Abstract
Plants have evolved protective mechanisms to ensure their survival when threatened by adverse environmental conditions during their transition to autotrophic growth. During germination, there is a 2- to 3-d period during which a plant can execute growth arrest when challenged by water deficit. This postgermination developmental checkpoint is signaled by the stress hormone abscisic acid (ABA), which induces the expression of the bZIP transcription activator ABI5. The growth arrest efficiency depends on ABI5 levels, and abi5 mutants are ABA-insensitive and unable to execute the ABA-mediated growth arrest. Here we show that a novel ABI5-interacting protein, designated as AFP, can form high molecular weight (Mr) complexes with ABI5 in embryo-derived extracts. Like ABI5, ABI five binding protein (AFP) mRNA and protein levels are induced by ABA during seed germination. Two different afp mutant alleles (afp-1 and afp-2) are hypersensitive to ABA, whereas transgenic plants overexpressing AFP are resistant; in these plants, AFP and ABI5 protein levels are inversely correlated. Genetic analysis shows that abi5-4 is epistatic to afp-1, indicating the ABA hypersensitivity of afp mutants requires ABI5. Proteasome inhibitor studies show that ABI5 stability is regulated by ABA through ubiquitin-related events. When expressed together, AFP and ABI5 are colocalized in nuclear bodies, which also contain COP1, a RING motif protein. Our results suggest that AFP attenuates ABA signals by targeting ABI5 for ubiquitin-mediated degradation in nuclear bodies.
Keywords: Abscisic acid, ABI5, postgermination growth arrest, protein degradation
The transition from heterotrophic to autotrophic growth is one of the most fragile phases during the life cycle of a flowering plant. Plants have evolved protective mechanisms to ensure their survival under adverse environmental conditions during this transition. Recently, we have identified a developmental checkpoint that occurs during germination within ∼2 d after stratification (Lopez-Molina et al. 2001). During this time interval, exposure to abscisic acid (ABA) or osmotic stress triggers a sustained but reversible growth arrest of germinated embryos (Lopez-Molina et al. 2001), which become osmo-tolerant (Lopez-Molina et al. 2001). Arabidopsis mutants deficient in ABA biosynthesis or ABA signaling are unable to execute this response (Koornneef et al. 1984; Finkelstein 1994; Laby et al. 2000; Lopez-Molina and Chua 2000; Quesada et al. 2000; Lopez-Molina et al. 2001; Gazzarrini and McCourt 2001; Finkelstein et al. 2002), suggesting that the postgermination growth arrest is an active process requiring ABA signaling. Although the molecular events underpinning the growth arrest are not known, the latter clearly represents a novel adaptive mechanism for plant survival under conditions of water deficit (Lopez-Molina et al. 2001).
Several Arabidopsis mutants have been isolated that either bypass this ABA-mediated growth arrest or show an enhanced response to ABA (for review, see Finkelstein et al. 2002). The latter category includes mutations in ERA1, which encodes a subunit of a farnesyl transferase (Cutler et al. 1996), and ABH1, which encodes a mRNA cap binding protein (Hugouvieux et al. 2001). Mutants bypassing the ABA-mediated growth inhibition were designated abi1, abi2, abi3, abi4, abi5, abi8 (for ABA-insensitive), cho1, and cho2 (Koornneef et al. 1984; Finkelstein 1994; Lopez-Molina and Chua 2000; Finkelstein et al. 2002; Nambara et al. 2002). The ABI8, CHO1, and CHO2 genes have not yet been cloned (Finkelstein et al. 2002; Nambara et al. 2002). ABI1 and ABI2 encode two homologous serine/threonine phosphatases of class 2C (Leung et al. 1994, 1997; Meyer et al. 1994; Rodriguez et al. 1998), and both abi1 and abi2 have identical Gly-to-Asp mutations in their phosphatase domains (Leung et al. 1994, 1997; Meyer et al. 1994; Rodriguez et al. 1998). Although the targets of these phosphatases are not known (for review, see Finkelstein et al. 2002), the wild-type (WT) proteins, ABI1 and ABI2, have been proposed as negative regulators of ABA signaling that operates during germination, as well as in vegetative tissues (Gosti et al. 1999). The ABI4 gene encodes an APETALA2 domain transcription factor (Finkelstein 1994; Finkelstein et al. 1998).
