,F; data not shown).
Figure 2.
A molecular analysis of branching morphogenesis in Hip1 mutant lungs. (A–D,G–H) Whole-mount in situ hybridization, using digoxigenin-labeled ribo-probes, on wild-type (A,C,G) and Hip1 mutant (B,D,H) lungs (ventral view) at 10.5 and 11.5 dpc. (A) Shh expression in the epithelium increases at the distal tips of the wild-type lung buds. The expression pattern of Shh in the Hip1 mutant lung buds (B) is similar to that of a wild-type embryo (A). In contrast, at 10.5 dpc, Ptch1 expression in the mesenchyme is significantly increased and expanded in the Hip1 mutant lung (D) as compared to that of the wild-type embryo (C). At 11.5 dpc, Ptch1 expression becomes localized to the mesenchyme at the tips of the most recently developed buds in the wild-type lungs (arrowheads point to interbud regions where Ptch1 expression is down-regulated; G), yet Ptch1 expression remains uniformly expressed throughout the mesenchyme in Hip1 mutant lung buds (H). As development proceeds, Ptch1 expression is further down-regulated in the wild-type lungs (I; data not shown), yet Ptch1 expression remains high in Hip1−/− lungs throughout embryonic development (J; data not shown). (E–F) β-Galactosidase staining of lungs (ventral view) dissected from Hip1 heterozygous (+/−) and homozygous (−/−) embryos. The bacterial LacZ gene, inserted into the Hip1 locus, is expressed under the Hip1 promoter. Cells transcribing the LacZ gene can be viewed by histochemical staining for β-galactosidase activity. In the heterozygous lungs (E), Hip1 transcripts are mainly confined to the mesenchyme at the tips of the lung buds, whereas in the Hip1 mutant lungs (F), there is a dramatic increase in signal throughout the mesenchyme of the buds. This increase in signal seems to be greater than that due to the presence of an additional copy of the LacZ gene in Hip−/− lungs and indicates an up-regulation of the LacZ transcript level (E,F; data not shown).
Defective secondary branching in Hip1 mutant lungs is due to the loss of Fgf10 signaling
Reciprocal interactions between the epithelium and the surrounding mesenchyme play a key role in inducing lung branching in a temporal- and spatial-specific manner (for reviews, see Hogan 1999; Warburton et al. 2000). Both loss-of-function and gain-of-function studies indicate that initial branching of the lung epithelium is controlled by a mesenchymal production of fibroblast growth factor 10 (Fgf10). Expression of Fgf10 is initiated in mesenchymal cells some distance from the epithelium at focal sites adjacent to where branches later emerge (Bellusci et al. 1997). Fgf10 knockout mice exhibit a lungless phenotype and die shortly after birth due to respiratory failure (Min et al. 1998; Sekine et al. 1999).
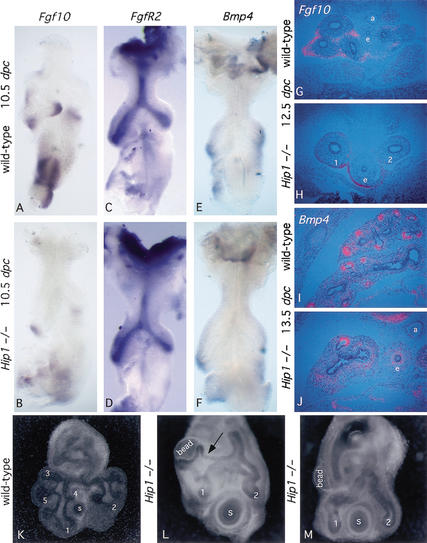
Analysis of Shh mutants indicates that Shh signaling plays an important role in the regulation of Fgf10 expression in the mesenchyme, and, consequently, of the branching process (Litingtung et al. 1998; Pepicelli et al. 1998). In the wild-type lung at 10.5 dpc, Fgf10 is expressed in mesenchyme cells at the distal tips of the primary lung buds and is localized to the prospective sites of lung bud formation (Bellusci et al. 1997; Fig. 3A,G). In Shh mutant lungs, Fgf10 expression is broadly up-regulated in the lung mesenchyme and expression is observed in mesenchyme cells immediately adjacent to the lung epithelium (Pepicelli et al. 1998). Thus, Shh signaling to the mesenchyme negatively regulates Fgf10 expression, and the resulting misregulation of Fgf10 in Shh mutant lungs most likely accounts for the observed failure of secondary branching.
Figure 3.
