Abstract
The PWI motif is a highly conserved domain of unknown function in the SRm160 splicing and 3′-end cleavage-stimulatory factor, as well as in several other known or putative pre-mRNA processing components. We show here that the PWI motif is a new type of RNA/DNA-binding domain that has an equal preference for single- and double-stranded nucleic acids. Deletion of the motif prevents SRm160 from binding RNA and stimulating 3′-end cleavage, and its substitution with a heterologous RNA-binding domain restores these functions. The NMR solution structure of the SRm160-PWI motif reveals a novel, four-helix bundle and represents the first example of an α-helical fold that can bind single-stranded (ss)RNA. Structure-guided mutagenesis indicates that the same surface is involved in RNA and DNA binding and requires the cooperative action of a highly conserved, adjacent basic region. Thus, the PWI motif is a novel type of nucleic acid-binding domain that likely has multiple important functions in pre-mRNA processing, including SRm160-dependent stimulation of 3′-end formation.
Keywords: RNA-binding domain, spliceosome, 3′-end cleavage
An important goal of studies in gene regulation is to understand the molecular basis by which multisubunit complexes recognize target substrates to facilitate the catalysis, coordination, and regulation of different reactions. RNA-binding proteins play central roles in the assembly and function of complexes at virtually every step in the gene expression pathway, including mRNA transcription, 5′-end capping, splicing, 3′-end processing (cleavage and polyadenylation), surveillance, turnover, transport, and translation. Discrete domains in these proteins are usually responsible for RNA binding and function in conjunction with one or more other domains that provide functionalities such as protein–protein interactions, RNA modification, and subcellular localization.
There are several well-characterized RNA-binding domains that play essential functions in gene expression (Burd and Dreyfuss 1994; Draper 1999; Antson 2000; Dreyfuss et al. 2002). These include the RNA Recognition Motif [RRM; also referred to as the RNA-binding domain (RBD) or RNP motif; Query et al. 1989], KH (Siomi et al. 1993), dsRBD (St Johnston et al. 1992), S1 (Suryanarayana and Subramanian 1979), Zinc finger (Miller et al. 1985), and different basic-rich motifs (Malim et al. 1990; Chen and Frankel 1994). In addition to facilitating the assembly of multisubunit complexes that perform critical functions in RNA synthesis and processing, RNA-binding domains such as these can serve important regulatory roles by altering RNA structure to promote or suppress interactions, for example, by positive- or negative-acting protein or RNA trans-acting factors (Siomi and Dreyfuss 1997). They can also serve to promote the transport of RNA molecules to specific intracellular locations by locating targeting signals (Yaniv and Yisraeli 2001). Such activities can be regulated in a cell-type and developmental stage-specific manner and are known to play key roles in many cell growth and differentiation pathways (Curtis et al. 1995). On/off regulation of RNA activity mediated by RNA-binding domains provides rapid responses to changing requirements in gene expression and can be regulated by various signaling pathways. Perhaps not surprisingly in light of their widespread and critical physiological roles, disruption of the activity of RNA-binding domains has been directly implicated in several human diseases (Siomi and Dreyfuss 1997).
Clearly, the identification and characterization of new domains involved in RNA recognition, as well as a more detailed understanding of existing domains, is important for elucidating the molecular basis of mechanisms in gene expression. The PWI motif is a highly conserved domain of unknown function, named after an almost invariant Pro-Trp-Ile signature located within its N-terminal region (Blencowe and Ouzounis 1999). The PWI motif is present in SRm160 (SR-related nuclear matrix protein of 160 kD) and several other known or putative pre-mRNA processing components. SRm160 functions as a coactivator of constitutive and exon enhancer-dependent splicing, by bridging interactions between important splicing factors that bind directly to pre-mRNA (Blencowe et al. 1998; Eldridge et al. 1999). It also stimulates the 3′-end cleavage of transcripts and is thought to mediate coupling between splicing and 3′-end formation (McCracken et al. 2002). After splicing, SRm160 remains bound to mRNA and forms a component of an exon junction complex that has been implicated in mRNA export and turnover by the nonsense-mediated decay pathway (Kataoka et al. 2000; Le Hir et al. 2000a,b; Zhou et al. 2000; Kim et al. 2001; Lykke-Andersen et al. 2001; Lejeune et al. 2002). In addition to SRm160, the PWI motif is present in other spliceosomal proteins, including mammalian homologs of the yeast Prp3p protein (PRP3), which is a component of the U4/U6 snRNP (Horowitz et al. 1997; Lauber et al. 1997; Wang et al. 1997), and two proteins of unknown function (S164/fSAP94 and PRO1777) recently identified in proteomic analyses of human splicing complexes (Rappsilber et al. 2002; Zhou et al. 2002). An intriguing feature of the PWI domain is that it always resides at the N or C terminus of a protein, never in the middle. A subset of PWI motif proteins also contain an RRM, providing additional evidence for a close link between these proteins and RNA metabolism (Fig. 1A).
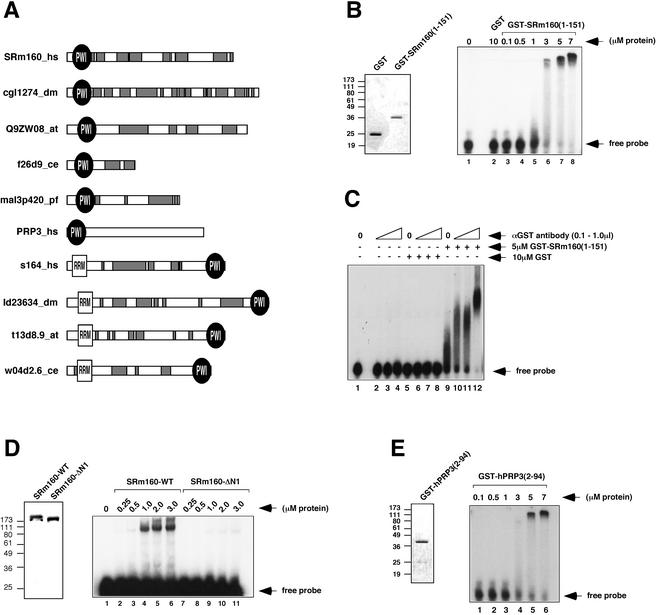
Figure 1.
PWI motif-containing domains of SRm160 and hPRP3 bind RNA. (A) Domain organization of selected PWI-motif proteins from the SMART database (Schultz et al. 1998). The approximate location of PWI and RNA recognition motifs (RRMs), as well as regions of low sequence complexity (gray boxes) are indicated. (B) RNA EMSA of GST-SRm160(1–151) and GST proteins with T7-MCS RNA (see Materials and Methods). The amounts of each protein in the EMSA experiment are indicated in micromolars. A Coomassie-stained gel containing the purified proteins is shown in the left panel. (C) Supershift assay with GST-SRm160(1–151). Five micromolars of GST-SRm160(1–151) or 10 μM of GST was incubated with the T7-MCS RNA and increasing amounts of a polyclonal antiserum specific for GST. RNA binding was analyzed by EMSA. Note that the complexes formed specifically between GST-SRm160(1–151) and RNA were only separated for a short time, in order to observe a supershift with the anti-GST antibody. (D) RNA-EMSA with purified, baculovirus-expressed, wild-type SRm160 (SRm160-WT) and SRm160 lacking residues 1–151 (SRm160-ΔN1). Binding to the T7-MSC-RNA was assayed as in B and C. A Coomassie-stained gel containing the purified SRm160-WT and SRm160-ΔN1 proteins is shown in the left panel. (E) RNA-EMSA with GST-hPRP3(2–94) and the T7-MCS RNA. A Coomassie-stained gel containing the GST-hPRP3(2–94) protein is shown in the left panel.
In this study we present a combined structural and functional characterization of the PWI motif. We demonstrate that it is a new type of nucleic acid-binding domain with dual specificity for RNA and DNA. We show that the RNA-binding activity of the PWI motif in SRm160 is important for the stimulation of mRNA 3′-end processing. The solution structure of this PWI motif reveals that it consists of a novel, four-helix bundle. Comparison with other solved structures reveals weak similarity to a DNA-binding region within Endonuclease III and other helix-hairpin-helix family glycosylases involved in DNA repair, but not to any existing structures of RNA-binding proteins. Structure-guided mutagenesis indicates that RNA- and DNA-binding requires the same surface of the PWI domain, as well as the cooperative activity of an adjacent, highly conserved, basic-rich region. Unlike any other α-helical RNA-binding domains previously characterized, the PWI domain has a similar binding affinity for single- and double-stranded nucleic acids. Our study therefore reveals the existence of a new mode of nucleic acid binding that is likely to have multiple important functions in gene expression, including the stimulation of mRNA 3′-end formation by SRm160.
