Abstract
An RNA interference (RNAi)-based genome-wide screen was performed to detect genes that contribute to genome stability in somatic cells of Caenorhabditis elegans. We identified 61 such genes; these also affect spontaneous mutagenesis in the germ line. Their sequence suggests a role in DNA repair and/or replication, in chromatin remodeling, or in cell cycle control; there are also many novel genes that are highly conserved from yeast to human. Because known mutator genes are causally involved in many hereditary and sporadic human cancers, it is likely that some of these new mutators are equally relevant in cancer etiology.
Keywords: Carcinogenesis, mutator genes, DNA repair, spontaneous DNA damage, C. elegans
The first genetic analyses of mechanisms that protect the genome against spontaneous mutations were performed in bacteria (Friedberg et al. 1995), leading to the identification of DNA mismatch repair genes and the elucidation of the mechanism by which the repair machinery recognizes a DNA replication error and repairs the damage (for review, see Kolodner 1996; Modrich and Lahue 1996). This field of research gained clinical relevance when a hereditary form of cancer, hereditary nonpolyposis colon cancer (HNPCC), was found to be the result of germ line mutations in human homologs of DNA mismatch repair genes (e.g., Fishel et al. 1993; Leach et al. 1993). Also, many if not all sporadic human cancer cells display a mutator phenotype, meaning that they have enhanced levels of mutagenesis of some type: point mutations, frame shifts, gross chromosomal rearrangements, or aneuploidy. This has led to the hypothesis that the conversion of a wild-type cell into a mutator cell is an early event in carcinogenesis and that a mutator phenotype is causal to the development of cancer; multiple loss-of-function mutations in a limited number of genes are required for a regular cell to become a cancer cell (Fearon and Vogelstein 1990; Loeb 1991, 2001). Although the chances of several mutations coinciding in one cell are very low (the product of the chances for each mutation), a mutator's enhancement of the general level of mutagenesis greatly enhances the chance of the combination of mutations to arise.
Here we used RNA interference (RNAi) to systematically screen the nematode Caenorhabditis elegans genome for genes that, when inactivated, increase the level of spontaneous mutagenesis. We identified 61 genes with presumed roles in DNA repair, replication, chromatin organization, and cell cycle control. These mutator genes protect the C. elegans genome against insertion and deletion mutations in both the soma and the germline, and are thus potential tumor suppressors in higher species.
Results and Discussion
Genome-wide RNAi screens became feasible recently with the generation of a library of bacterial strains that each produce double-stranded RNA (dsRNA) for an individual nematode gene. The current library contains 16,757 bacterial strains that will target ∼86% of the 19,427 current predicted genes of the C. elegans genome, and the loss-of-function phenotype when performing systemic RNAi on a genome-wide scale is estimated to be ∼65% (Fraser et al. 2000; Kamath et al. 2003). Animals fed these bacteria induce RNAi in almost all tissues of the nematode (Timmons and Fire 1998; Kamath et al. 2001).
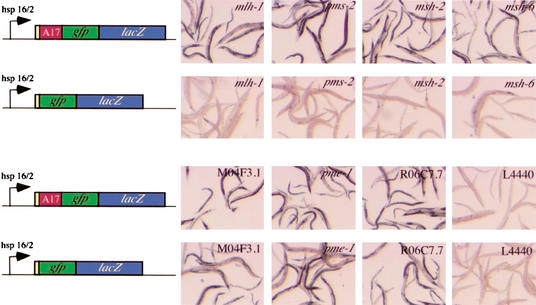
As a readout for increased DNA instability, we used a recently developed reporter assay that allows the detection of frameshifts and small insertions or deletions in somatic cells of C. elegans (Tijsterman et al. 2002). In brief, a gfp-LacZ reporter construct is put out of frame by an A17 mononucleotide DNA repeat cloned directly between the initiation ATG and the gfp-LacZ open reading frame (ORF; Fig. 1). The presence of such a DNA repeat will enhance the chances of a frameshift mutation to occur and therefore increase the sensitivity of the reporter system.
Figure 1.
RNAi-mediated DNA instability in C. elegans. Strains that contain heat shock promoter-driven out-of-frame gfp-lacZ reporter gene fusions were assayed on dsRNA homologous to mlh-1, pms-2, msh-2, msh-6, M04F3.1, Y71F9AL.18 (pme-1), R06C7.7, and the empty vector control L4440. Two different out-of-frame constructs were tested, one containing a mononucleotide (A17) repeat placed immediately downstream of the ORF's ATG (this reporter has been used to screen the RNAi library), the other identical except that it lacks the mononucleotide repeat.
