Abstract
p190RhoGAP and Rho are key regulators of oligodendrocyte differentiation. The gene encoding p190RhoGAP is located at 19q13.3 of the human chromosome, a locus that is deleted in 50%–80% of oligodendrogliomas. Here we provide evidence that p190RhoGAP may suppress gliomagenesis by inducing a differentiated glial phenotype. Using a cell culture model of autocrine loop PDGF stimulation, we show that reduced Rho activity via p190RhoGAP overexpression or Rho kinase inhibition induced cellular process extension, a block in proliferation, and reduced expression of the neural precursor marker nestin. In vivo infection of mice with retrovirus expressing PDGF and the p190 GAP domain caused a decreased incidence of oligodendrogliomas compared with that observed with PDGF alone. Independent experiments revealed that the retroviral vector insertion site in 3 of 50 PDGF-induced gliomas was within the p190RhoGAP gene. This evidence strongly suggests that p190 regulates critical components of PDGF oncogenesis and can act as a tumor suppressor in PDGF-induced gliomas by down-regulating Rho activity.
Keywords: p190RhoGAP, glioma, RhoA
The Rho family of GTPases plays a key role in actin cytoskeletal regulation. In addition, Rho proteins have been implicated in cellular transformation and cell cycle control. Activated Rho has been shown to be necessary for Ras-induced cellular transformation (Olson et al. 1998) and for oncogenesis downstream of a mutant Epidermal Growth Factor Receptor (EGFR; Boerner et al. 2001). In addition, increased endogenous Rho activity has been observed in Ras-transformed fibroblasts compared with wild-type controls (Sahai et al. 2001). Transfection of dominant-negative forms of Rho, Rac, and Cdc42 results in inhibition of serum-independent proliferation in Ras-transformed fibroblasts. Moreover, activated Ras and Rho cooperate in growth factor-regulated cell cycle progression in nontransformed NIH 3T3 cells (Danen et al. 2000). Rho has been shown to induce the down-regulation of the cell cycle inhibitor p21, thereby allowing cell cycle progression (Sahai et al. 2001).
Several Rho family members have been implicated as oncogenes in human cancers. For example, RhoC is overexpressed in 90% of inflammatory breast cancer tumor samples, and human mammary epithelial cells stably transfected with RhoC are tumorigenic (van Golen et al. 2000). Moreover, RhoA is overexpressed in the human breast cancer cell line MDA-MB-231 (Denoyelle et al. 2001).
Rho signaling is down-regulated by p190RhoGAP, a 190-kD GTPase-activating protein that enhances the intrinsic hydrolysis of RhoGTP to RhoGDP (Settleman et al. 1992). Because Rho plays a key role in cytoskeletal rearrangements and cell cycle control, it is likely that regulators of Rho such as p190RhoGAP are also essential players in these processes. p190RhoGAP has been shown to be involved in actin cytoskeletal rearrangements induced by Src (Chang et al. 1995) as well as integrin stimulation (Sharma 1998; Arthur and Burridge 2001). Moreover, p190 interacts with p120 RasGAP in a tyrosine phosphorylation-dependent manner. The p190/p120 complex has been implicated in cytoskeletal changes such as embryonic morphogenesis (Dupont and Blancq 1999) and cell migration (Kulkarni et al. 2000). In addition, p190 has been implicated in metastasis (Zrihan-Licht et al. 2000). Both the GTPase domain and the GAP domain of p190 act as tumor suppressors in Ras-dependent transformation (Wang et al. 1997).
p190 maps to 19q13.3 on the human chromosome (Tikoo et al. 2000), a region that is deleted in 50%–80% of oligodendrogliomas (Smith and Jenkins 2000). This finding suggests that loss of p190 may play a unique role in gliomagenesis. Oligodendrogliomas are comprised of cells resembling oligodendrocyte progenitors. These tumors vary in malignancy, with the most aggressive having a mean survival of merely 12 mo, even with combined treatment modalities of surgery, radiation therapy, and chemotherapy (Holland 2000). Advances in understanding the molecular basis of these tumors have identified the combined deletion of chromosomal regions 1p36 and 19q13.3 (Bello et al. 1994; Reifenberger et al. 1994). These deletions characterize a subset of oligodendrogliomas that respond better to present chemotherapy (Cairncross et al. 1998). The putative tumor suppressors in these chromosomal locations have not yet been identified.