Analysis of major signaling pathways involved in lung branching in Hip1 mutant lungs. (A–F) Whole-mount in situ hybridization, using digoxigenin-labeled ribo-probes, on wild-type (A,C,E) and Hip1 mutant (B,D,F) lungs (ventral view) at 10.5 dpc. Fgf10 expression in the mesenchyme at the prospective sites of secondary bud formation in wild-type lungs (A) is lost in the Hip1 mutant lung (B). FgfR2 is expressed in the epithelium of both the wild-type (C) and Hip1 mutant (D) lungs with no apparent differences. The dynamic pattern of Bmp4 expression in the distal epithelial cells of the terminal buds and in the adjacent mesenchyme is indistinguishable between wild-type (E) and Hip1 mutant (F) lungs. (G–J) Isotopic section in situ using 33P-UTP-labeled ribo-probes performed on sections of wild-type (G,I) and Hip1 mutant (H,J) thoracic cavities at 12.5 and 13.5 dpc. Dorsal side is down. These images focus on the entire lung in both wild-type and Hip1 mutant sections at 12.5 dpc. However, at 13.5 dpc, only images of the right lung are shown. The expression pattern of Fgf10 (G,H) and Bmp4 (I,J) correspond to the patterns described in the whole-mount in situ earlier, in which Fgf10 expression is decreased at the distal tips of the Hip1 mutant (H) primary buds (numbered 1 and 2) and lost from the future sites of secondary bud formation, but there are no changes in Bmp4 expression between wild-type (I) and Hip1 mutant (J) lungs. e, esophagus; a, aorta. (K–M) Induction of epithelial branching in Hip1 mutant lungs by FGF10. (L) Dissected Hip1 mutant lungs at 10.5 dpc in culture (primary buds are numbered 1 and 2), with beads soaked in recombinant FGF10 protein (labeled bead) implanted in the mesenchyme, show significant epithelial budding toward the bead at 84 h of culture. (M) In contrast, Hip1 mutant lungs implanted with control (PBS) beads (labeled bead) show no sign of budding toward the bead at the corresponding time point. Wild-type lungs (buds are numbered 1–5, as seen in Fig. 1E), at 48 h (K) and 80 h (data not shown) of culture, show significantly more budding compared with those of the Hip1 mutant. The right primary bud is to the left. Note that the branch (L, arrow) that grows toward the FGF10 bead represents a novel bud that arises at a more proximal position within the primary lung bud than the wild-type branches. s, stomach.
In contrast to delocalized Fgf10 expression in Shh mutant lungs, Fgf10 expression was slightly down-regulated at the distal tips of the primary lung buds in Hip1−/− lungs at 10.5 dpc, but completely absent from the mesenchyme where secondary branching normally initiates (Fig. 3B). At 11.5 dpc, outgrowth of a single secondary branch from both primary buds of Hip1 mutant lungs (Fig. 1H) correlated with low levels of Fgf10 expression (data not shown). The loss of secondary branching in Hip1 mutant lungs was most likely a direct consequence of the failure of Fgf10 expression at the prospective sites of secondary bud formation. Thus, whereas both Shh and Hip1 mutants exhibit a deficiency in lung branching, the phenotype results from opposite actions of these factors. Loss of Shh-mediated inhibition of Fgf10 results in a broad, mesenchymal up-regulation of Fgf10 expression, leading to the loss of the focal sources of Fgf10 that normally trigger branching events. In contrast, enhanced Shh signaling in Hip1 mutants leads to a nearly complete repression of normal Fgf10 expression in the early lung, resulting in a failure of Fgf10-mediated initiation of secondary branching. Whereas Fgf10 expression was clearly Hip1 dependent, expression of FgfR2, the receptor for Fgf10, was unaltered (Fig. 3C,D), as was the expression of several genes that encode other signaling molecules that are implicated in the regulation of lung development, such as Bmp4 (Fig. 3F,J), Fgf9, Wnt7b, and Wnt2 (data not shown). Thus, modulating Fgf10 expression may be the principal role of Shh signaling during lung development.
Hh signaling is not absolutely required for primary bud formation, as revealed by the presence of two stunted primary buds in Shh mutant lungs (Litingtung et al. 1998; Pepicelli et al. 1998). Primary buds also developed in Hip1 mutants, a possible reflection of the time lag between Hh signaling and Hip1 induction. In this model, any effects due to the lack of Hip1 will not be apparent until after the outgrowth of the two primary buds when significant levels of Hip1 transcripts have accumulated in response to Shh signaling such that Hip1 levels can now effectively modulate Shh action.