The transgene is integrated in the C. elegans genome as a multicopy array. Only constructs that acquire a mutation that brings the reporter LacZ gene back in frame will be visualized by a β-galactosidase staining. The advantage of LacZ over other colorimetric indicators (such as gfp) is that prolonged staining can visualize relatively low levels of expression. Therefore we can detect a frameshift in only one of the transgenic copies of the integrated high copy array. This shows up as a blue cell, or—if the frameshift occurred early during development—a patch of blue cells. Previously, we found that frequent blue patches, as a result of restoration of LacZ expression, are visualized in mismatch repair-deficient animals (msh-6 mutants). These patches were not observed in repair-proficient animals (Tijsterman et al. 2002).
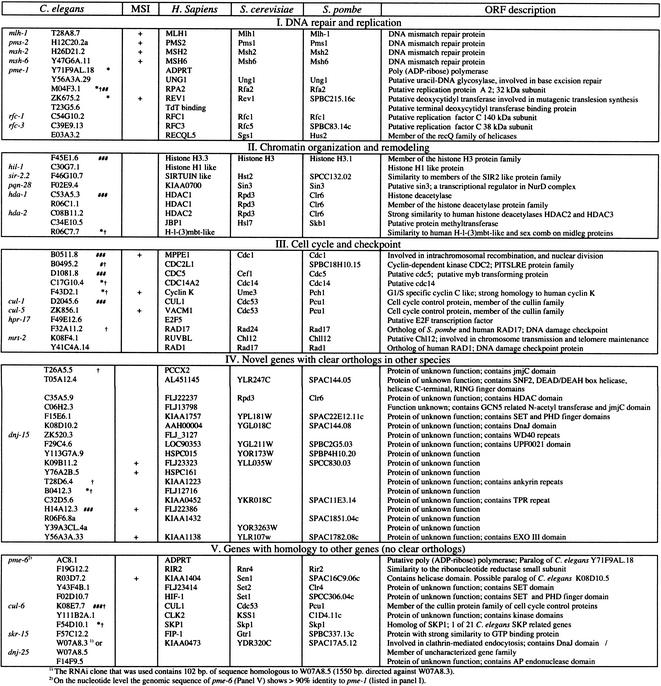
Using this assay we tested 16,606 genes by RNAi and found 61 genes whose knock-down phenotype results in somatic blue patches similar to those seen in msh-6-deficient animals (Table 1), a pattern indicative of a mutator phenotype. Among the identified genes are the four orthologs of human DNA mismatch repair genes, msh-2, msh-6, mlh-1, and pms-2 (Fig. 1; note that these were recognized in blinded procedure, without the investigator knowing the gene identity). This four-out-of-four hit rate suggests a good coverage in this screen. The sequence of the identified genes was subsequently inspected for domain structures, and we searched the sequenced genomes of Homo sapiens, Drosophila melanogaster, Saccharomyces cerevisiae, and Schizosaccharomyces pombe for homologous genes. We termed gene-pairs orthologous if the reverse BLASTP identifies the original C. elegans sequence. Based on their sequence homology, we tentatively classified the identified putative mutator genes in five categories: (I) DNA repair and replication, (II) chromatin remodeling, (III) cell cycle, (IV) novel genes with orthologs in many species, and (V) genes that have no clear orthologs in other species.
Table 1.
Genes that protect the C. elegans genome against mutations
The genes were identified on the basis of a mutator phenotype, as measured by the reversion of LacZ expression, upon growth on dsRNA homologous to the cognate C. elegans gene. MSI (microsatellite instability) indicate the situation where the reversion to proper LacZ expression depended on the presence of the mononucleotide repeat. Based on protein domain structure and sequence homology to known proteins, the identified genes were placed into five categories (homologous proteins in Homo sapiens, S. cerevisiae and S. pombe, are depicted). Panels I (DNA repair and replication), II (chromatin organization and remodeling) and III (cell cycle and check point) contain genes which at some level have been described in other species. Panel IV: Novel genes with clear orthologs in mammalian species. Panel V: Genes that have some level of homology to genes in other species or to C. elegans family members, but at present lack clear orthologous genes. Other notable RNAi induced phenotypes, i.e., a reduced broodsize (*), embryonic lethality (#10–30%; ##30–60%, ###60–100%), and slow growth (†) are also indicated. The absence of such marking reflects that this phenotype has not been observed despite careful inspection.