Two other ABA-INSENSITIVE genes, ABI3 and ABI5, encode a putative acidic domain transcription factor (Giraudat et al. 1992) and a basic leucine zipper (bZIP) transcription factor (Finkelstein and Lynch 2000; Lopez-Molina and Chua 2000), respectively. Both abi3 and abi5 mutants were initially recovered by virtue of their ability to germinate in the presence of ABA (Koornneef et al. 1984; Finkelstein 1994; Lopez-Molina and Chua 2000). Subsequently, it was demonstrated that both ABI5 and ABI3 mRNA and protein levels are stringently up-regulated by ABA or osmotic stress, but only during the time period when the postgermination growth arrest can occur (Lopez-Molina et al. 2001, 2002). The identification of this molecular signature further supports the existence of a development checkpoint. As ABI3 plays a central role in the establishment of desiccation tolerance and dormancy during zygotic embryogenesis (Koornneef et al. 1984; Finkelstein 1994; Parcy et al. 1994, 1997), the osmo-tolerance of the postgermination arrested embryos is likely acquired through the recruitment by ABI3 of embryogenesis pathways implicated in desiccation tolerance (Lopez-Molina et al. 2002). ABI3 has also been shown to be required for appropriate ABI5 expression (Finkelstein and Lynch 2000; Soderman et al. 2000; Lopez-Molina et al. 2002). By using transgenic plants, we demonstrated that restoration of ABI5 levels in abi3 mutants could reestablish ABA-dependent growth arrest and expression of late embryogenesis genes (Lopez-Molina et al. 2002). This result, together with those of others (Soderman et al. 2000), led to the notion that ABI5 acts downstream of ABI3.
The efficiency of the postgermination ABA-dependent growth arrest is directly dependent on ABI5 protein levels. (Lopez-Molina et al. 2001; Brocard et al. 2002). We have demonstrated that ABA has two effects on the ABI5 protein: (1) it activates ABI5 as a growth repressor, including an increased promoter occupancy on its target promoters (Lopez-Molina et al. 2002), and (2) the hormone also signals the inhibition of ABI5 protein degradation via the 26S proteasome (Lopez-Molina et al. 2001). The mechanism of ABI5 activation and the factors that regulate its ubiquitin-mediated proteolysis are unknown.
To gain further insight into the molecular mechanisms of ABA signaling that leads to postgermination growth arrest, we used yeast two-hybrid assays to isolate proteins that may interact with ABI5. Among all the cDNA clones recovered, 15% encoded the same protein, which we designated as ABI five binding protein (AFP). Here, we present a molecular and genetic characterization of AFP and Arabidopsis mutants (afp) deficient in this protein. Our results indicate that AFP is a novel negative regulator of ABA signaling by facilitating the degradation of ABI5.
Results
Yeast two-hybrid screen identifies a novel ABI5 binding protein (AFP)
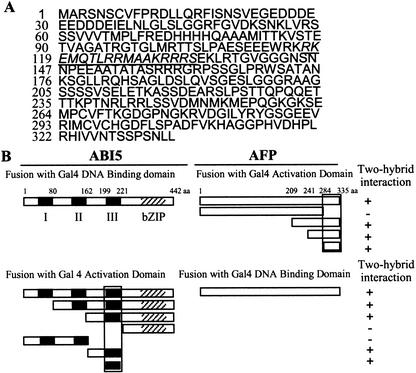
We used yeast two-hybrid assays to isolate and identify ABI5 interacting proteins by using a cDNA library made from mRNAs derived from a mixture of different Arabidopsis tissues (see Materials and Methods). Fifteen percent of the cDNA clones recovered encode the same protein, which was designated AFP. AFP is located on chromosome 1 (accession no. AAF67775) and encodes a protein with a predicted molecular weight (Mr) of 36,383 (Fig. 1A); however, this protein migrates with an apparent mass of 42 kD on SDS gels (see below). There are three genes (accession nos. NP_2;189598, NP_2;566163, AAL07175) encoding AFP-related proteins in the Arabidopsis genome, and this small AFP protein family appears to be plant-specific. By using yeast two hybrid assays, we found that the C-terminal region of AFP (amino acids 284–335) and domain III of ABI5 (amino acids 199–221) were necessary and sufficient for the interaction of the two proteins (Fig. 1B). Each of these two domains is conserved within its own gene family (61%–77% and 81% amino acid identity for AFP and ABI5, respectively).
Figure 1.
Isolation and expression of an ABI5-binding protein. (A) Deduced amino acid sequence encoded by the ABI five binding protein (AFP) gene. The putative nuclear localization motif is underlined. (B) Delineation of the AFP and ABI5 interaction domains (black boxes) by yeast two-hybrid experiments. (Left) ABI5 cDNA constructs fused to sequences encoding either GAL4 DNA binding or activation domain as indicated. (Right) Same as left panel but with AFP cDNA constructs. + indicates interaction; − indicates no interaction. Roman numerals refer to the three conserved ABI5 domains (Finkelstein and Lynch 2000; Lopez-Molina and Chua 2000). bZIP indicates basic leucine-zipper domain; BD indicates DNA binding domain. For each tested DNA construct, Western blot analyses were performed to confirm protein expression in yeast cells (data not shown).