Results
The highly conserved N-terminal domain of SRm160 binds RNA
The N-terminal region of SRm160, spanning residues 1–151, represents the most highly conserved domain of the protein (Blencowe et al. 1998). For example, it shares 52% identity between human and Caenorhabditis elegans SRm160 homologs (Longman et al. 2001). To investigate the function of this region, we performed interaction assays using amino acid residues 1–151 as bait in a yeast two-hybrid screen of a HeLa cDNA library. The same region, fused to GST, was also used to isolate interacting proteins by affinity chromatography of HeLa nuclear extract proteins. Both assays revealed interactions with proteins known to bind directly to RNA (J. Bowman and B.J. Blencowe, unpubl.). In GST pull-down assays, these interactions were completely lost after RNase pretreatment of the nuclear extract, suggesting that SRm160(1–151) and these RNA-binding proteins primarily associate through an interaction bridged by RNA. This raised the interesting possibility that the conserved N-terminal domain of SRm160 might function in RNA binding.
To test whether the conserved N-terminal domain of SRm160 binds RNA, we performed electrophoretic mobility shift assays (EMSAs) using GST-SRm160(1–151) and different RNA substrates. Consistent with an RNA-binding function, increasing concentrations of GST-SRm160(1–151) resulted in the decreased mobility of a radiolabeled RNA transcript of 47 nucleotides, derived from the multiple cloning site of pBS (T7-MCS RNA; Fig. 1B). Approximately 4–7 μM of the fusion protein was sufficient to quantitatively shift the input RNA into a discrete complex (Fig. 1B, lanes 7,8). At 3 μM of the fusion protein, the RNA substrate was distributed in a smear in the gel, indicating either weak binding or possibly the multimerization of the fusion protein on the transcript (Fig. 1B, lane 6; see below). Similar results were obtained with a transcript of distinct sequence (derived from the bacteriophage lambda Nut site) which consists of a stable hairpin-loop structure (data not shown). In experiments with either substrate, levels of GST (up to 10 μM) did not result in any change in the mobility of the input RNA (Fig. 1B, lane 2). These results indicate that the N terminus of SRm160 can bind RNA with relatively low sequence specificity, and with an apparent affinity in the micromolar range.
To confirm that the shift in gel mobility of the RNA substrates is due to binding of GST-SRm160(1–151) and not minor contaminating proteins in the preparation, we performed a supershift experiment using a polyclonal antibody specific for GST (αGST Ab; Fig. 1C). Addition of increasing amounts of αGST Ab resulted in a slower migration of GST-SRm160(1–151)-containing complexes (Fig. 1C, lanes 10–12), whereas addition of equivalent amounts of αGST Ab did not alter the migration of the T7-MCS RNA in the presence (Fig. 1C, lanes 6–8) or absence (Fig. 1C, lanes 2–4) of GST alone. The progressively slower migrating complexes probably reflect multivalent interactions between the polyclonal αGST Ab and GST (note that the complexes in Fig. 1C were resolved on a lower percent gel in order to observe a supershift). These results confirm that GST-SRm160(1–151) binds RNA.
RNA-binding activity of SRm160 requires the highly conserved N-terminal domain
In splicing reactions performed in vitro, the association of SRm160 with pre-mRNA is initially dependent on other splicing factors that bind the substrate directly (Blencowe et al. 1998; Eldridge et al. 1999). However, SRm160 can be cross-linked to spliced mRNA, suggesting that it is either in close proximity or else in direct contact with mRNA at a later stage of the spliceosome cycle (Le Hir et al. 2000b). This transition suggests that the RNA-binding activity associated with SRm160(1–151) may initially be masked by other components that interact with SRm160. To investigate whether SRm160 can bind RNA in the absence of other factors and whether such an activity requires the conserved N-terminal domain of the protein, we next compared the ability of highly purified, baculovirus-expressed, wild-type SRm160 (SRm160-WT), and a deletion derivative missing residues 1–151 (SRm160-ΔN1), to bind RNA (Fig. 1D). Notably, SRm160-WT, but not SRm160-ΔN1, significantly reduced the mobility of a fraction of the RNA (T7-MCS) in gel shift assays (Fig. 1D, cf. lanes 2–6 and 7–11). The binding affinity of the SRm160-WT protein was somewhat less than that of GST-SRm160(1–151), because a lower yield of gel-shifted complexes was obtained (cf. Fig. 1B and D). This could be attributed to a fraction of inactive protein in the preparation and/or the presence of a higher concentration of salt (300 mM vs. 100 mM) present in the binding reaction, which was necessary to maintain the solubility of the SRm160-WT and SRm160-ΔN1 proteins. Nevertheless, the results demonstrate that SRm160, in the absence of other splicing components, can bind RNA and that, consistent with the results described above, this activity depends on the presence of the highly conserved N-terminal domain of the protein.
PWI motif-containing proteins bind RNA
An interesting possibility raised by the results described thus far is that the PWI motif within the conserved N-terminal domain of SRm160 is responsible for RNA-binding activity. If this is the case, the PWI motif of an otherwise completely unrelated splicing factor should also bind RNA. To test this, a GST fusion protein containing amino acids 2–94 of hPRP3 [GST-PRP3(2–94)] was initially assayed for RNA-binding activity (Fig. 1E). This region primarily comprises a PWI motif (residues 3–76) that is only 35% identical with the analogous region in SRm160 and bears no sequence similarity with SRm160 outside of the conserved motif residues (Blencowe and Ouzounis 1999). Significantly, GST-PRP3(2–94) bound the T7-MCS transcript with remarkably similar affinity as SRm160(1–151), shifting essentially all of the RNA in the 5–7 μM concentration range (cf. Fig. 1B, lanes 5–8, and 1E, lanes 3–6). This suggests that the RNA-binding activities of GST-SRm160(1–151) and GST-PRP3(2–94) involve the PWI motifs of these proteins, a conclusion that was later confirmed by structure-guided mutagenesis of these domains (see below).
To further characterize the properties of the PWI motif region of SRm160, we next asked whether this domain possesses preferential binding for single- versus double-stranded (ds)RNA, and also whether it can bind ssDNA and dsDNA. Gel mobility-shift assays were performed using GST-SRm160(1–151) and each type of nucleic acid substrate, corresponding in sequence to the T7-MCS RNA. In each case, a similar binding affinity was measured, indicating that the PWI motif has dual RNA- and DNA-binding properties as well as a similar preference for either double- or single-stranded nucleic acid substrates (data not shown).
The RNA-binding function of the PWI domain of SRm160 facilitates the 3′-end cleavage of transcripts
The RNA-binding function of the PWI domain of SRm160 could potentially act at multiple steps in pre-mRNA processing, because SRm160 is important for splicing and also promotes the 3′-end processing and export of transcripts. We recently demonstrated that deletion of the N-terminal domain of SRm160 containing the PWI motif prevents SRm160 from stimulating 3′-end cleavage in vivo (McCracken et al. 2002). Taken together with the results of the present study, this suggests that the 3′-end cleavage-stimulatory function of SRm160 requires the RNA-binding function provided by the PWI motif. To test this, we asked whether substitution of the PWI motif region of SRm160 with a heterologous RNA-binding domain can rescue its cleavage-stimulatory function (Fig. 2).