Genes in the first class are involved in DNA repair and replication. Besides the mismatch repair genes, this category also includes replication protein A2 (M04F3.1), which is known to act in multiple DNA metabolic processes including DNA replication, nucleotide excision repair, and double-strand-break repair (e.g., New et al. 1998; Araujo et al. 2000; Walter and Newport 2000), pme-1 (Y71F9AL.18), the C. elegans Parp [poly(ADP-ribose) polymerase], a polymerase involved in DNA damage response (Burkle 2001), and the C. elegans homolog of REV1, which has been shown to function in mutagenic translesion synthesis (Wang 2001).
The second class of genes is involved in chromatin organization and remodeling. Although at present not much is known about the role of chromatin condensation in protecting DNA against damage, a role for such genes in genome stabilization has been described (Celeste et al. 2002). Note also from this list that three proteins that are known to function in a complex are independently recognized: SIN3, HDAC1, and HDAC2 (Zhang et al. 1997); all show the mutator phenotype. This is also seen in the third class of genes, which have a putative role in cell cycle control. For example cdc5 and cdc14 work together in a mitotic checkpoint, and cell cycle arrest upon DNA damage is dependent on these genes (Stegmeier et al. 2002 and references therein).
In addition, there are 29 genes that have not been described previously (Table 1, categories IV and V). Genes in Table 1, category IV, have clear orthologs in other species and many of them contain recognizable sequence motifs, for example, an exonuclease, which could point to a role in DNA repair and/or DNA synthesis. Other domains such as the GCN5 (GNAT) histone acetyltransferase domain, JmjC domain, and SET domains suggest that they are involved in chromatin remodeling (e.g., Kuo et al. 2000; Lachner and Jenuwein 2002), indicating that the genes categorized in Table 1, category IV, are not a separate class, but based on putative function, could be involved in the same DNA metabolic processes as the genes listed in categories I to III.
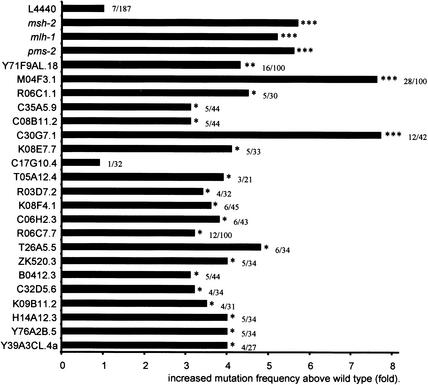
Can we be sure that the blue patched phenotype originates from frameshift mutations in the DNA and does not occur as shifts at the transcriptional or translational level? One clue is that most worms have a different pattern of blue patches. If shifts occurred at the transcriptional or translational level, one would expect that all worms would have the same pattern, for example, completely blue worms or one specific tissue. The variable pattern indicates a stochastic event in development. Another way to establish that these mutator genes indeed act at the level of mutations in DNA is to investigate whether RNAi of these genes results in a germline mutator phenotype, as one would expect that mutator genes act in the germline as well as in the soma to protect progeny from harmful mutations. We performed two different assays to validate a possible germline mutator phenotype. (1) For three genes, we quantified the frequency of induced recessive lethal mutations in a balanced region of the genome (Rosenbluth et al. 1983): RNAi against M04F3.1 (encoding replication protein A2), pme-1, and R06C7.7 resulted in 14-, 9- and 8-fold elevated germline mutation frequencies above the wild-type control (data not shown), indicating that these genes protect the germline of C. elegans against mutations. (2) For 24 genes, we determined the mutation frequency using an endogenous monitor gene (Greenwald and Horvitz 1980; see also below). As shown in Figure 2, 23 genes have a significantly increased germline mutation frequency compared to the wild-type control, indicating that these genes (and thus probably 96% of the genes that were identified) also protect the germline against mutations. Of this subset, only C17G10.4(RNAi), the putative ortholog of cdc14, does not show an increased germline mutation frequency, suggesting that this gene may only act in the soma to protect the DNA against mutations.
Figure 2.