AFP and ABI5 have similar temporal expression patterns in response to ABA
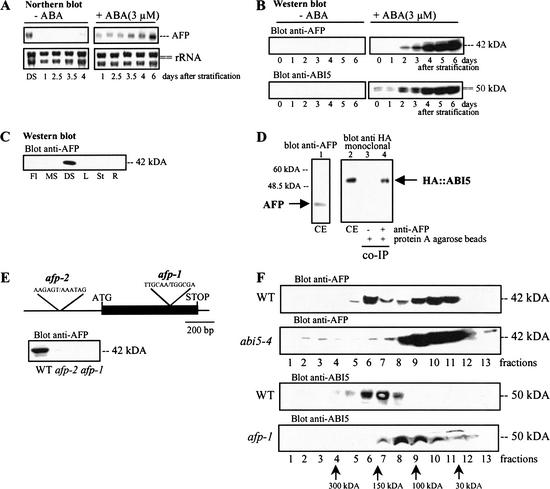
Like ABI5, AFP mRNA and protein levels increased during seed development and desiccation reaching plateau values in mature seeds (Fig. 2A; data not shown). After stratification and during germination, AFP mRNA and protein levels were induced by ABA within the developmental window when growth arrest can occur (Fig. 2A,B). However, once seedling autotrophic growth was established, AFP was no longer detectable in all tissues examined (Fig. 2C). These results, which are very similar to those obtained with ABI5 and ABI3 (Lopez-Molina et al. 2001, 2002), prompted us to examine whether ABI5 and AFP interact in vivo.
Figure 2.
ABA regulates AFP expression and AFP and ABI5 form complexes in vivo. (A) Wild-type (WT) seeds of the Wassilewskija (Ws) ecotype were stratified and treated with or without 3 μM ABA for the indicated times before RNA extraction. DS indicates dry seeds. Expression of AFP was analyzed by Northern blot (5 μg RNA/lane), and rRNA expression levels were used as loading controls. (B) WT seeds were stratified and treated with or without 3 μM ABA for the indicated times before protein extraction. Protein expression was analyzed by Western blots (10 μg protein/lane) by using a polyclonal antibody to AFP or ABI5. (C) AFP expression in different plant tissues was analyzed by Western blots (5 μg protein/lane). Fl, flower; MS, mature silique; DS, dry seed; L, true leaf; St, stem; R, root. (D) Total protein extract (CE) from 5-day-old WT/35S-HA∷ABI5 seedlings treated with ABA (5 μM) were immunoprecipitated with antibody to AFP (see Materials and Methods). Western blots were analyzed with a monoclonal antibody to HA to detect coimmunoprecipitated HA-ABI5 protein. (E, top) T-DNA insertion sites of two independent T-DNA insertion lines (afp-1, afp-2). (E, bottom) Western blot (10 μg/lane) analyses of AFP protein levels in WT, afp-1, and afp-2. (F) Protein extracts from WT, abi5-4, and afp-1 were loaded on a linear 5%–25% glycerol gradient to fractionate protein complexes (see Materials and Methods). After centrifugation, equal aliquots of gradient fractions were analyzed for ABI5 and AFP levels by Western blots (see Materials and Methods).
AFP and ABI5 interact in vivo
Coimmunoprecipitation experiments using extracts of germinating embryos treated with ABA showed that antibody to AFP was able to coimmunoprecipitate HA∷ABI5 (Fig. 2D). To explore the mode of interaction between the two proteins and to study the function of AFP, we identified two independent T-DNA insertions (Sussman et al. 2000) with lesions in the AFP locus. The mutant afp-1 has a T-DNA insertion in the coding sequence at codon 221, whereas afp-2 has an insertion in promoter sequences (Fig. 2E, upper panel). Western blot analysis showed that afp-1 is a null allele, whereas afp-2 is leaky (Fig. 2E, lower panel). Glycerol gradient fractionation of extracts from ABA-treated WT embryos showed that AFP (42 kD) was distributed in two high Mr peaks in fractions 6 and 7 (∼150 kD) and 9 and 10 (∼60 kD; Fig. 2F). Similarly, ABI5 (50 kD) was detected predominantly in fractions 6 and 7, although not in the low-molecular-weight fractions. These results, which support the notion of high-molecular complexes of AFP/ABI5, are consistent with the coimmunoprecipitation data (Fig. 2D). As expected, in abi5-4 extracts, which lack ABI5, most of the AFP protein was found in the lower-molecular-weight fractions. A similar shift in AFP distribution was found in afp-1 extracts (Fig. 2F).