One of the most powerful tools used to decipher tumor biology is genetic modeling of cancer in mice. Mouse models have been developed to characterize molecular lesions that participate in glioma formation (Holland 2001). One such model uses a transgenic mouse that expresses the avian leukosis virus (ALV) cell surface receptor, tv-a, under the control of either the nestin (Ntv-a) or the GFAP (Gtv-a) promoter. A gene of choice is cloned into the RCAS vector, which expresses the protein in the context of ALV, thereby allowing for cell-specific infection of a gene of interest (Holland et al. 1998). Because GFAP is expressed only in differentiated astrocytes and nestin is expressed in neural precursors that give rise to neuronal and glial lineages, the oncogenic effects of specific signaling pathways in cells at particular stages of differentiation can be studied (Holland et al. 2000).
PDGF signaling regulates the differentiation of glia and blocks progression of oligodendroglial precursors to oligodendrocytes (Bogler et al. 1990). Furthermore, overexpression of PDGF is a well-defined lesion of oligodendrogliomas (Guha et al. 1995). To model PDGF autocrine stimulation in astrocytes and glial progenitors in culture and in vivo, the RCAS tv-a system has been used to constitutively express PDGF. In glial culture, PDGF stimulation promotes a rapidly proliferating, undifferentiated cell type. These effects are reversible by pharmacologic blockade of the PDGF receptor kinase. In vivo, PDGF autocrine stimulation induces the formation of low-grade oligodendrogliomas in 40% of injected Gtv-a animals and in 60% of injected Ntv-a mice (Dai et al. 2001). The above data imply that overexpression of PDGF encourages tumor growth by inducing a dedifferentiation of GFAP+ astrocytes or by blocking the differentiation of nestin+ glial progenitors. In addition, the data support a more far-reaching hypothesis that gliomagenesis is a dedifferentiation process, thereby implicating molecules that induce glial differentiation as potential tumor suppressors.
In another mouse glioma model (Uhrbom et al. 1998), brain tumors are induced at a high frequency by a recombinant Moloney mouse leukemia virus (MMLV) coding for platelet-derived growth factor (PDGF-B). The majority of these tumors resemble human glioblastoma multiforme (GBM), the most malignant form of glioma. These tumors are likely to evolve through a combination of autocrine PDGF receptor stimulation and proviral insertional mutagenesis. Thus, this model can be used to identify genes that cooperate with PDGF in the genesis of malignant glioma; such genes will be tagged by the integrated proviral DNA.
We have recently shown that p190 plays a critical role in oligodendrocyte differentiation by down-regulating Rho activity (Wolf et al. 2001). Our working hypothesis is that oligodendrogliomas are induced by a dedifferentiation event, and that p190 is part of the molecular machinery of oligodendrocyte differentiation that is lost in a large percentage of these tumors. To test this hypothesis, PDGF overexpression was selected as a model for study because it has been shown to involve a dedifferentiation process. We now present evidence from both in vitro and in vivo experiments that strongly suggests that p190 is involved in differentiation and cell cycle control that inhibits glioma formation.