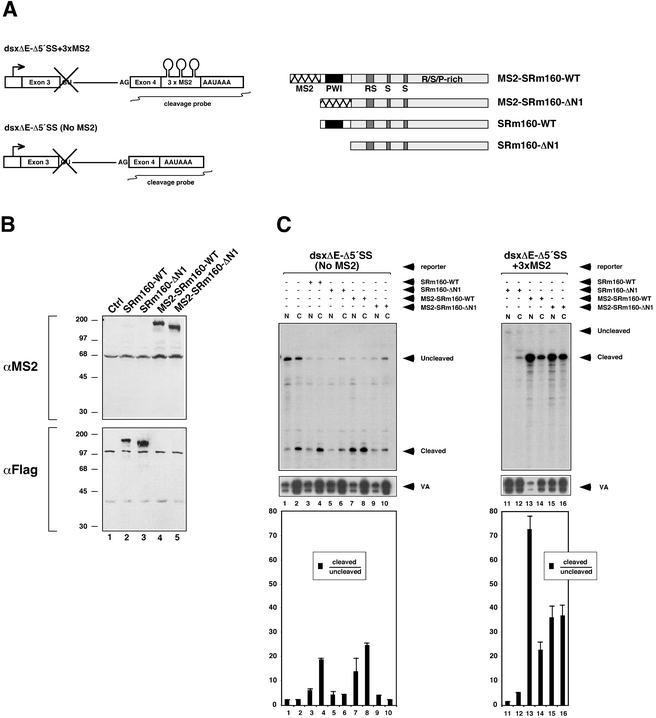
Figure 2.
A heterologous RNA-binding domain can functionally substitute for the PWI domain of SRm160. (A) Diagram of pre-mRNA reporters and test proteins. Pre-mRNA reporters were derived from exons 3 and 4 of the Drosophila doublesex (dsx) gene and contained a deletion in the 5′-splice site to eliminate splicing activity. One of the reporters contains three tandem RNA-binding sites for the phage MS2 coat protein in exon 4, as indicated. The position of the RNase protection probes used to monitor 3′-end cleavage of transcripts from each reporter is indicated. Test proteins corresponding to SRm160-WT and SRm160-ΔN1 (Fig. 1D legend), with or without an MS2 RNA-binding domain fused to the N terminus, were analyzed. All four proteins contained an N-terminal Flag-epitope for detection. (B) Immunoblots of proteins from cells transfected with or without expression plasmids for the four test proteins shown in A. The immunoblot in the top panel was probed with the anti-MS2 antibody, and the immunoblot in the bottom panel was probed with the anti-Flag antibody. Note: Both MS2 fusion proteins were detected with the anti-Flag antibody after prolonged exposure (data not shown). (C) RNase protection analysis of reporter transcripts with the 3′-end protection probes illustrated in panel A. Cells transfected with each reporter and test protein combination are indicated. A pol III reporter (pSPVA) was cotransfected in each case as an internal control for transfection efficiency and RNA recovery. RNase protections were performed on RNA isolated from both nuclear (N) and cytoplasmic (C) fractions from transfected cells, as indicated. Ratios of cleaved to uncleaved RNA in each fraction are indicated in the bar graphs with standard deviations, as determined from three separate analyses. Note that the two gel panels are from the same experiment and were separated only to facilitate labeling the positions of the different size RNA products.
We replaced the N-terminal domain of SRm160 with the MS2 bacteriophage coat protein, which binds with high affinity and specificity to a defined stem-loop structure, the MS2-binding site (Carey et al. 1983). Expression plasmids encoding Flag-epitope-tagged versions of wild type (fSRm160-WT) and SRm160 lacking residues 1–151 (fSRm160-ΔN1), in each case with or without an MS2 domain fused N-terminally (fMS2-SRm160-WT and fMS2-SRm160-ΔN1), were transfected into human 293 cells with one of two versions of a model, two-exon, pre-mRNA reporter containing the SV40-late cleavage and polyadenylation signal (Fig. 2A). These reporters differed only by the presence or absence of three tandem MS2-binding sites (3xMS2) in the 3′-exon. The 5′ splice site was deleted in the reporters in order to separate possible effects of SRm160 expression on 3′-end cleavage from effects on splicing, because these steps are normally coupled and can influence one another (Wassarman and Steitz 1993; Nesic and Maquat 1994; Lutz et al. 1996; Gunderson et al. 1998; Lou et al. 1998; Vagner et al. 2000). An RNA pol III-transcribed viral-associated (VA) RNA reporter, which is not influenced by SRm160 levels, was included in each transfection as an internal control for transfection efficiency and RNA recovery. For each transfection condition, harvested cells were separated into nuclear and cytoplasmic fractions to assess whether the expressed protein influences the nuclear:cytoplasmic ratio of the reporter transcripts, as well as their level of 3′-end cleavage.
The levels of all RNAs were measured by quantitative RNase protection using 32P-UTP-radiolabeled antisense probes; the probes used to quantify cleavage levels of the pre-mRNA reporter transcripts are depicted in Figure 2A. The levels of proteins from the transfected expression plasmids were monitored by immunoblotting with either anti-Flag or anti-MS2 protein antibodies (Fig. 2B). This revealed comparable expression levels between fSRm160-WT and fSRm160-ΔN1 proteins, and between fMS2-SRm160-WT and fMS2-SRm160-ΔN1 proteins. The MS2 fusion proteins were expressed at considerably lower levels than the SRm160 proteins lacking the MS2 domain (Fig. 2B, cf. lanes 4,5 and 2,3; see below), and could only be detected with the anti-Flag antibody after prolonged exposure of the blot shown in Figure 2B (data not shown).
In transfections with the pre-mRNA reporter lacking MS2-binding sites, expression of fSRm160-WT resulted in an approximately sixfold increase in the ratio of cleaved to uncleaved RNA, whereas expression of fSRm160-ΔN1 did not significantly alter the ratio of cleaved:uncleaved RNA, compared to the transfection using the control, empty expression plasmid (Fig. 2C, cf. lanes 3–6 and 1,2). In the case of the substrate lacking MS2-binding sites, expression of fMS2-SRm160-WT and fMS2-SRm160-ΔN1 proteins resulted in similar relative levels of cleavage compared to the corresponding proteins lacking the MS2 domain (Fig. 2C, cf. lanes 7–10 and 3–6). However, the presence of the MS2 domain in the context of wild-type SRm160 caused an increased level of cleaved RNA appearing in the nuclear as well as the cytoplasmic fraction, whereas SRm160-WT without the MS2 domain resulted in an increased level of cleaved RNA appearing primarily in the cytoplasmic fraction (Fig. 2C, cf. lanes 7 and 3; see below). Despite the considerably lower levels of expression of the fMS2-fusion proteins, compared to fSRm160-WT and fSRm160-ΔN1 proteins (Fig. 2B, cf. lanes 4,5 and 3,4), fMS2-SRm160-WT still resulted in an approximately sixfold stimulation of cleavage of the dsx pre-mRNA reporter lacking MS2 sites (Fig. 2C, lane 8). This suggests that nonspecific binding activity attributed to the MS2 domain synergizes with the RNA-binding activity of the PWI domain, allowing for relatively efficient targeting of the fMS2-SRm160-WT protein to transcripts lacking MS2-binding sites.
Parallel transfections performed with both fMS2-SRm160-WT and fMS2-SRm160-ΔN1 expression plasmids, together with the pre-mRNA reporter containing MS2-binding sites, both resulted in significant increases in the cleaved:uncleaved ratios, compared to the corresponding ratios observed when the same proteins were expressed with the reporter pre-mRNA lacking MS2-binding sites (Fig. 2C, cf. lanes 13–16 and 7–10). Expression of fMS2-SRm160-WT resulted in an approximately fourfold further increase in cleavage efficiency over fSRm160, indicating that more efficient RNA binding provided by the interaction between the MS2 domain and its cognate binding sites enhances the cleavage-stimulatory activity of SRm160 (Fig. 2C, cf. lanes 13,14 and 7,8). Importantly, expression of fMS2-SRm160-ΔN1 resulted in an ∼10-fold increase in the cleaved:uncleaved RNA ratio compared to when this protein was expressed with the dsxΔE-Δ5′SS reporter lacking MS2-binding sites (Fig. 2C, cf. lanes 15,16 and 9,10), or when fSRm160-ΔN1 was expressed in combination with the dsxΔE-Δ5′SS+3xMS2 pre-mRNA (Fig. 2C, cf. lanes 15,16 and 11,12). When the fMS2-SRm160 derivatives were expressed with the reporter containing binding sites, the increases in cleaved RNA were observed in the nuclear as well as the cytoplasmic fractions, indicating that the relatively high-affinity RNA-binding interaction provided by the MS2 domain causes transcript retention in the nucleus, in addition to stimulation of 3′-end cleavage (Fig. 2C, lanes 13,15).