To test the hypothesis that the loss of branching in Hip1 mutants reflects the absence of Fgf10 signaling, we implanted FGF10-soaked beads into the mesenchyme of Hip1 mutant lungs at 10.5 dpc. FGF10 induced considerable epithelial budding toward the FGF10 bead (arrow in Fig. 3L), whereas PBS soaked beads failed to induce epithelial budding (Fig. 3M). Thus, loss of Fgf10 expression, as a result of elevated Hh signaling, most likely explains the lack of secondary bud formation in Hip1 mutant lungs.
Hip1 and Ptch1 share redundant roles in lung branching
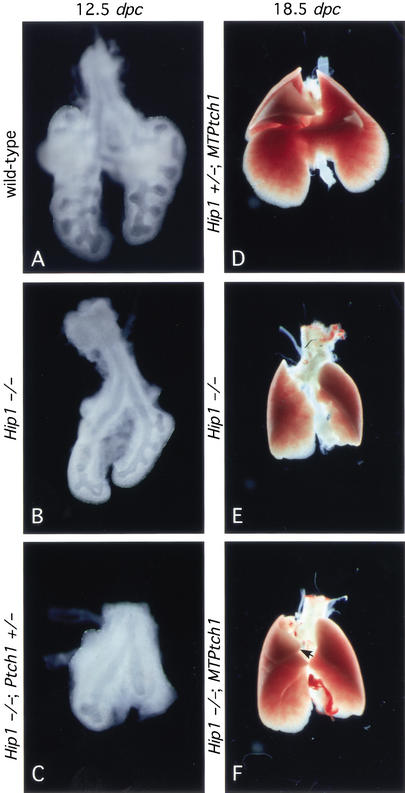
The fact that Hip1 mutant mice do not display overt phenotypes in multiple tissues that require Hh signaling for proper patterning may reflect a functional redundancy between Ptch1 and Hip1 because the expression of Ptch1 overlaps with that of Hip1 and both appear to function as negative regulators of Hh signaling. Preliminary studies support this model because attenuating Ptch1 activity (Ptch1+/−) in a Hip1 mutant background leads to an accelerated lethality around 11.5–12.5 dpc (data not shown). Lungs from Hip1−/−; Ptch1+/− animals (Fig. 4C) (n = 5) at this stage are consistently smaller in size (although the mesenchyme appears to be thicker) than Hip1 mutant lungs (Fig. 4B) and also exhibit more severe branching defects from the two primary buds (Fig. 4, cf. B and C). If Hip1 and Ptch1 play a similar role in modulating Hh signaling, overexpression of Ptch1 might rescue the lung defects in Hip1 mutants. To test this hypothesis, we introduced a Ptch1 transgene (MTPtch1; Milenkovic et al. 1999), in which Ptch1 is expressed at a basal level under the control of the metallothionein promoter, into the Hip1−/− mutant background. Hip1 mutant animals carrying MTPtch1 (n = 3) exhibit a slight increase in lung size (Fig. 4, cf. E and F), whereas lungs from Hip1+/− animals carrying MTPtch1 (Fig. 4D) cannot be distinguished from those of wild-type animals. In addition, a third right lobe (arrow in Fig. 4F), which is partially fused to the main right lobe, can also be observed in Hip1−/−; MTPtch1 animals. These results indicate a modest rescue of lung defects in the Hip1 mutant by MTPtch1. Taken together, these genetic studies reveal a functional redundancy between Hip1 and Ptch1 in lung branching and also suggest that similar functional interactions exist between Hip1 and Ptch1 in other tissues.
Figure 4.
Hip1 and Ptch1 share redundant roles in lung branching. Ventral view of lungs dissected at 12.5 dpc (A–C) and 18.5 dpc (D–F). At 12.5 dpc, five distinct lobes are apparent in wild-type lungs (A), whereas only two lobes are generated in lungs from Hip1 mutants (B). (C) Lungs from Hip1−/−; Ptch1+/− animals at this stage are smaller in size (although the mesenchyme appears to be thicker) than Hip1 mutant lungs and also exhibit more severe branching defects from the two primary buds. (D) Animals carrying a Ptch1 transgene (MTPtch1), in which Ptch1 is under the control of the metallothionein promoter, do not exhibit any lung defect. When MTPtch1 is expressed in Hip1−/− mutants, lungs from these animals (F) appear to be slightly larger in size, compared with lungs from Hip1−/− embryos (E). In addition, a third lobe (F, arrow), which is partially fused to the right lobe, can also be observed.