RNAi-mediated germline mutator phenotype in C. elegans. An endogenous mutational target, unc-93 (e1500), in which mutations can be recognized based on altered movement of the animal (Greenwald and Horvitz 1980) is used to determine the level of spontaneous mutagenesis levels in the C. elegans germline. This assay was performed for 24 genes (Y-axis), several from each category listed in Table 1. As a control, the empty vector L4440 was assayed. The X-axis represents the number of independent unc-93 (e1500) cultures that showed revertant worms. *, p < 0.05; **, p < 0.001; ***, p < 0.0001; using the Chi-square test. The absolute number of cultures that contained revertants versus the total number of cultures analyzed is also indicated. The mutational loads in msh-2(RNAi), mlh-1(RNAi), and pms-2(RNAi) being 18/187, 16/183, and 16/168, respectively, were determined in a separate experiment with a reduced background level of mutations: 3/177.
With this experimental approach, we expected to identify genes whose loss-of-function phenotype is the frequent occurrence of frameshifting mutations in particular DNA substrates (note the presence of the mono A repeat in the reporter) and which thus display not only a mutator phenotype but also a so-called microsatellite instability (MSI) phenotype. To test this presumption, we tested for all clones whether the reversion to proper LacZ expression depended on the presence of the mononucleotide repeat or whether this reversion would also occur in an out-of-frame lacZ lacking a repeat. As shown in Figure 1, RNAi against the mismatch repair genes mlh-1, pms-2, msh-2, and msh-6 does not result in a blue-patched phenotype in the absence of this repeat, which is in good agreement with the MSI phenotype of tumor cells associated with a defect in DNA mismatch repair (MMR). In addition to these four, nine other RNAi clones (Table 1) induced the MSI phenotype, making these genes logical candidates to constitute the class of yet unidentified genes that are causally mutated in HNPCC or HNPCC-like patients.
Additionally, the screen resulted in 48 clones, for example, M04F3.1, pme-1, and R06C7.7 that induced reversion to correct lacZ expression independent of the presence of the mononucleotide repeat in the reporter (Fig. 1), suggesting that the molecular lesion that leads to such reversion to in-frame LacZ expression occurs at alternative DNA substrates. Perhaps small insertions or deletion mutations are induced in DNA sequences that are positioned between the ATG and the LacZ coding region; the reporter contains an upstream GFP ORF fused to the LacZ ORF. To test this possibility directly, we determined the mutation spectrum for three of these genes, M04F3.1, Y71F9AL.18 (pme-1), and R06C7.7. RNAi against these genes was performed on unc-93 worms that carry a dominant mutation in the unc-93 gene rendering these worms immobile and egg-laying defective (Greenwald and Horvitz 1980). Loss-of-function mutations in unc-93 results in wild-type movement and egg laying, and thus loss-of-function mutations in this mutational target can easily be isolated and subsequently identified by sequencing the unc-93 ORF.
Table 2 lists the molecular nature of the mutations that are induced in M04F3.1(RNAi), Y71F9AL.18(RNAi), and R06C7.7(RNAi), which are primarily small deletion or insertion events. Strikingly, these DNA alterations did not preferentially occur at sequences that have a repeated nature, which explains why, in the somatic mutator assay, LacZ expression is restored in transgenes that lack the mononucleotide repeat (Fig. 1). This outcome differs from the frameshift-resulting mutations in mismatch repair-deficient animals—those occur solely in repeated nucleotide tracts present in the unc-93 mutational target (Tijsterman et al. 2002)—and suggests that the processes (or their disturbance) that can trigger such mutational events are functionally different for these different types of mutator genes.
Table 2.