These results demonstrate that the high-affinity RNA binding activity of the MS2 protein can functionally substitute, and even further enhance, the role of the highly conserved PWI motif-containing domain of SRm160 in facilitating 3′-end cleavage. Taken together with the data demonstrating the RNA-binding activity of this domain, our results indicate that reduced 3′-end cleavage activity upon deletion of the N terminus of SRm160 is caused by the loss of the RNA-binding function associated with the PWI motif region. We conclude, therefore, that the RNA-binding function provided by this region is important for SRm160 to promote 3′-end cleavage.
Structure of the PWI motif reveals a novel nucleic acid-binding domain
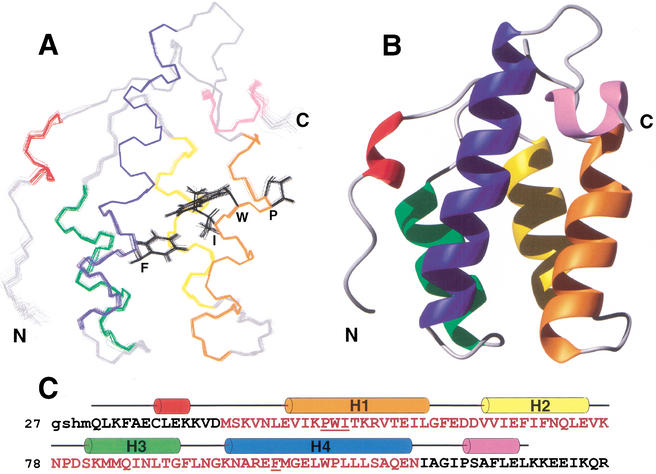
To better understand how the PWI domain interacts with nucleic acids, we determined its three-dimensional solution structure. Limited proteolysis of the 151-residue N-terminal construct of SRm160 used in the initial analyses, followed by mass spectrometry, identified a protease-resistant domain spanning residues 27–128, which encompasses the entire conserved PWI motif. NMR analysis of several constructs containing this domain indicated that it was a soluble, globular, and independently folded domain amenable to NMR analysis. The solution structure of SRm160(27–134) demonstrates that the PWI motif comprises a four-helix bundle, with structured N- and C-terminal elements (Fig. 3). The ordered N-terminal strand (residues 27–39) packs against the core motif, and residues 33–37 fold into a 3–10 helix (Fig. 3A). C-terminal residues 120–125 form an α-helix that is orthogonal to the core helices. Residues 127–134, which aid in the solubility of the PWI motif, are disordered, consistent with their susceptibility to proteolysis. The conserved region that defines the PWI motif includes all of helices (H) 1–4, and only five amino acids N-terminal of H1 (Fig. 3B,C). The signature Pro-Trp-Ile sequence, for which the PWI motif is named, resides in H1, and appears to be primarily involved in hydrophobic interactions within the core of the protein (Fig. 3A). The Trp and Ile residues pack against the highly conserved phenylalanine 101 in H4. Inspection of the surface properties of the PWI motif revealed an unusual feature for a nucleic acid-binding protein: there are no surface regions rich in basic residues (Fig. 4A). In fact, the pI of the PWI motif from SRm160 is acidic in nature (pI = 4.6). Further investigation revealed that PWI domains generally have acidic pI values, ranging from 4.3 to 6.8, with a median of 4.6. The PWI domain of PRP3 is the only exception, with a basic pI of 8.4 (see below).
Figure 3.
Solution structure of the PWI motif from SRm160. (A) Best-fit superimposition of the backbone atoms from the 20 lowest-energy structures of SRm160 residues 27–126. Residues 127–134, which are disordered, are excluded from the figure for clarity. Helical regions are colored, and nonhelical regions are in gray. Locations of the signature Pro (50), Trp (51), and Ile (52) residues, as well as the highly conserved Phe (101) residue, are indicated in black. (B) Ribbon representation of the lowest-energy calculated structure. The orientation and color scheme are the same as in A. (C) Sequence of the polypeptide used in structure determination. Residues labeled in brown correspond to those that constitute the PWI domain, and the black letters indicate those found in the flanking sequences. Colored cylinders above the sequence indicate α-helices, and follow the same color scheme used in A and B.
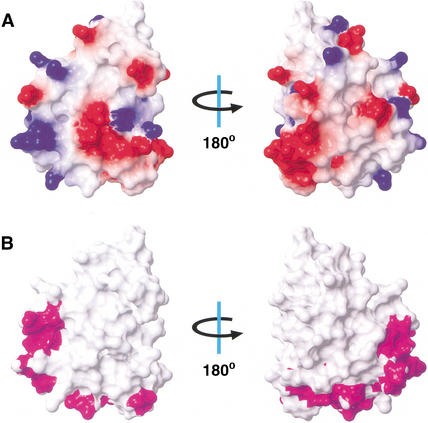
Figure 4.
Surface renderings of the PWI motif structure. (A) Electrostatic surface potential map of SRm160(27–126). Red represents regions of negative charge, blue shows positive charges, and white is neutral. The molecule on the left is in the same orientation as in Figure 3, and the molecule on the right is rotated by 180° about the indicated axis. (B) Map of the residues in SRm160(18–134) that have an altered amide chemical shift upon binding dsDNA (magenta). White surfaces correspond to residues that do not have altered chemical shifts, or ambiguous results due to spectral overlap.
A search for proteins with similar three-dimensional structures using the DALI database (Holm and Sander 1993) indicated that the PWI motif does not resemble any known, small, independently folded nucleic acid-binding motif or domain, and therefore appears to be a new nucleic acid-binding module. However, the search did reveal weak structural similarity to subregions of Endonuclease (Endo) III and MutY (Thayer et al. 1995; Guan et al. 1998), members of the helix-hairpin-helix (HhH) superfamily of DNA repair glycosylases (Z scores of 5.2 and 5.1 respectively; Nash et al. 1996). Interestingly, the regions within these proteins that are similar to the PWI motif are involved in DNA binding (Thayer et al. 1995; Guan et al. 1998; Hollis et al. 2000).
Structure-guided mutagenesis reveals important contact residues for nucleic acid binding and function
To understand how the PWI motif interacts with nucleic acids, we performed an NMR titration of SRm160(18–134) with a 31-bp length of double-stranded DNA and monitored changes in the NMR amide resonance frequencies. For this experiment, an SRm160 fragment consisting of residues 18–134 was used, because it included several additional N-terminal basic residues which were suspected to contribute to nucleic acid binding (see below). Figure 4B shows the residues in the PWI motif whose backbone amide resonances shifted upon addition of DNA. The largest effects localize to the regions near the loops, between H1 and H2, and H3 and H4. Interestingly, this region of the PWI motif corresponds to the DNA-binding surfaces in EndoIII and MutY that have weak structural similarity to the PWI domain. In addition to the residues shown in Figure 4B, we observed substantial resonance changes in the basic N-terminal region spanning residues 18–27, suggesting that this region of SRm160 is also involved in nucleic acid binding.
Based on the results of the NMR titration, we designed several mutants of the PWI motif in which residues suspected of interacting with DNA/RNA were mutated to alanine. Interestingly, none of the individual or combinations of point mutations in the core PWI motif completely abrogated binding to ssRNA or dsDNA, although mutation of Phe92 to Ala reduced binding slightly (data not shown). However, simultaneous mutation of three basic residues in the N-terminal region [Lys 20, Lys 22, and Lys 23; GST-SRm160(18–134)-TM (“Tail Mutant”); Fig. 5A], significantly reduced the affinity of SRm160(18–134) for both DNA and RNA (Fig. 5C, cf. lanes 9–13 and 3–8). Only a very minor level of binding to nucleic acids was observed for this mutant, even at the 20 μM concentration range (data not shown). This result indicated that the basic residues in the region 18–27 might play an important role in the nucleic acid-binding activity of SRm160(18–134). To investigate this, we prepared a GST-fusion protein containing SRm160 residues 18–26. This GST-fusion protein interacted very weakly, in the 6–20 μM range, with ssRNA and dsDNA, and the interaction was easily disrupted by increasing the concentration of NaCl (Fig. 5D, cf. lanes 10–13 and 3–8; data not shown). In comparison, the complex between SRm160(18–134) and dsDNA persisted at 480 mM NaCl (data not shown). A GST-fusion protein lacking the N-terminal basic region but containing just the PWI motif (residues 27–134) also did not bind stably to DNA or RNA (Fig. 5D, lanes 15–20; data not shown). This demonstrates that the basic region (18–26) and the PWI motif (27–134) of SRm160 are both required for full nucleic acid-binding activity (Fig. 5D, cf. lanes 3–8 and 9–20).