AFP is a negative regulator of ABA signaling
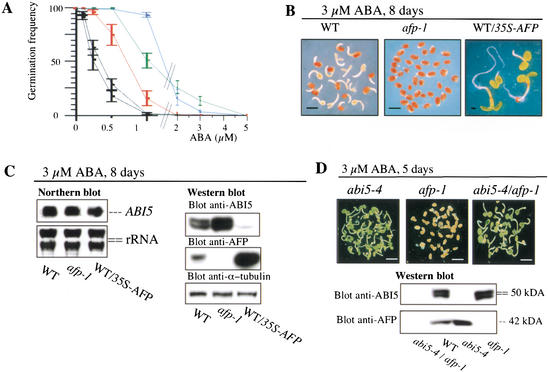
The identification of an AFP/ABI5 complex suggests that AFP may regulate ABI5 functions. Therefore, we analyzed ABA responses of afp-1 and afp-2 mutant plants, as well as of 35S-AFP transgenic plants. Western blot analyses indicated that AFP is not detectable in afp-1, whereas afp-2 still contains low levels of AFP (Fig. 2E, lower panel). In contrast, AFP levels in the 35S-AFP plants were 5- to 10-fold higher compared with WT plants (see below). In germination assays, afp-1 and afp-2 were hypersensitive to ABA compared with WT, whereas WT/35S-AFP transgenic plants were more resistant (Fig. 3A). The ABA-hypersensitive phenotype of afp-1 could be complemented with 35S-AFP in transgenic lines expressing AFP at WT levels (data not shown). Because ABA-induced accumulation of AFP reached a maximum 5 d after stratification, we also examined the formation of growth-arrested germinated embryos after a prolonged exposure to the hormone. After incubation in the presence of 3 μM ABA for 8 d, >95% of the afp-1 embryos did not germinate, whereas >85% of WT embryos germinated and displayed arrested growth (Fig. 3B). In contrast, WT/35S-AFP embryos fully germinated into green seedlings, even in the presence of 3 μM ABA. These results demonstrate that AFP is a negative regulator of ABA responses during germination.
Figure 3.
AFP is a negative regulator of ABA responses. (A) Inhibition of germination by exogenous ABA in WT (red), abi5-4 (green), afp-1 (black, lower curve), afp-2 (black, upper curve), and WT/35S-AFP (blue) seeds. Radical emergence was scored 3.5 d after stratification (see Materials and Methods). Standard error bars are shown (n = 3). (B) Phenotype of WT, afp-1, and WT/35S-AFP plants treated with 3 μM ABA for 8 d. For clarity, empty seed coats were removed. (C) WT, afp-1, and WT/35S-AFP seeds were stratified and grown on 3 μM ABA for 8 d before total RNA and protein extraction. rRNA and α-tubulin were used as loading controls in Northern (5 μg/lane) and Western (10 μg/lane) blots, respectively. (D) afp-1, abi5-4, and abi5-4/afp-1 double mutant seeds were stratified and grown on 3 μM ABA for 5 d before protein extraction and Western blot analysis (10 μg/lane). Pictures show seedlings before protein extraction. Bars, 1.2 mm.
AFP acts upstream of ABI5 to regulate negatively ABI5 protein levels
To see whether AFP function as a negative regulator depends on ABI5, we investigated the relationship between ABA responses and ABI5 levels. In WT, afp-1, afp-2, and a WT/35S-AFP line, ABI5 levels were inversely correlated with AFP levels at all ABA concentrations examined (Fig. 3C; data not shown). These differences could not be directly attributed to differences in ABI5 mRNA levels, as the latter were very similar (Fig. 3C). Because the afp-1/abi5-4 double mutant displayed ABA responses identical to those of abi5-4 (Fig. 3D), the ABA hypersensitive phenotype of afp mutants must require ABI5. These results led us to conclude that the negative regulatory function of AFP is executed through its effects on ABI5 protein levels.