Results
Constitutive PDGF expression induces down-regulation of p190
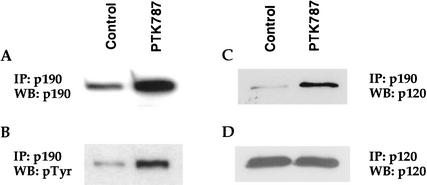
We used the Gtv-a/PDGF cell culture system to decipher the molecular events that contribute to glioma formation. Immunoprecipitation of p190 from Gtv-a/PDGF cells, followed by immunoblotting with anti-p190 antibody, revealed the presence of p190 in these cells. However, inhibition of PDGF signaling by PTK787 caused a threefold increase in p190 expression levels (Fig. 1A). This implies that chronic stimulation with PDGF causes down-regulation of p190. Because previous studies showed that tyrosine phosphorylation of p190 increases its GAP activity (Arthur et al. 2000) and is a crucial step in the differentiation of oligodendrocytes (Wolf et al. 2001), the phosphorylation status of p190 was tested. Immunoprecipitation of p190 followed by immunoblotting with phosphotyrosine showed that PTK787 treatment induced a threefold increase in phosphorylated p190. The increase in tyrosine phosphorylation of p190 corresponded to the increase in protein expression (Fig. 1B), suggesting that the additional p190 that was expressed was also phosphorylated.
Figure 1.
Expression patterns of p190 and p120 in PDGF/Gtv-a cells. PDGF/tv-a cells were treated with or without the PDGF receptor inhibitor PTK787, and cell lysates were analyzed by immunoprecipitation (IP) and Western blotting (WB) with the indicated antibodies. (A) Inhibition of PDGF signaling led to an increase in p190 levels. (B) Phosphorylation of p190 increased in conjunction with the increase in protein expression. (C) Immunoprecipitation of the cell lysates with anti-p190 antibody and subsequent immunoblotting with an anti-p120 antibody demonstrated an increase in the complex formation between these two proteins. (D) Immunoprecipitation of p120 followed by immunoblotting for p120 demonstrated no increase in p120 protein levels.
Interaction between p190 and its binding partner p120 is dependent on the phosphorylation status of p190. To examine p190/p120 complex formation in glial cells, lysates from Gtv-a/PDGF cells that were treated with or without PTK787 were subjected to immunoprecipitation with an anti-p190 antibody followed by immunoblotting with an anti-p120 antibody (Fig. 1C). The amount of p190/p120 complex was greatly increased in the PTK787-treated cells, without a demonstrable increase in the amount of p120 (Fig. 1D). Because the p190/p120 complex is dependent on tyrosine phosphorylation of p190, these data provide further evidence that an increase in phosphorylated p190 accompanies PDGFR inhibition.
Expression of p190 results in process extension
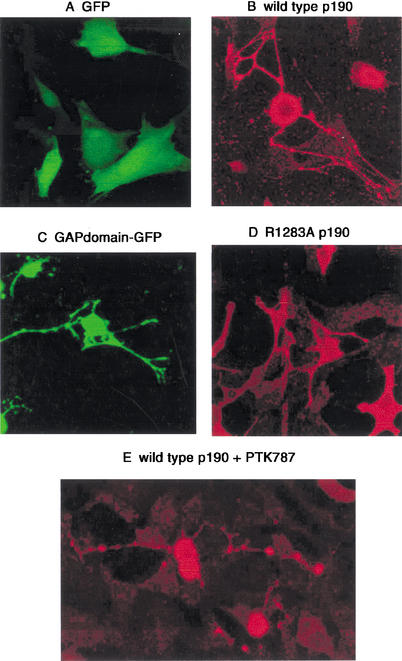
The data presented so far are consistent with the hypothesis that a portion of the dedifferentiation effect of PDGF may be mediated by a decrease in p190 levels. We therefore tested whether exogenous expression of p190 could reverse the dedifferentiated phenotype by examining cell morphology. Expression of a control protein (GFP) had no effect on cell morphology (Fig. 2A). In contrast, Gtv-a/PDGF cells transfected with p190 developed striking cellular extensions (Fig. 2B). The same phenotype was seen when the GAP domain of the protein fused to GFP was expressed (Fig. 2C). GAP activity was required for the development of this phenotype, because the processes were not extended when a GAP inactive mutant (R1283A) of full-length p190 was expressed (Fig. 2D). Cells expressing p190 RhoGAP that were treated with the PDGFR inhibitor PTK787 appeared indistinguishable from untreated cells, indicating that the effects of p190 are downstream of PDGF signaling (Fig. 2E).