A model of Hip1's role in lung branching morphogenesis
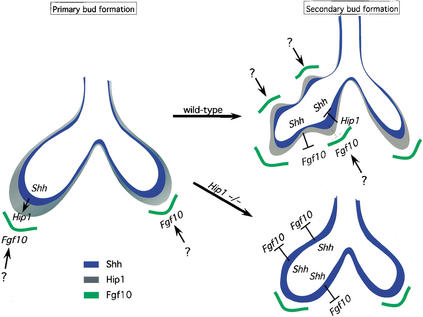
The analysis of Hip1 mutant lungs provides further insights into the interactions between Hh and Fgf signaling in lung branching morphogenesis. The mechanism by which Fgf10 expression is initiated, thereby triggering lung bud outgrowth, is not clear, but analysis of both Shh and Hip1 mutants indicates that neither is essential for this process. Localized Fgf10 expression is essential for secondary bud formation. Hh signaling is not responsible for the activation of Fgf10 expression associated with secondary bud formation but facilitates the correct spatial localization of Fgf10 in the lung mesenchyme. This could be achieved by Hh inhibition of Fgf10 expression in the interbud regions after Fgf10 expression has been initiated (Fig. 5). However, Shh expression is up-regulated at sites of secondary bud formation where Fgf10 expression is actually maintained. How then is Fgf10 expression maintained in the presence of high levels of Shh expression (and presumably high levels of Hh signaling) if Hh signaling also inhibits Fgf10 expression at the sites of secondary buds?
Figure 5.
A model of Hip1's role in lung branching morphogenesis. During lung branching morphogenesis, the two primary buds initially elongate and later bulge to generate the secondary buds. Localized Fgf10 (green) expression in the distal mesenchyme induces primary bud formation. The mechanisms by which Fgf10 expression is initiated and localized are not clear. Shh (blue) is expressed in the epithelium and is up-regulated at the distal tips of the primary buds. In this model, in the wild-type lung, secondary bud formation is also induced by localized Fgf10 expression in the mesenchyme corresponding to the future sites of secondary bud formation. It is likely that similar mechanisms (indicated by a question mark in the diagram) are used to localize Fgf10 to these sites. Epithelial expression of Hh may help restrict Fgf10 expression to the sites of secondary bud formation by inhibiting Fgf10 expression. The Hh pathway transcriptionally activates Hip1 (gray) and Ptch1 (not shown), antagonists of Hh signaling, to down-regulate the Hh pathway at the sites of bud formation. As a result, the signals that activate Fgf10 expression overcome the mild inhibition (broken line), if any, by Hh signaling. In contrast, in the interbud regions, the Hh pathway effectively inhibits Fgf10 expression. In the Hip1 mutant lung, there is a uniform up-regulation of the Hh pathway along the entire epithelium due to the absence of negative regulation by Hip1. Consequently, high levels of Hh inhibit Fgf10 expression and disrupt secondary bud formation. Thus, Hip1's negative regulation of the Hh pathway is necessary to ensure proper secondary bud formation.
We envisage two possible mechanisms. In the first, higher levels of Shh at the bud tips may not actually correspond to enhanced levels of Shh signaling, because negative feedback control on Shh signaling might effectively inhibit the signaling pathway. Once the amount of Ptch1 exceeds that required for Hh signaling, Ptch1, together with Hip1, sequester the Shh protein. This may quickly result in decreased Shh signaling, and as a result, less Shh signaling may result in regions of branching compared with the interbud regions. Thus, at the tips of the lung buds, Fgf10 expression can be maintained where Ptch1 and Hip1 are up-regulated, but Fgf10 expression is inhibited in the interbud region where initial signaling is not sufficient to maximally activate Ptc1 and Hip1 transcription. Although this type of mechanism is consistent with other feedback systems, it should be noted that in Shh-mediated induction of ventral cell identities in the neural tube and anterior-posterior digit identities in the limb, the highest threshold requirement for Shh signaling correlates with cells that appear to undergo maximal activation of Ptc1 expression.
A second model supposes that broad Shh signaling from the distal epithelium establishes a general proximal zone of Fgf10 repression in adjacent mesenchyme. As a result, Fgf10 activation can only occur at some distance from the underlying epithelium, thereby providing a distant source of ligand to trigger local, directed epithelial outgrowth. Up-regulation of Shh in the branching epithelium may eventually inhibit the focal sources of Fgf10 expression as the branching tips approach these groups of cells. Whatever the exact mechanism, the dynamic interaction between Shh and Fgf10 most likely extends beyond the initiation of secondary, lateral branches into later stages of branching morphogenesis because there is a marked reduction of airways in the single left lobe of the Hip1 mutant lung. Interestingly, Fgf4 expression in the limb is induced by Shh (Laufer et al. 1994; Niswander et al. 1994), and recent work suggests a link between Fgf and Hh signaling in expansion of the diencephalic primordium of the mouse brain (Ishibashi and McMahon 2002). Thus, the interaction between these two signaling pathways plays an important regulatory role in the morphogenesis of several distinct structures in the developing vertebrate embryo.