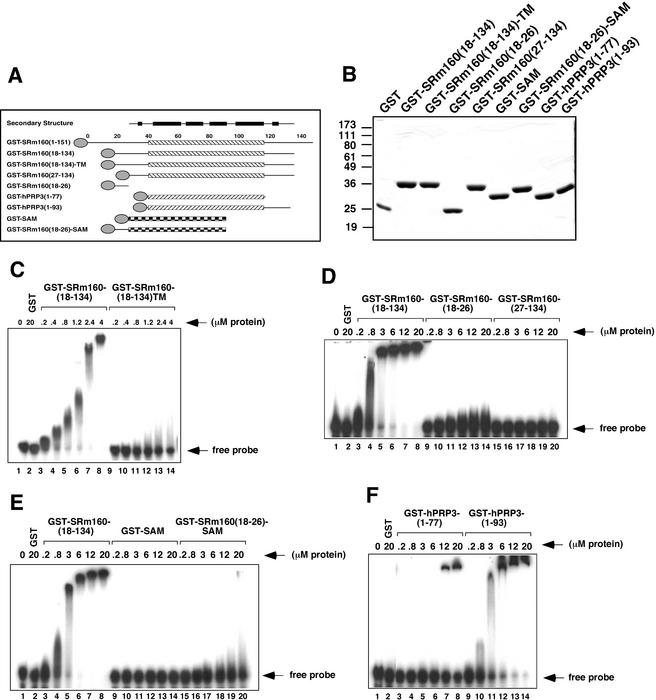
Figure 5.
Cooperative roles of core PWI motifs and conserved, adjacent, basic domains in RNA binding. (A) Schematic diagram of GST-fusion proteins used in RNA-EMSA experiments shown in B–F. Regions corresponding to the conserved PWI motifs in the SRm160 and PRP3 constructs are indicated by the hatched boxes. The structured region of the p73 SAM domain is represented by the checkered box. The residue numbering for SRm160 and the construct used in structure determination, including secondary structure, is depicted above the other constructs. Approximate locations of the three lysine-to-alanine substitutions (Lys 20, Lys 22, and Lys 23) in the GST-SRm160(18–134)-TM (“Tail Mutant”) protein are indicated by small triangles. (B) Coomassie-stained gel of GST-fusion proteins used for gel shift analysis. Masses of the molecular weight markers are indicated. (C) Simultaneous mutation of Lys 20, Lys 22, and Lys 23 to alanines in the GST-SRm160(18–134) protein leads to an abrogation of RNA binding (cf. lanes 9–14 and 3–8). (D) The N-terminal basic domain is required but not sufficient for RNA-binding activity of the PWI motif. Whereas the GST-fusion protein that consists of residues 18–134 binds to the T7-MCS RNA (see Materials and Methods) in the same manner as the N-terminal fragment used in Figure 1 (lanes 3–8), subfragments 18–26 and 27–134, in which the structure of the PWI domain is maintained, are not able to bind RNA in a similar manner. Interactions of the 18–26 fragment (lanes 9–14) with RNA are reduced, and the 27–134 fragment does not appear to interact at all. Similar results were observed in all cases for dsDNA (data not shown). GST itself does not shift the RNA (lane 2). (E) The N-terminal basic region specifically cooperates with the PWI domain to bind RNA. Substitution of the PWI motif with the SAM domain from p73, another helical bundle protein with a similar pI (lanes 15–20), does not rescue binding activity provided by the PWI motif (lanes 3–8) and results in a similar level of binding as observed for the basic region alone (cf. D, lanes 9–14). The SAM domain on its own also does bind RNA (lanes 9–14). (F) Efficient RNA binding of the hPRP3 PWI motif requires a C-terminal basic domain. Removal of 17 amino acid basic sequence from the C terminus of hPRP3 (lanes 3–8) leads to a considerable reduction in binding relative to the GST-PRP3(1–93) fusion protein (lanes 9–14).
The strong contribution played by the N-terminal basic “tail” prompted us to investigate whether this region can confer nucleic acid-binding ability to another helix bundle, or whether this activity is specific to the PWI helix bundle. We prepared a GST fusion with the basic region fused to the N terminus of another small helix bundle, the sterol-alpha-motif (SAM) domain of p73 (Fig. 5E; Chi et al. 1999). By itself, the SAM domain did not bind RNA or DNA (Fig. 5E, lanes 9–14; data not shown). Moreover, when fused at its N terminus to SRm160(18–26), only a minor level of RNA or DNA binding was detected, similar to the levels observed for GST-SRm160(18–26) (cf. Fig. 5E, lanes 15–20, and Fig. 5C, lanes 9–14). These results demonstrate that the PWI motif specifically functions in RNA or DNA binding, and that another compact helical bundle cannot substitute for this function. Moreover, the N-terminal basic region of SRm160 is necessary, but not sufficient, for nucleic acid binding of the SRm160 PWI domain. These results therefore indicate that the N-terminal basic region and PWI motif specifically function together in a cooperative manner to provide for stable binding to nucleic acids.
Conserved basic regions flanking PWI motifs cooperate in nucleic acid binding
The results described above raise the question of whether the presence of a basic region adjacent to the PWI motif is a general requirement for the nucleic acid-binding function of these domains. An alignment of different PWI motifs and adjacent regions reveals that all PWI domains have one of two adjacent, conserved, N-terminal basic-rich sequences (Fig. 6). The only exceptions are the mammalian PRP3 homologs, in which the PWI motif begins at residue 3. Examination of the C-terminal sequences of PRP3 proteins, however, revealed a basic-rich region that could, in principle, function in an analogous manner to the N-terminal basic region adjacent to other PWI motifs. To test this, we compared the RNA- and DNA-binding activities of GST-fusion proteins consisting of the hPRP3 PWI domain, with (residues 1–93) or without (residues 1–77) the flanking basic region (Fig. 5F). Whereas GST-PRP3(1–77) bound weakly to RNA and DNA (Fig. 5F, lanes 3–20; data not shown), similar to the results shown in Figure 1, GST-PRP3(1–93) bound relatively efficiently, quantitatively shifting RNA or DNA in the 3–6 μM range (Fig. 5F, lanes 9–14). Thus, although the hPRP3 PWI motif is sufficient for weak binding to nucleic acids, similar to the SRm160 PWI motif, stable binding also requires the cooperative action of an adjacent, basic-rich region.
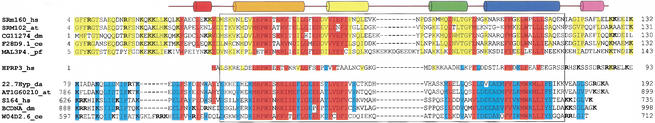
Figure 6.
Multiple alignment of PWI motifs and conserved adjacent basic domains. A representative set of 11 PWI motif proteins are shown aligned. The set represents proteins with either an N-terminal PWI motif (top five sequences) or a C-terminal PWI motif (bottom five sequences). Highly conserved residues among all PWI motifs are colored in red. Residues that are specifically conserved in proteins containing an N-terminal motif are indicated in yellow, and residues specifically conserved in proteins with a C-terminal PWI motif are indicated in blue. Only residues that are similar in at least four out of five of the grouped sequences are colored. Basic amino acids located in the flanking regions are in bold type. Note that hPRP3, which lacks an N-terminal basic region, has a disproportionately higher number of basic residues C-terminal to the PWI motif. The region corresponding to the core PWI motif is boxed, and the secondary structure of the corresponding regions in the SRm160 motif is indicated by cylinders above the sequences.
Taken together, our results highlight the conserved and cooperative role of a flanking basic region for binding of PWI motifs to nucleic acids. Although these basic domains are necessary, they are insufficient for nucleic acid binding, because full binding activity requires the conserved PWI motif residues. Thus, the four-helical bundle comprising the PWI motif and a flanking basic region, whether at the N or C terminus, constitutes a new type of nucleic acid-binding module, with important roles in gene expression, including the stimulation of 3′-end cleavage.