Loss of function of M04F3.1, Y71F9AL.18 and R06C7.7 results in frequent insertion/deletion mutations
| RNAi against gene
|
Frameshift mutationsa
|
Mutation type (del =deletions; ins = insertions)
|
Mutated sequence: unc-93 mutational target
|
|---|---|---|---|
| M04F3.1 | 10/12 | del. nt 21 | TGGCTTTTTCCAGATactcttctatgatccctgcttCAATATTCTGTGGAA |
| del. nt 27 | ATATTTGACGCGCaatttcatatctcctctcaggtttggcCACTAAATGCAGC | ||
| del. A | TATGTAGGTATGTAGGaTATGTAGGTATGTA | ||
| G→ACT | TCTACATTTTGCTA(G→ACT)TTGACAACTTT | ||
| TA→ACT | ACTAACCTTTGTA(TA→ACT)AACTCCAACT | ||
| G→AT | GACCAATTTATTAC(G→AT)TGGAAGTCTTC | ||
| ins. A | AAAATACTTTGGGCTCACTCGAAAAACTCT | ||
| ins. A | AACTGATTTTTTTGATATTTTAATTATTTTC | ||
| ins. T | CATGCATTCGGTAGATTGATGAAGATAATG | ||
| ins. T | AAACATCGGTGGCCGTAGTTTTCCAATTTTC | ||
| Y71F9AL.18 | 6/9 | del. nt 1184→ins. nt 523 | AAATTTGAA(del. nt 2147–3331→ins. nt 4358–4880)GGCCACCAC |
| del. AAAA | TTTGGAGCGATTCCAaaaaAGAAGGATGATAG | ||
| TA→GAGCCCAAA | CGAGGAGCCCAAA(TA→GAGCCCAAA)TTTTCCTTTCGAG | ||
| ins. AC | TTCAGTGGGAAGTTCACGAAATGATCGTCGT | ||
| ins. A | TTTCTCTTCCTGTTCAACTGCATTTAACGGAC | ||
| ins. A | AAGAGATTCCCAAAAATCGATATTTAGTGAA | ||
| R06C7.7 | 7/10 | del. CATCAT | TATCACTTGCCATATcatcatTATTTGTTCCATC |
| del. GTCATT | TATTTTCTATACATTgtcattAATTTGAGGCCGA | ||
| del. AAAC | AATTGGTCACAATGCaaacTCTCATGCATTCG | ||
| del. TCT | TTTTTCCAGATACTCTtctATGATCCCTGCTTC | ||
| del. N10→ins. GT | AAAACTTCGATT(catcttttgc→GT)AAAAATTTG | ||
| del. N10→ins. T16 | TCATATCTCC(tctcaggttt→TTTTTTTTTTTTTTTT)GGCCACTAAA | ||
| ins. T | CTACATACCTACATATTCTACAAACCTACAT |
The remainder are single base pair substitutions (not depicted).
In this study we systematically screened the C. elegans genome for genes that protect the genetic information against mutations. This screen was certainly not saturated: the RNAi library covers 87.2% of all predicted genes, and RNAi does not always efficiently result in a loss-of-function phenotype [estimates suggest knock-out phenotypes for 65% of the genes if all phenotypes are taken into account (Fraser et al. 2000; Kamath et al. 2003)]. The screen we describe here does not depend on the silencing of a gene in a given tissue or stage of development: as long as some sublineages are hit, one will see blue patches. Thus it would not be surprising if the success rate is >65%. To evaluate the hit rate of this screen further, we retested a collection of genes (in triplicate) that are known or expected to be involved in various types of DNA metabolic processes (Boulton et al. 2002 and references therein; e.g., base excision repair, nucleotide excision repair, homologous and nonhomologous recombination, DNA damage checkpoint, and DNA replication). Of 73 genes tested, 17 clones scored positive: 13 were previously identified in the genome-wide RNAi screen, but 4 were initially missed, confirming the notion of a certain degree of interexperimental variation in high-throughput genome-wide RNAi screens. Also note that the effectiveness of RNAi in knocking down gene expression can be variable for individual genes, and the mutation frequency for the genes we analyzed are therefore not the mutation frequency of null alleles per se. In support, the elevated mutation frequency by msh-6(RNAi) is lower than the increase in mutation frequency as a result of a genetic knockout: msh-6(pk2504).
One advantage of screening an RNAi library is that essential genes can also be recognized (Table 1); these are usually overlooked by normal forward screens. In these cases, the blue patches are seen either in sick or sterile animals, or in “escapers”: animals that have escaped the embryonic lethal phenotype but have inherited the RNAi effect, leading to reduction of function of the relevant gene in somatic tissues. In the complete set of genes we found 19 mutator genes that also cause embryonic lethality, reduced brood size, or a severe growth delay (Table 1).
In total, 61 genes were identified. Of these, 82% have clear orthologs in the human genome. This is much higher than the estimated value of all genes that have a counterpart encoded by the human genome (only 53% of the C. elegans genes that have been studied genetically or biochemically show homology to genes in other species; Reinke et al. 2000), confirming that the genes protecting the genome's integrity are ancient and well conserved. Forward mutator screens have been done with bacteria and yeast. An advantage of screening with C. elegans is that as a multicellular organism, it can have supplemental mechanisms for DNA protection, which may not exist in unicellular organisms. Indeed, we recognized several genes that are present in human and fly but not in yeast (Table 1), suggesting that multicellular organisms have indeed evolved additional mechanisms to protect the integrity of the DNA.