ABI5 is ubiquitinated in vivo
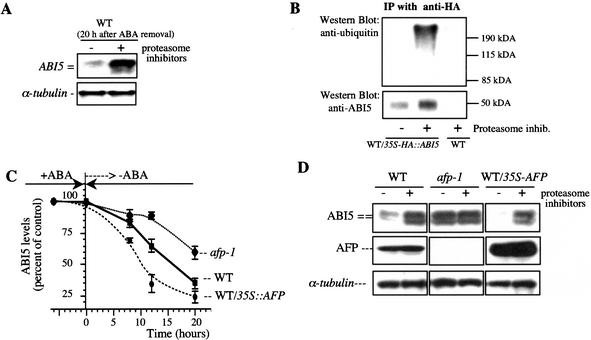
Previous work has shown that in extracts of germinating embryos, ABI5 protein stability is increased by ABA, but the protein is rapidly degraded in vitro via 26S proteasomes on ABA removal (Lopez-Molina et al. 2001). To see whether ABI5 is also degraded by 26S proteasomes in vivo, we directly investigated the effects of proteasome inhibitors on ABI5 protein levels in living plants. Figure 4A shows that, after ABA removal, proteasome inhibitors blocked ABI5 degradation. Thus, ABI5 protein levels are subject to stringent regulation via the 26S proteasome during the establishment and maintenance of, and exit from, the ABA-dependent growth arrest. We next used a transgenic line constitutively expressing HA∷ABI5 to study posttranslational regulation of ABI5 by ABA. As expected, proteasome inhibitors promoted the accumulation of poly-ubiquitinated ABI5 in vivo (Fig. 4B). This result confirms and extends previous in vitro data (Lopez-Molina et al. 2001).
Figure 4.
AFP promotes ABI5 protein degradation. (A) ABI5 protein degradation in vivo can be inhibited by proteasome inhibitors. WT seeds were stratified and grown in 3 μM ABA for 8 d. Seedlings were then transferred to a medium without ABA in presence or absence of proteasome inhibitors (MG132, MG115, PS1; 25 μM each; Calbiochem) for 20 h. ABI5 and α-tubulin protein levels were analyzed by Western blots (10 μg/lane); (B) WT/35S-HA∷ABI5 and WT control seeds (Landsberg ecotype) were grown in the absence of ABA for 8 d and treated for 2 h in the presence or the absence of proteasome inhibitors before immunoprecipitation using anti-HA antibodies (Lopez-Molina et al. 2001). (Top) Ninety percent of the immunoprecipitate was used to detect ubiquitinated forms of ABI5 by using an antibody to ubiquitin (Santa Cruz). (Bottom) The remaining 10% was used to quantify the amount of HA∷ABI5 using anti-ABI5 antibody. (C) Apparent ABI5 protein half-life in WT, afp-1, and WT/35S-AFP after ABA removal. Seedlings were treated with 3 μM ABA for 6 d and transferred to a medium without ABA. ABI5 protein levels were analyzed by Western blots and normalized by using α-tubulin levels. Standard errors are shown (n = 3). (D) WT, afp-1, and WT/35S-AFP seeds were treated as described in A. ABI5, AFP, and α-tubulin protein levels were measured by Western blots (10 μg/lane).
AFP enhances ABI5 proteolysis
The inverse correlation between AFP and ABI5 protein levels and the degradation of ABI5 by 26S proteasomes suggests that one function of AFP could be to enhance ubiquitin-mediated proteolysis. To test this hypothesis, we first assayed the apparent ABI5 protein half-life by comparing the time course of decrease in ABI5 protein levels after ABA removal in WT, afp-1, and WT/35S-AFP embryos. Figure 4C shows that the apparent half-life of ABI5 was 21 h for afp-1 and 9 h for WT/35S-AFP compared with 14 h for WT.
We next examined whether these differences in half-lives reflected differences in 26S proteasome degradation rates. For this purpose, we used proteasome inhibitors to block ABI5 degradation in the three genetic backgrounds. In WT and a WT/35S-AFP transgenic line, treatment with proteasome inhibitors substantially increased ABI5 levels by 5- to 10-fold (Fig. 4D). A greater increase was observed in a WT/35S-AFP transgenic line than in WT plants primarily because, in the absence of the inhibitors, lower ABI5 levels were detected in the former. In contrast, ABI5 levels in afp-1 mutants were already as high as those in inhibitor-treated WT and a WT/35S-AFP transgenic line, and only a very small increase was seen on inhibitor treatment (Fig. 4D). Our results suggest that ubiquitin-mediated proteolysis of ABI5 is critically dependent on the level of AFP, which appears to facilitate ABI5-regulated proteolysis.
Our results thus far suggest that AFP could prevent ABI5 over-accumulation in the presence of ABA (Fig. 3C) and promote ABI5 degradation to terminate the stress signal on ABA removal so that growth can resume (Fig. 4C,D). Consistent with the latter, in WT plants, the apparent half-life of AFP is significantly longer than that of ABI5 such that AFP levels remain virtually unchanged during the period of ABI5 degradation (data not shown).