Discussion
Function of the PWI motif
Our study reveals that the PWI motif represents a new type of nucleic acid-binding domain with an important function in gene expression. Interestingly, although the motif is highly conserved among several eukaryotic proteins, it is not known to exist in prokaryotes, consistent with it having roles associated specifically with pre-mRNA processing. Moreover, more proteins contain the motif in higher eukaryotes than in lower eukaryotes. For example, it is present in mammalian but not yeast or C. elegans homologs of the otherwise highly conserved PRP3 splicing factor (Blencowe and Ouzounis 1999). This suggests that the motif may have evolved to provide RNA-binding functions associated with the more complex requirements for pre-mRNA processing in higher eukaryotes, which include the recognition of relatively frequent, yet poorly conserved pre-mRNA processing signals, as well as the more extensive regulation and coupling of different steps involved in pre-mRNA processing. In the present study, we provide evidence that the PWI domain facilitates the stimulation of 3′-end formation by allowing the binding of SRm160 to transcripts.
While our studies provide evidence that the RNA-binding activity of the SRm160-PWI motif participates in 3′-end formation, other functions of the motif in pre-mRNA processing are also possible. Experiments involving titration of different levels of WT-SRm160 and SRm160-ΔN1 proteins in reactions did not result in any differential effects on splicing of two distinct substrates (J. Bowman, and B.J. Blencowe, unpubl.), arguing that the SRm160-PWI motif may not be important for spliceosome formation. However, the presence of the motif in other mammalian spliceosomal proteins, including PRP3 and two uncharacterized proteins, S164/fSAP94 and PRO1777 (Rappsilber et al. 2002; Zhou et al. 2002), suggests that it might participate in splicing. For example, the PWI motifs in these proteins could facilitate one or more of the multiple transitions in RNA–RNA or RNA–protein interactions during the course of the spliceosome cycle. The presence of an N-terminal RRM in a subset of PWI motif proteins (Fig. 1A; Blencowe and Ouzounis 1999), including S164/fSAP94, suggests that these two domains function in a coordinated manner to link separate RNA molecules or possibly RNA/protein complexes. Similar to SRm160, the PWI motifs of these other proteins may also facilitate steps in pre-mRNA processing that are coupled to spliceosome formation, such as 3′-end formation.
Our experiments revealed relatively low-affinity binding to RNA and DNA, in the μM range. This is comparable to the binding affinities of some RRM/RBDs of hnRNP-type and SR family proteins, which have relatively weak specificity for sequences in pre-mRNA (Abdul-Manan and Williams 1996; Tacke et al. 1997; Liu et al. 1998). This property of the SRm160 PWI motif suggests that it might function to provide transient binding to RNA in a semisequence-specific or sequence-independent manner. A candidate step involving this type of role is the formation of a splicing-dependent complex on mRNA. SRm160 forms a splicing-dependent cross-link 20–24 nucleotides upstream of exon-exon junctions, a region that lacks any sequence conservation (Le Hir et al. 2000a). Thus, it is possible that the PWI motif provides a nonsequence-specific binding platform that facilitates the formation of complexes on mRNA, which participate in steps in gene expression that are influenced by splicing, such as mRNA 3′-end formation, nonsense-mediated decay, and export. The relatively weak binding interaction would provide flexibility by allowing rapid dissociation of the SRm160–mRNA complex. Such an event might be important for allowing transport of mRNA, because SRm160 does not efficiently shuttle between the nucleus and the cytoplasm and is thought to have a limited trajectory in the cytoplasm (Lykke-Andersen et al. 2000; Lejeune et al. 2002). Consistent with this view, expression of the MS2-SRm160 fusion proteins (Fig. 2), which bind with relatively high affinity to their cognate sites, caused increased retention of RNA in the nucleus. The relatively weak binding affinity of the PWI motif may therefore be important for release of transcripts into the cytoplasm, as well as facilitating 3′-end formation.
Structure of the PWI motif: A new nucleic acid-binding module
In an effort to better understand the possible spatial orientations of the globular PWI domain and the flanking basic region when bound to DNA or RNA, we examined the structure of the PWI domain with the flexible basic region (residues 18–26) modeled onto the N terminus in a variety of orientations. Nucleic acid was docked along the magenta surface indicated in Figure 4B, using the structurally similar subregion that binds DNA in the repair-glycosylases AlkA and Endo III as a guide. This arrangement indicated that the flexible basic region would be in an excellent position to interact either with the phosphate backbone or lie along the minor groove (dsDNA) or major groove (dsRNA) in the case of double-stranded nucleic acids. Thus, it appears that it is sterically possible for the basic region to interact with a nucleic acid that is simultaneously bound to the “bottom” of the PWI domain helix bundle. In the case of the PWI domain of PRP3, a similar model can be envisioned. Although the basic region of PRP3 is C-terminal to the PWI domain, it is also predicted to interact with dsDNA. The sequence corresponding to the helix that is C-terminal to the PWI motif in SRm160 is not conserved in hPRP3. These residues in PRP3 are not predicted to be helical or pack against the globular portion of the protein. This would allow plenty of conformational freedom for the basic residues in this region to contact nucleic acid. In contrast, even though the analogous C-terminal region in SRm160 contains basic amino acids, monitoring of the amide chemical shift changes upon addition of DNA indicated that these unfolded residues are not involved in nucleic acid binding.
Our data show that the PWI domain interacts equally well with ssDNA, dsDNA, ssRNA, and dsRNA. This suggests that it is primarily the sugar-phosphate backbone that is recognized by the protein and that the bipartite binding module is flexible enough to accommodate multiple types of nucleic acid structures. A mechanism by which both the globular and the flexible regions of the module have weak affinity for nucleic acids is consistent with this notion. To our knowledge, such a mode of interaction has not been reported previously for RNA-binding proteins. Indeed, our structural results for the PWI domain do not conform directly to any known nucleic acid-binding domains, but rather combine features from different systems.
In recent years, the elucidation of the three-dimensional structures for a number of RNA-binding domains has revealed several different modes of RNA binding, some of which employ design principles that are also used by DNA-binding domains (for reviews, see De Guzman et al. 1998; Cusack 1999; Draper 1999; Antson 2000; Perez-Canadillas and Varani 2001). A reoccurring theme in many such motifs, for example the OB fold found in a variety of proteins (Murzin 1993), is the presence of antiparallel β-sheets that have conserved aromatic residues on their surface. This type of structure can serve as a nonsequence-specific binding platform, where general stacking interactions between bases and aromatic side chains are important for stabilizing the RNA–protein complex (Oubridge et al. 1994; Allain et al. 1996). Although the SRm160 PWI domain does not have any β-sheets, it does have several surface exposed aromatic residues positioned at the nucleic acid-binding surface that could potentially interact with base pairs. These residues, however, are not highly conserved among PWI domains, and the individual mutation of the surface exposed phenylalanines 62 and 92 to alanine had only a minor effect on the binding properties of the SRm160 PWI domain (B. Szymczyna, S. McCracken, C. Arrowsmith, and B. Blencowe, unpubl.).
The structural and nucleic acid-binding properties of the PWI domain are also inconsistent with the “groove-binding” proteins and peptides, another important class of RNA-binding motifs (Puglisi et al. 1992, 1995; Battiste et al. 1995). The major and minor grooves of RNA often contain bulges and mismatches that alter canonical groove dimensions, and are thought to be important for configuring an RNA helix to provide the contact surface for an α-helix or other polypeptide conformations. The flexible basic region of the PWI module has some similarities to the small arginine-rich peptides that are capable of adopting a variety of conformations within such sequence-dependent grooves of RNA, but such a mode of interaction would not be consistent with the single-stranded nucleic acid-binding properties of SRm160 and hPRP3. Similarly, chemical shift mapping of the SRm160 PWI domain did not reveal evidence that any of the helices lie within the major groove of DNA, but instead suggest that the turns between helices 2 and 3 and between 4 and 5 form the nucleic acid-binding surface (Fig. 4B). In other RNA-binding domains, such as the dsRBD and KH motifs, conserved loops extending from α-helices are thought to play an important and direct role in RNA binding (Ryter and Schultz 1998; Lewis et al. 2000). The interhelical loops on the binding surface of the PWI domain, however, are neither highly conserved nor long enough to extend into the major or minor groove of RNA. Rather, it appears that the entire lower surface of the globular portion of the PWI domain may be required for the interaction.