Normal somatic cells transform into malignant cancer cells after multiple gene inactivations. These gene inactivations are probably inflicted by the mutator phenotype that many or all cancer cells have. In, for example, mismatch repair-deficient tumors, there is an increased mutation frequency that is often seen as DNA repeat instability. There are still tumor suppressor genes to be discovered; we suggest that among the genes identified in this screen are somatic mutators that cause human cancers.
Materials and methods
Genome-wide RNAi screen
The RNAi library and the RNAi feeding protocol are described by Ahringer and coworkers (Kamath et al. 2001). E. coli clones containing dsRNA producing feeding constructs were grown overnight at 37°C in LB containing ampicillin. Agar plates were seeded with these cultures, incubated overnight at room temperature and then seeded with 2–4 worms of strain NL3400: pkIs1604 [rol-6(su1006) hsp16/2∷ATG(A)17gfp/lacZ (out of frame)] at the L3/L4 developmental stage. Nematodes were grown at 20°C for 5 d, washed off the plates, fixed, and stained for the presence of β-galactosidase with X-gal. Positive feeding clones were tested again in triplicate and with strain NL3401: pkIs1605 [rol-6(su1006) hsp16/2∷gfp/lacZ(out of frame)]. For all clones that induced the mutator phenotype, plasmids were isolated and the dsRNA encoding DNA sequence was sequenced.
unc-93 reversion assay
On 50 assay plates (9-cm Petri dishes seeded with RNAi food), 30 unc-93 (e1500) animals of the L1 stage were placed, grown until food deprivation (the F1 was also directly inspected for revertants to exclude spontaneous mutations in earlier generations; none were found), and subsequently transferred twice to fresh RNAi assay plates (similar numbers of worms were transferred). Then, cultures were scored for the presence of worms that had reverted to a wild-type phenotype. The number of plates that contain such revertants was taken as a measurement for the germline mutation frequency. For RNAi against genes M04F3.1, Y71F9AL.18, and R06C7.7, this procedure was repeated three times to obtain a sufficient number of mutations that are within the unc-93 gene (note that inactivation of at least five different loci suppress the unc-93 phenotype). In all animals, the unc-93 gene was sequenced.
Acknowledgments
We thank members of the Plasterk lab for discussions and the Caenorhabditis Genetic Stock Centre for providing some of the strains used in this study.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Corresponding authors.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1060703.
References
- Araujo SJ, Tirode F, Coin F, Pospiech H, Syvaoja JE, Stucki M, Hubscher U, Egly JM, Wood RD. Nucleotide excision repair of DNA with recombinant human proteins: Definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes & Dev. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Gartner A, Reboul J, Vaglio P, Dyson N, Hill DE, Vidal M. Combined functional genomic maps of the C. elegans DNA damage response. Science. 2002;295:127–131. doi: 10.1126/science.1065986. [DOI] [PubMed] [Google Scholar]
- Burkle A. Poly(APD-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett. 2001;163:1–5. doi: 10.1016/s0304-3835(00)00694-7. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington, DC.: Am. Soc. Microbiol.; 1995. [Google Scholar]
- Greenwald IS, Horvitz HR. unc-93(e1500): A behavioral mutant of Caenorhabditis elegans that defines a gene with a wild-type null phenotype. Genetics. 1980;96:147–164. doi: 10.1093/genetics/96.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:research0002.1–research0002.10. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, le Bot N, Moreno S, Sohrman M, et al. Systematic functional analysis of the C. elegans genome using RNAi. Nature. 2003;421:286–287. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes & Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- Kuo MH, vom Baur E, Struhl K, Allis CD. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- ————— A mutator phenotype in cancer. Cancer Res. 2001;61:3230–3239. [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Rosenbluth RE, Cuddeford C, Baillie DL. A rapid eukaryotic mutagen test system using the reciprocal translocation, eT1(III;V) Mutat Res. 1983;110:39–48. [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Tijsterman M, Pothof J, Plasterk RH. Frequent germline mutations and somatic repeat instability in DNA mismatch-repair-deficient Caenorhabditis elegans. Genetics. 2002;161:651–660. doi: 10.1093/genetics/161.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Wang Z. Translesion synthesis by the UmuC family of DNA polymerases. Mutat Res. 2001;486:59–70. doi: 10.1016/s0921-8777(01)00089-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]