Figure 2.
Exogenous expression of p190 RhoGAP and related constructs affects cellular morphology. Gtv-a cells were transfected with the indicated constructs. Protein expression and cell morphology were monitored as described in Materials and Methods. (A) GFP expression did not affect cell morphology. (B) Expression of full-length wild-type p190 caused contraction of the cell body and development of process-like formations. (C) The GAP domain of p190 produced a phenotype resembling that of the full-length protein. (D) GAP inactive R1283A p190 did not alter cell morphology. (E) Exposure of the transfected cells to PTK787, a PDGF receptor inhibitor, did not alter the striking morphological changes caused by expression of wild-type 190, indicating that the effects of p190 are downstream of PDGF receptor signaling.
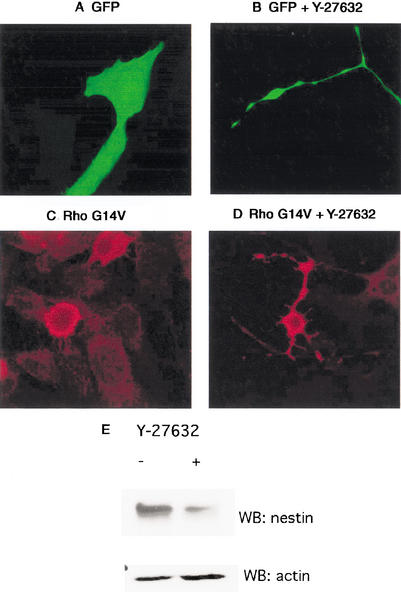
Because p190 likely exerts its phenotypic effects by down-modulating RhoGTP signaling, one would predict that pharmacologic inhibition of Rho signaling would have effects similar to p190 expression. Indeed, cells treated with the Rho kinase inhibitor Y-27632 exhibited a similar morphology to those transfected with p190 RhoGAP (Fig. 3B) as compared with control cells (Fig. 3A). In contrast, expression of constitutively active Rho (G14V) caused a retraction of the cell body without the appearance of cellular extensions (Fig. 3C). When cells expressing constitutively active Rho were treated with Y-27632, however, cellular extensions appeared (Fig. 3D). These data indicate that Rho signaling through Rho kinase causes retraction of the cell body and/or processes. When Rho signaling is inhibited, either by inhibition of Rho kinase or by down-regulation of RhoGTP, extension of cellular processes predominates.
Figure 3.
Cells treated with Rho kinase inhibitor develop a differentiated phenotype. Gtv-a/PDGF ells were transfected with GFP and were either untreated (A) or exposed to 50 μM Y-27632 (B). Treated cells were demonstrably more spindly than untreated cells. (C) Constitutively active Rho G14V caused the cell body to contract without the appearance of cell processes. (D) The phenotype induced by expression of Rho G14V was reversed by exposing the cells to Y-27632, a Rho kinase inhibitor. This indicates that the effects of constitutively active Rho occur at least in part through the action of Rho kinase. (E) Lysate from control and Y-27632-treated cells was electrophoresed on an 8% gel and immunoblotted with anti-nestin antibody. Treated cells demonstrated a two- to threefold decrease in the nestin neuronal progenitor marker. Western blotting of the lysate with an anti-actin antibody served as a loading control.
Rho kinase inhibition resulted in morphological changes that resemble process extension during differentiation. Lysates from cells treated with or without Y-27632 were therefore immunoblotted with antibodies to the progenitor marker nestin. A marked down-regulation of nestin levels was seen in cells treated with Y-27632 (Fig. 3E), whereas no changes were detected for the actin loading control. These data indicate that inhibition of Rho signaling promotes development of a more differentiated molecular phenotype. Y-27632-treated cells did not express markers of oligodendrocyte differentiation (MAG, MBP), but remained positive for GFAP. Because no new markers appeared, the precise nature of the differentiated cell cannot be defined at present.