Although the structures of several RNA-binding domains composed entirely of α-helices have been solved (Predki et al. 1995; Dubnau and Struhl 1996; Berglund et al. 1997), none of these have a three-dimensional structure similar to the PWI motif. Moreover, in contrast to domains consisting of β-sheets which serve important roles in ssRNA recognition, structures consisting entirely of helices are not known to bind ssRNA. Although we cannot exclude the possibility that small regions of secondary structure in our ssRNA probes exist, possibly induced by the PWI motif, our binding studies did not reveal a significant difference in binding affinity between ssRNA and dsRNA. The equal affinity for either type of nucleic acid and the apparent nonsequence-specific binding property of the PWI domain suggest primary contacts with the sugar-phosphate backbone, rather than extensive base-specific contacts. Regardless of the precise mechanism of binding, our results demonstrate that the PWI domain and its flanking basic domain represent a new type of nucleic acid-binding module that is likely to have numerous important roles in pre-mRNA processing, including the stimulation of 3′-end formation by SRm160.
Materials and methods
Preparation of recombinant PWI-containing proteins
GST-fusion protein constructs were prepared by PCR amplification of DNA fragments from full-length SRm160, hPRP3, and p73 cDNAs. Fragments corresponding to residues 1–151, 18–134, 18–26, or 27–134 of SRm160; residues 1–77 and 1–93 of hPRP3; and residues 491–554 of p73 were subcloned into the Nde1 and BamH1 restriction sites of the pET15b expression vector, or a modified pGEX-2TK expression vector containing an Nde1 site engineered immediately upstream of the BamHI site. The GST-SRm160(18–26)-SAM fusion construct was made by joining the nine-amino-acid SAM sequence to the 5′ end of the p73 construct (residues 491–554) by PCR, and then subcloning this fragment into the modified pGEX-2TK expression vector. The QuikChange XL Site Directed Mutagenesis Kit from Stratagene was used to generate alanine mutants of the 18–134 construct: K20A, K22A, K23A; K54A, R55A; F62A; F92A; and N94A, K96, N97A (data not shown). Proteins were expressed in E. coli BL21-Gold (DE3) cells (Stratagene). The GST-fusion proteins were purified using Glutathione Sepharose 4B (Amersham Pharmacia Biotech) using standard protocols, but also washed with a series of high-salt solutions (1M, 2M, 1M NaCl) and DnaK elution buffer (50 mM Tris at pH 8, 2 mM ATP, 10 mM MgSO4). Samples were stored at 4°C in ACB buffer (10 mM Tris at pH 7.5), 10 mM HEPES at pH 7.5, 100 mM NaCl, 10 mM β-mercaptoethanol) with 10% glycerol, 1 mM PMSF, 10 mM DTT, and 1× Complete EDTA-free Protease Inhibitor Cocktail Tablet solution (Boehringer Mannheim). Purification of the histidine-tagged proteins for NMR studies was described (Szymczyna et al. 2002). Baculovirus-expressed wild-type and ΔN1 versions of SRm160 were prepared as described by McCracken et al. (2002).
Pre-mRNA reporters, expression plasmids, and transfection assays
Transfection of human 293 cells and RNase protection assays were performed as described by McCracken et al. (2002). The pre-mRNA plasmid reporter dsxΔE-Δ5′SS+3xMS2 was constructed by insertion of an EcoRI fragment from pSP73-MS2–3 (blunt-ended using dNTPs and Klenow fragment) into the blunt-ended ClaI site of the pre-mRNA reporter dsxΔE-Δ5′ss, which was constructed by deletion of a KpnI-Bst-XI fragment containing the 5′-splice site of pAd2MLP-dsxΔΕ (McCracken et al. 2002).
Expression plasmids pcDNA3-fMS2, pcDNA3-fMS2-SRm160- WT and pcDNA3-fMS2-SRm160-ΔN1 were constructed by insertion of the BamH1 digestion product of a PCR amplification reaction of pC1-neo-hMS2 using primers Bam-MS2-5′ fwd, CGGGATCCAATGGGCGCCTCCAACTTCAC; and Bam-MS2-3′ rev, CGGGATCCCCGCCGCCGTAGATGCCG. The MS2 coding sequence in these constructs was “humanized” to improve expression. This was achieved using a set of eight partially overlapping oligonucleotides and a series of PCR amplification steps in order to incorporate human codon-usage biases. The full-length ORF was cloned into the Xho and EcoRI sites of pcI-Neo to create pCI-neo-hMS2. The resulting “humanized” MS2 protein is expressed at significantly higher levels than that from the original MS2 ORF from the MS2 bacteriophage (data not shown).
PWI domain mapping
Purified histidine-tagged SRm160(1–151) was partially proteolyzed with 5 μg/mL trypsin, chymotrypsin, and proteinase A over a 24-h period, and samples of the reactions were quenched with the addition of gel loading buffer at regular intervals. Protease-resistant fragments were separated by electrophoresis and analyzed by MALDI-TOF mass spectrometry using a Bruker Daltonics Biflex III. Based on these results, six different SRm160 constructs were designed (18–128, 27–128, 40–128, 18–134, 27–134, 40–134) and tested for protein expression and solubility and assayed for suitability for NMR structure determination using 15N-HSQC experiments.
NMR spectroscopy and structure analysis
NMR experiments were recorded at 25°C on a Varian INOVA 600MHz spectrometer,. For data processing and analysis, the NMRPipe/NMRDraw (Delaglio et al. 1995), SPSCAN (R.W. Glaser and K. Wüthrich, http://gaudi.molebio.uni-jena.de/∼rwg/spscan) and XEASY (Bartels et al. 1995) programs were used. Sequence-specific resonance assignments were obtained using reduced dimensionality experiments, as described (Szymczyna et al. 2002).
Calculations of the protein structure were performed with the program DYANA (Güntert et al. 1997) using a torsion angle dynamics protocol and the structural statistics listed in Table 1. Interproton distance restraints were obtained from a simultaneous 15N and 13C edited NOESY spectrum with a mixing time of 150 msec (Pascal et al. 1994). Peak analysis of the spectra was accomplished with interactive peak picking in the program XEASY, and the cross-peaks were assigned using a combination of automatic and manual methods. An initial fold of the protein was calculated based on unambiguously assigned NOEs, and the NOAH module in DYANA was used to aid in the assignment of the remaining NOE cross-peaks (Guntert et al. 1997). Backbone dihedral angle restraints were obtained from 1Hα and 13Cα chemical shifts using TALOS (Cornilescu et al. 1999). Hydrogen bond restraints, two for each hydrogen bond (NH-O = 2.0Å; N-O = 3Å), were applied to residues that have α-helical properties (dihedral angles and NOESY patterns). MOLMOL (Koradi et al. 1996) was used to analyze the 20 energy minimized conformers, calculate the electrostatic surface, and prepare figures of the structures. The coordinates for the 20 lowest energy structures are deposited in the Protein Data Bank under accession no. 1MP1.
Table 1.
Statistics for the ensemble of structures calculated for the PWI motif of SRm160a
| Distance restraints: | |
| Intraresidue | 679 |
| Sequential (|i - j| = 1) | 773 |
| Medium range (2 ≤ |i - j| ≤ 4) | 744 |
| Long range (4 < |i - j|) | 845 |
| Hydrogen bonds | 22 × 2 |
| Dihedral angle restraints | |
| All | 106 |
| φ, ϑ | 53, 53 |
| Pairwise R.M.S.D. | |
| All residuesb | |
| Backbone atoms | 0.27 + 0.07 |
| All heavy atoms | 0.85 + 0.07 |
| Ordered regionsc | |
| Backbone atoms | 0.21 + 0.06 |
| All heavy atoms | 0.80 + 0.09 |
aEnsemble of the 20 lowest energy structures out of 200 calculated.
bRMSD values for residues 27–126.
cRMSD values for residues in α-helices.
15N-HSQC spectra of SRm160(18–134) were recorded both in the presence and absence of the desalted dsDNA used in the gel shift assays and synthesized by Midland Certified Reagent Co. Both spectra were recorded in identical solution conditions: 25 mM phosphate buffer at pH 7.0, 300 mM NaCl, 2 mM MgCl2, 1 mM DTT, 1 mM PMSF, 1× Complete EDTA-free Protease Inhibitor Cocktail Tablet 10% D2O/90% H2O. Changes were assessed by considering changes in both chemical shift and peak intensity.