Materials and methods
Standard molecular biology techniques were performed as described (Sambrook and Russell 2001).
Generation of Hip1 null mice
A complete description of the targeting vector construct, chimera production, and allele identification is provided in the Supplemental Material.
Skeletal preparations, histology, and in situ hybridization
Skeletal preparations were performed as described (Hogan et al. 1994). Histological analysis, whole-mount in situ hybridization using digoxigenin-labeled probes, and section in situ hybridization using 33P-labeled ribo-probes were performed as described (Wilkinson and Nieto 1993). For whole mount in situ hybridization, at least five mutant embryos were examined for each probe and consistent results were observed. For section in situ hybridization, at least two mutant embryos were examined for each probe and consistent results were obtained.
Lung organ culture
Heparin beads (Sigma) were cut with tungsten needles, washed three times in phosphate-buffered saline (PBS), soaked in recombinant human FGF10 (R&D Systems, Inc.) for 4–5 h at room temperature, and stored at 4°C. Prior to use, the FGF10 beads were washed three times in PBS (Park et al. 1998), then implanted into the mesenchyme of lungs isolated from 10.5 dpc mouse embryos dissected in L15 medium supplemented with 1% serum. The lungs were placed on nucleopore polycarbonate filters (Whatman) and cultured in DMEM:F12 (1:1) supplemented with 10% fetal calf serum (Hyclone), 1× penicillin/streptomycin, and 1× L-glutamine for up to 90 h.
Acknowledgments
We thank all those who supplied probes, Matthew Scott for the gift of MTPtch1 mice, Jill McMahon for blastocyst injection, and Yong-Mei Hu for genotyping. We thank members of the Chuang laboratory for discussion, and Joy Alcedo, Matthias Hebrok, Shaun Coughlin, and Tom Kornberg for critical reading of the manuscript. Work in the laboratory of A.P.M. was supported by grants from the NIH (NS 33642 and DK 56246). P.-T. Chuang was a fellow of the Leukemia Society of America. Work in the Chuang laboratory was supported by the Sandler Family Supporting Foundation, the HHMI Biomedical Research Support Program, and a grant from the NIH (HL67822).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Corresponding authors.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1026303.
References
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- Chuang P-T, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Johnson RV, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: Induction of a mouse patched gene by Hedgehog. Genes & Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Morphogenesis. Cell. 1999;96:225–233. doi: 10.1016/s0092-8674(00)80562-0. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes & Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, McMahon AP. A Sonic hedgehog-dependent signaling relay regulates growth of diencephalic and mesencephalic primordial in the early mouse embryo. Development. 2002;129:4807–4819. doi: 10.1242/dev.129.20.4807. [DOI] [PubMed] [Google Scholar]
- Laufer E, Nelson CE, Johnson RL, Morgan BA, Tabin C. Sonic hedgehog and Fgf-4 act through a signaling cascade and feedback loop to integrate growth and patterning of the developing limb bud. Cell. 1994;79:993–1003. doi: 10.1016/0092-8674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Litingtung Y, Lei L, Westphal H, Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998;20:58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- McMahon, A.P., Ingham, P.W., and Tabin, C.J. 2003. The developmental roles and clinical significance of Hedgehog signaling. Curr. Top. Dev. Biol. (In press). [DOI] [PubMed]
- Milenkovic L, Goodrich LV, Higgins KM, Scott MP. Mouse patched1 controls body size determination and limb patterning. Development. 1999;126:4431–4440. doi: 10.1242/dev.126.20.4431. [DOI] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes & Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswander L, Jeffrey S, Martin GR, Tickle C. A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature. 1994;371:609–612. doi: 10.1038/371609a0. [DOI] [PubMed] [Google Scholar]
- Park WY, Miranda B, Lebeche D, Hashimoto G, Cardoso WV. FGF-10 is a chemotactic factor for distal epithelial buds during lung development. Dev Biol. 1998;201:125–134. doi: 10.1006/dbio.1998.8994. [DOI] [PubMed] [Google Scholar]
- Pepicelli CV, Lewis PM, McMahon AP. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol. 1998;8:1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, et al. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Warburton D, Schwarz M, Tefft D, Flores-Delgado G, Anderson KD, Cardoso WV. The molecular basis of lung morphogenesis. Mech Dev. 2000;92:55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]