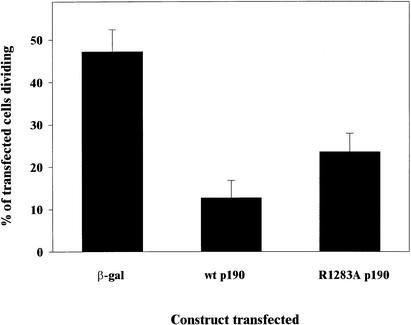
Given that differentiation and withdrawal from the cell cycle are inextricably linked, we examined whether p190 regulates exit from the cell cycle. Gtv-a/PDGF cells were transfected with either β-galactosidase as a negative control or HA-tagged wild-type (wt) or R1283A p190. Cells were incubated with BRDU, to allow incorporation into the nucleus during cell division. Cells were then fixed, permeabilized, and stained with an antibody against the transfected protein as well as an antibody against BRDU. The percentage of transfected cells that incorporated BRDU was determined. As depicted in Figure 4, transfection of p190 resulted in a fourfold decrease in the number of dividing cells. When the R1283A GAP inactive mutant was transfected, a twofold increase in the number of dividing cells was observed compared with wild-type p190. These data indicate that p190 expression can either slow down or halt entry into the cell cycle, in a manner mediated by its GAP activity as well as additional functional domains within the p190 molecule.
Figure 4.
Cells expressing p190 do not divide as frequently as control cells. G-tva/PDGF cells transfected with β-galactosidase or wild-type or R1283A p190 were labeled with BRDU for 6–7 h and then stained for β-galactosidase or p190 and BRDU. The percentage of dividing cells was calculated and plotted. Data were collected from four experiments.
Role of p190 in glioma formation
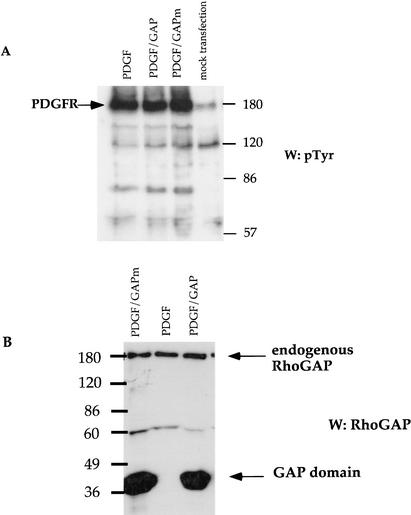
The data described above strongly suggested that p190 is involved in the differentiation and cessation of proliferation of glia. The next two sets of experiments were designed to determine whether these properties allowed p190 RhoGAP to inhibit gliomagenesis. An RCAS vector was constructed that directed expression of both PDGF and an HA-tagged GAP domain of p190. Because the RCAS vector can only accommodate 3 kb of DNA, subcloning the coding sequence for the full-length p190 protein was not feasible. As a negative control, an RCAS vector expressing PDGF and a GAP inactive mutant (R1283A) was constructed. We first verified that both proteins were expressed. To assess the levels of PDGF expression, a bioassay to monitor PDGF receptor activation was performed. RCAS vectors were transfected into 293T cells, and conditioned media was then added to serum-starved 3T3 cells. The extent of phosphorylation of the PDGF receptor was approximately the same for each RCAS vector (Fig. 5A), indicating that the vectors expressed equal levels of PDGF. The ALV packaging cell line DF-1 was then transfected with the RCAS constructs. Immunoblotting of the DF-1 lysates with anti-p190RhoGAP antibody demonstrated a protein ∼40 kD in size, corresponding to the predicted size of the HA-tagged GAP domain (Fig. 5B).
Figure 5.
Expression of PDGF and the GAP domain from RCAS vectors. (A) RCAS vectors were tested for expression of PDGF. 293T cells were transiently transfected with the RCAS vectors. Forty-eight hours posttransfection, supernatants of the 293T cells were incubated with serum-starved 3T3 cells for 20 min. The 3T3 cells lysates were then immunoblotted for pTyr. The PDGF receptor (180-kD band) was phosphorylated to the same extent using all three constructs. (B) DF-1 cells transfected with the three RCAS vectors were lysed, and expression of the GAP domain was assayed by immunoblotting with anti-p190 antibody. Wild-type and mutant GAP domains were expressed.