AFP and ABI5 colocalize in nuclear bodies
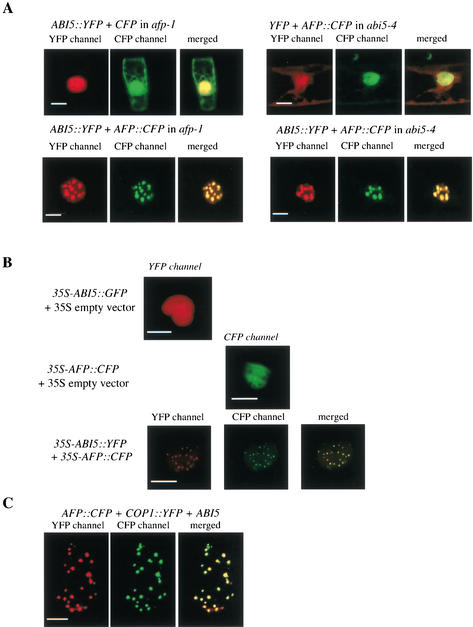
To determine the intracellular localization of AFP and ABI5, we transiently expressed 35S-ABI5∷YFP (or ABI5∷GFP) and 35S-AFP∷CFP fusion genes, separately or together, in onion epidermal cells or Arabidopsis afp-1 and abi5-4 mutant plants treated with 3 μM ABA for 7 d. Figure 5A shows that when individually expressed, both AFP∷CFP and ABI5∷YFP were distributed in the nucleoplasm in abi5-4 and afp-1 mutants, respectively. However, when expressed together, the two fusion proteins became colocalized in nuclear bodies (Fig. 5A). Similar results were obtained when the same constructs were expressed in onion epidermal cells (Fig. 5B). Nuclear body formation was not detected after coexpression with an ABI5 mutant lacking the D3 domain (data not shown), which is needed for AFP interaction (Fig. 1B). As Arabidopsis COP1, a RING motif protein, is known to be localized in nuclear bodies (von Arnim and Deng 1993; Ang et al. 1998), we investigated whether AFP/ABI5 might also reside in the same nuclear bodies when cobombarded with COP1 in onion epidermal cells. Figure 5C shows that this was indeed the case.
Figure 5.
Colocalization of AFP, ABI5, and COP1 in nuclear bodies. (A) Subcellular localization of ABI5∷YFP and AFP∷CFP in different Arabidopsis genetic backgrounds. Cells were cobombarded with equal amounts of the DNA constructs indicated. Living cells were imaged through CFP and YFP filter channels. (B) Subcellular localization of ABI5∷YFP (or ABI5∷GFP) and AFP∷CFP in onion epidermal cells. Onion peels were bombarded with 35S-ABI5∷YFP (or 35S-ABI5∷GFP), 35S-AFP∷CFP, or the two constructs together. Florescence images were collected through the YFP or the CFP filter channel. (C) Subcellular nuclear localization of ABI5, AFP∷CFP and COP1∷YFP in onion epidermal cells. Note that a few AFP/ABI5 nuclear bodies do not appear to contain COP1. Epidermal cells, cobombarded with all three constructs, were imaged through CFP and YFP filter channels. Bars, 5 μm.
Discussion
So far, few mutations affecting negative regulators of the ABA-mediated growth arrest in Arabidopsis have been isolated. These regulators include ERA1, a subunit of a farnesyl transferase (Cutler et al. 1996), and ABH1, a mRNA Cap binding protein (Hugouvieux et al. 2001). However, the targets of these regulators have not yet been identified. Here, we show that AFP is a novel negative regulator of ABA responses and that it executes its function through the bZIP transcription activator ABI5. Two lines of genetic evidence support this conclusion: (1) mutants lacking (afp-1), or deficient (afp-2) in, AFP are hypersensitive to ABA (Fig. 3A,B); and (2) the abi5-4 mutation is epistatic to afp-1 (Fig. 3D). This latter genetic interaction is confirmed by the observed physical interaction between ABI5 and AFP in coimmunoprecipitation experiments (Fig. 2D). Moreover, glycerol fractionation experiments (Fig. 2F) indicate that ABI5 and AFP form a complex of ∼200 kD. We speculate that this complex likely contains an ABI5 leucine zipper dimer (100 kD), with one AFP molecule binding to each ABI5 monomer. The inverse correlation in ABI5 and AFP protein levels observed in afp mutants and 35S∷AFP transgenic plants suggests that AFP can regulate negatively ABI5 protein levels, despite having similar ABI5 mRNA levels (Fig. 3C). Therefore, this regulation must be posttranslational. In afp mutants, high ABI5 levels are little affected by 26S proteasome inhibitors (Fig. 4D), suggesting that ABI5 is more stable in an afp mutant background. Conversely, ABI5 appears to be less stable in 35S∷AFP transgenic plants compared with WT, because in these transgenic plants, ABI5 levels show the highest increase on treatment with proteasome inhibitors (Fig. 4D). Taken together, our results support the notion that AFP facilitates ubiquitin-mediated proteolysis of ABI5. The negative regulation of ABI5 serves to attenuate ABA signals, as well as to ensure rapid degradation of ABI5 after stress removal.