RNA-binding assays
Protein samples were diluted in 10 mM HEPES at pH 7.5, 100 mM NaCl, 5 mM EDTA, 1 mM DTT, 20% glycerol. Binding was performed in 10-μL reactions containing 200 fmol of 32P-UTP-labeled T7-RNA probes or 15-μL reactions containing 1 pmol of dsDNA 5′-end labeled with Cy5 (data not shown). RNA probes were prepared by T7 run-off transcription from pBSKS− digested with XhoI, resulting in a 34-base transcript, GGGAA CAAAAGCTGGGTACCGGGCCCCCCCTCGA. Desalted oligodeoxynucleotides for the dsDNA gel shifts were purchased from ACGT, CCATTACCATATATGGACTTCCGGTGCTA CC and GGTAGCACCGGAAGTCCATATATGGTAATGG. The final binding conditions were 10 mM Tris at pH 7.5, 10 mM HEPES at pH 7.5, 100 mM NaCl, 0.1% Triton X-100, 2 mM MgCl2, 1.5 mM DTT, 7% glycerol. After samples were incubated at 4°C for 30 min, they were run at 100 or 150 V on a preelectrophoresed 5% polyacrylamide gel containing 0.25× TBE and 10% glycerol for 4–6 h at 4°C.
Acknowledgments
We thank Emanuel Rosonina for help with two-hybrid assays, Montegomery Gill and Lea Harrington for help with baculovirus expression, and Peter Stockley and Jenny Baker for providing anti-MS2 antibody. We thank Alan Cochrane, Rick Collins, Aled Edwards, Henry Siu, Christos Ouzounis, and Emanuel Rosonina for helpful discussions and comments on the manuscript. This research was supported by operating grants from the Canadian Institutes for Health Research (CIHR), the National Cancer Institute of Canada (C.H.A. and B.J.B.) and the NIH (B.R.G.). B.J.B. also thanks the Aronovitz and Coblenz Families Foundation for their support. B.R.S. is the recipient of a CIHR studentship, C.H.A. and B.J.B. are recipients of the Premier's Research Excellence Award, B.J.B. is a CIHR Scholar, and C.H.A. is a CIHR Scientist.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Corresponding authors.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1060403.
References
- Abdul-Manan N, Williams KR. hnRNP A1 binds promiscuously to oligoribonucleotides: Utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain FH, Gubser CC, Howe PW, Nagai K, Neuhaus D, Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- Antson AA. Single-stranded-RNA binding proteins. Curr Opin Struct Biol. 2000;10:87–94. doi: 10.1016/s0959-440x(99)00054-8. [DOI] [PubMed] [Google Scholar]
- Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- Battiste JL, Tan R, Frankel AD, Williamson JR. Assignment and modeling of the Rev Response Element RNA bound to a Rev peptide using 13C-heteronuclear NMR. J Biomol NMR. 1995;6:375–389. doi: 10.1007/BF00197637. [DOI] [PubMed] [Google Scholar]
- Berglund H, Rak A, Serganov A, Garber M, Hard T. Solution structure of the ribosomal RNA binding protein S15 from Thermus. Nat Struct Biol. 1997;4:20–23. doi: 10.1038/nsb0197-20. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Ouzounis CA. The PWI motif: A new protein domain in splicing factors. Trends Biochem Sci. 1999;24:179–180. doi: 10.1016/s0968-0004(99)01387-0. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Issner R, Nickerson JA, Sharp PA. A coactivator of pre-mRNA splicing. Genes & Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Carey J, Cameron V, de Haseth PL, Uhlenbeck OC. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983;22:2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- Chen L, Frankel AD. An RNA-binding peptide from bovine immunodeficiency virus Tat protein recognizes an unusual RNA structure. Biochemistry. 1994;33:2708–2715. doi: 10.1021/bi00175a046. [DOI] [PubMed] [Google Scholar]
- Chi SW, Ayed A, Arrowsmith CH. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 1999;18:4438–4445. doi: 10.1093/emboj/18.16.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Curtis D, Lehmann R, Zamore PD. Translational regulation in development. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- Cusack S. RNA–protein complexes. Curr Opin Struct Biol. 1999;9:66–73. doi: 10.1016/s0959-440x(99)80009-8. [DOI] [PubMed] [Google Scholar]
- De Guzman RN, Turner RB, Summers MF. Protein–RNA recognition. Biopolymers. 1998;48:181–195. doi: 10.1002/(SICI)1097-0282(1998)48:2<181::AID-BIP7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Draper DE. Themes in RNA–protein recognition. J Mol Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- Eldridge AG, Li Y, Sharp PA, Blencowe BJ. The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc Natl Acad Sci. 1999;96:6125–6130. doi: 10.1073/pnas.96.11.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd S, Tainer JA. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Horowitz DS, Kobayashi R, Krainer AR. A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA. 1997;3:1374–1387. [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Lauber J, Plessel G, Prehn S, Will CL, Fabrizio P, Groning K, Lane WS, Luhrmann R. The human U4/U6 snRNP contains 60 and 90kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA. 1997;3:926–941. [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000a;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon–exon junctions. Genes & Dev. 2000b;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- Liu H-X, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes & Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D, McGarvey T, McCracken S, Johnstone IL, Blencowe BJ, Caceres JF. Multiple interactions between SRm160 and SR family proteins in enhancer-dependent splicing and development of C. elegans. Curr Biol. 2001;11:1923–1933. doi: 10.1016/s0960-9822(01)00589-9. [DOI] [PubMed] [Google Scholar]
- Lou H, Neugebauer KM, Gagel RF, Berget SM. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Murthy KG, Schek N, O'Connor JP, Manley JL, Alwine JC. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes & Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- ————— Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- McCracken S, Lambermon M, Blencowe BJ. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol Cell Biol. 2002;22:148–160. doi: 10.1128/MCB.22.1.148-160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for nonhomologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash HM, Bruner SD, Scharer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- Nesic D, Maquat LE. Upstream introns influence the efficiency of final intron removal and RNA 3′-end formation. Genes & Dev. 1994;8:363–375. doi: 10.1101/gad.8.3.363. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Pascal SM, Muhandiram DR, Yamazaki T, Forman-Kay JD, Kay LE. Simultaneous acquisition of 15N-edited and 13C-edited NOE spectra of proteins dissolved in H20. J Magn Reson. 1994;103:197–201. [Google Scholar]
- Perez-Canadillas JM, Varani G. Recent advances in RNA–protein recognition. Curr Opin Struct Biol. 2001;11:53–58. doi: 10.1016/s0959-440x(00)00164-0. [DOI] [PubMed] [Google Scholar]
- Predki PF, Nayak LM, Gottlieb MB, Regan L. Dissecting RNA–protein interactions: RNA–RNA recognition by Rop. Cell. 1995;80:41–50. doi: 10.1016/0092-8674(95)90449-2. [DOI] [PubMed] [Google Scholar]
- Puglisi JD, Ton R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA–arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- Puglisi JD, Chen L, Blanchard S, Frankel AD. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide–RNA complex. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- Query CC, Bentley RC, Keene JD. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter JM, Schultz SC. Molecular basis of double-stranded RNA–protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of. Proc Natl Acad Sci. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryanarayana T, Subramanian AR. Functional domains of Escherichia coli ribosomal protein S1. Formation and characterization of a fragment with ribosome-binding properties. J Mol Biol. 1979;127:41–54. doi: 10.1016/0022-2836(79)90458-3. [DOI] [PubMed] [Google Scholar]
- Szymczyna BR, Pineda-Lucena A, Mills JL, Szyperski T, Arrowsmith CH. 1H, 13C, and 15N resonance assignments and secondary structure of the PWI domain from SRm160 using reduced dimensionality NMR. J Biomol NMR. 2002;22:299–300. doi: 10.1023/a:1014904502424. [DOI] [PubMed] [Google Scholar]
- Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain. Proc Natl Acad Sci. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes & Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- Wang A, Forman-Kay J, Luo Y, Luo M, Chow YH, Plumb J, Friesen JD, Tsui LC, Heng HH, Woolford JL, Jr, et al. Identification and characterization of human genes encoding Hprp3p and Hprp4p, interacting components of the spliceosome. Hum Mol Genet. 1997;6:2117–2126. doi: 10.1093/hmg/6.12.2117. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Steitz JA. Association with terminal exons in pre-mRNAs: A new role for the U1 snRNP? Genes & Dev. 1993;7:647–659. doi: 10.1101/gad.7.4.647. [DOI] [PubMed] [Google Scholar]
- Yaniv K, Yisraeli JK. Defining cis-acting elements and trans-acting factors in RNA localization. Int Rev Cytol. 2001;203:521–539. doi: 10.1016/s0074-7696(01)03015-7. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]