In contrast to ABI5, AFP levels appear to be only slightly elevated by proteasome inhibitors (Fig. 4D), suggesting that this protein is more stable and consistent with its role as a signal attenuator. We note that AFP continues to be induced by ABA in the abi5-4 mutant (Fig. 3D). This result indicates that ABA-induction of AFP can occur via an ABI5-independent pathway.
Subcellular localization studies showed that in the absence of AFP, ABI5 is diffusely distributed in the nucleoplasm, but AFP/ABI5 complexes are localized to discrete nuclear bodies. The ABI5 domain III is needed for interaction with AFP (Fig. 1B) and for colocalization in nuclear bodies (data not shown). The molecular mechanism that underlies this interaction-dependent colocalization remains to be elucidated. As both afp mutant alleles are hypersensitive to ABA, localization in nuclear bodies appears to not be required for ABI5 function. Indeed, these subnuclear structures may represent sites of ABI5 degradation, which is promoted by AFP. This conclusion is supported by the finding that the AFP/ABI5 nuclear bodies also contain COP1, a RING motif protein that is a putative E3 ligase (von Arnim and Deng 1993; Hardtke and Deng 2000). Nuclear bodies containing COP1 have been proposed to be sites of protein inactivation and degradation (Wang et al. 2001). Future experiments will determine whether ABI5 is a substrate of COP1 or other E3 ligases in nuclear bodies.
We have previously shown that ABA can inhibit ABI5 degradation in vegetative tissues of transgenic plants constitutively over-expressing ABI5. AFP and its family members share a highly homologous C-terminal domain, which interacts with domain III of ABI5 (Fig. 1B). It should be pointed out that domain III of ABI5, is highly conserved among ABI5 homologs. These include the ABF transcription factors in Arabidopsis, which have been shown to regulate the responses of a plant to osmotic stress (Choi et al. 2000; Kang et al. 2002; Kim et al. 2002). Therefore, our work opens the way to study the function of AFP homologs, which might play similar roles in ABA and stress signaling pathways operating in vegetative tissues.
Materials and methods
Plant material, plant transformation, growth conditions, and plasmid constructions
Plant material, growth conditions, and ABA-treatment were as described in Lopez-Molina et al. (2001). Germination frequencies were obtained by scoring radical emergence (n = 3, ∼200 seeds per experiment). Plants were grown under identical conditions before seed harvesting. pBA002 (Kost et al. 1998) and pBin19 (Clontech) were used as binary vectors. T-DNA insertion mutants in AFP genes were obtained from the Arabidopsis Wisconsin-Madison knockout facility following the recommended procedure (Sussman et al. 2000). The PCR primers used in the screen were 5′-TGGATCGTGGCATAAAAATCTGAT TACTG-3′ and 5′-TAGAGGATTTGTACAGCAGCAAACAA GTG-3′.
Yeast two-hybrid experiment
Yeast two-hybrid experiments were performed as described (Xie et al. 2002).
RNA extraction, AFP antibody production, and Northern and Western blot analyses
RNA extraction and Northern blot hybridizations were performed as described in Vicient and Delseny (1999) and Lopez-Molina and Chua (2000). Polyclonal anti-AFP antiserum was obtained from rabbits immunized with HIS-tagged AFP expressed and purified from Escherichia coli (Stratagene). Western blot analyses were performed according to Lopez-Molina et al. (2001).
Coimmunopurification and glycerol gradient fractionation of extracts
Eight-day-old WT/35S∷HA-ABI5 transgenic seedlings were grown on solidified medium containing 0.5 μM ABA. Proteins were extracted in 50 mM Tris-HCl (pH 7), 150 mM NaCl, 10 mM MgCl2, and 0.5% NP40 supplemented with 1 mM PMSF and a mixture of anti-protease (complete Mini, Amersham Pharmacia). AFP was immunoprecipitated with rabbit antibody to AFP for 2 h at 4°C. Protein A agarose beads were then added for 1 h. Beads were washed three times with the same buffer. An equal volume of SDS-loading buffer was added to stop the reactions. Proteins were analyzed by Western blots by using mouse monoclonal antibody to HA as the first antibody. Glycerol gradient fractionation was performed by using extracts prepared from 6-day-old ABA-treated (3 μM) WT, afp-1, abi5-4 seedlings. Extraction buffer contained protease inhibitors as described above and 0.1% Triton X-100, 5% glycerol, 100 mM NaCl, and 50 mM Tris-HCl (pH 7.5). Extracts were loaded onto 5%–25% glycerol gradients and centrifuged for 16 h (250,000g). Fractions were collected, and proteins were precipitated with 10% TCA and resuspended in SDS gel sample buffer (Laemmli 1970) before Western blot analysis.
Subcellular localization experiments
Six-day-old Arabidopsis seedlings treated with 3 μM ABA or onion epidermal layers were bombarded according to standard procedures (Seki et al. 1998). ABI5∷YFP, AFP∷CFP, and YFP∷COP1 gene fusions were under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Ballesteros et al. 2001).
Acknowledgments
We thank Dr. Peter Hare for critical reading of the manuscript, Dr. Qi Xie for his generous gift of a cDNA library, Dr. X.-W. Deng for COP1 cDNA, Dr. Eashwar Narayanan for help in protein sequence analysis, Drs. Alison North and Bjorn Dittmer-Roche of the Rockefeller Bio-Imaging Resource Center for their assistance with confocal imaging, and Drs. Tim Tellinghuisen and Beate Kümmerer for advice with glycerol gradient fractionation. L.L.M. was supported by fellowships of the Human Frontier Science Program and the Swiss National Science Foundation. N.K. was a visiting student from Tokyo University, Japan. This work was supported by DOE grant DE-FG02-94ER20142 to N.H.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL chua@rockvax.rockefeller.edu; FAX (212) 327-8327.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1055803.
References
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsisdevelopment. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH. LAF1: A MYB transcription activator for phytochrome A signaling. Genes & Dev. 2001;15:2613–2625. doi: 10.1101/gad.915001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR. Regulation and role of the Arabidopsisabscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002;129:1533–1543. doi: 10.1104/pp.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs: A family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the abi3mutation. Plant J. 1994;5:765–771. [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsisabscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsisabscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P. Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr Opin Plant Biol. 2001;4:387–391. doi: 10.1016/s1369-5266(00)00190-4. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the ArabidopsisABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;11:1897–1910. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Deng XW. The cell biology of the COP/DET/FUS proteins: Regulating proteolysis in photomorphogenesis and beyond? Plant Physiol. 2000;124:1548–1557. doi: 10.1104/pp.124.4.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. Arabidopsisbasic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Ma J, Perret P, Li Z, Thomas TL. ArabidopsisABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 2002;130:688–697. doi: 10.1104/pp.003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61:377–383. [Google Scholar]
- Kost B, Spielhofer P, Chua NH. GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI. The Arabidopsissugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2000;23:587–596. doi: 10.1046/j.1365-313x.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. ArabidopsisABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The ArabidopsisABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:541–547. doi: 10.1093/pcp/41.5.541. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin D, Chait B, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:1–12. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P. A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics. 2002;161:1247–1255. doi: 10.1093/genetics/161.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J. Regulation of gene expression programs during Arabidopsisseed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell. 1994;6:1567–1582. doi: 10.1105/tpc.6.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Misera S, Giraudat J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsisseed development. Plant Cell. 1997;9:1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL. Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics. 2000;154:421–436. doi: 10.1093/genetics/154.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E. ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 1998;421:185–190. doi: 10.1016/s0014-5793(97)01558-5. [DOI] [PubMed] [Google Scholar]
- Seki M, Lida A, Morikawa H. Transient expression of foreign genes in tissues of Arabidopsis. In: Martinez-Zapater JM, Salinas J, editors. Arabidopsis protocols. Totowa, NJ: Humana Press; 1998. pp. 219–226. [DOI] [PubMed] [Google Scholar]
- Soderman EM, Brocard IM, Lynch TJ, Finkelstein RR. Regulation and function of the ArabidopsisABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000;124:1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S. The Arabidopsisknockout facility at the University of Wisconsin-Madison. Plant Physiol. 2000;124:1465–1467. doi: 10.1104/pp.124.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Delseny M. Isolation of total RNA from Arabidopsis thalianaseeds. Anal Biochem. 1999;268:412–413. doi: 10.1006/abio.1998.3045. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. Ring finger motif of Arabidopsis thalianaCOP1 defines a new class of zinc-binding domain. J Biol Chem. 1993;268:19626–19631. [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsiscryptochromes with COP1 in light control development. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature. 2002;419:167–170. doi: 10.1038/nature00998. [DOI] [PubMed] [Google Scholar]