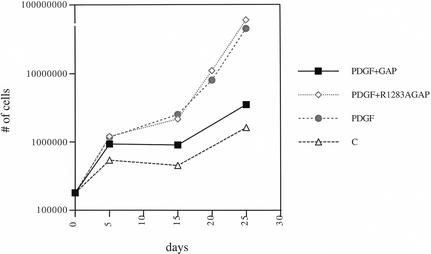
Supernatants from the viral producing DF-1 cells were used to infect Ntv-a primary brain cultures in vitro. Growth rates were monitored after infection to determine the effect of expression of the GAP domain on PDGF-induced cell growth. As depicted in Figure 6, the growth rate of cells expressing PDGF and the GAP domain was strongly inhibited compared with that of cells expressing PDGF and GFP. Cells expressing PDGF and the mutant GAP domain, however, maintained PDGF-induced growth. Control cells that were mock-infected remained relatively static in their growth.
Figure 6.
In vitro growth curves. Primary brain cultures from nestin/tv-a mice were either uninfected or infected with RCAS-PDGF-IRES-GFP, RCAS-PDGF-IRES-GAP, or RCAS-PDGF-IRES-mutantGAP. Cells were allowed to grow in culture, and were periodically trypsinized and counted. The graph represents the extent of growth of each cell type. Expression of the wild-type GAP domain inhibits PDGF-induced proliferation, but the mutant GAP domain does not have this effect.
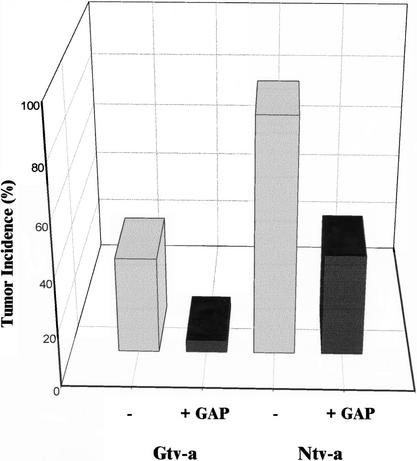
The RCAS vector system was then used to determine the effects of p190 on glioma development in vivo. DF-1 cells producing RCAS-PDGF alone or RCAS-PDGF and the p190 GAP domain were injected into the frontal cortex of neonatal Gtv-a or Ntv-a mice. Tumor formation was determined after killing the animals at 12 wk of age, or earlier if animals developed hydrocephalus or paralysis. Expression of the GAP domain reduced PDGF-induced tumor formation in both animal strains (Fig. 7). In the Gtv-a mouse, tumor incidence decreased from 35.7% (5/14) to 4.3% (1/23) with the introduction of the GAP domain of p190. In the Ntv-a mouse, tumor incidence decreased from 87.5% (7/8) to 37.5% (3/8). These data strongly suggest that p190 RhoGAP can inhibit PDGF-induced glioma formation.
Figure 7.
In vivo tumor generation. DF-1 cells producing RCAS-PDGF or RCAS-PDGF + p190 GAP domain were injected into Gtv-a or Ntv-a mice. All mice were killed at 12 wk or earlier if symptoms appeared. Coexpression of PDGF and the GAP domain of p190 significantly reduced tumor incidence in the Gtv-a (p < 0.01) and Ntv-a (p < 0.05) mice compared with PDGF expression alone.
Proviral insertion in p190RhoGAP
The above-described line of investigation could be potentially biased by our present knowledge of glial cell development and our assumption of the critical role of differentiation in glioma formation. We therefore present independent and unbiased evidence that further validates the interrelation between PDGF-induced glioma formation and p190RhoGAP. For these experiments, a different retroviral model for PDGF-induced gliomas was used. This vector encodes PDGF-B on an MLV-based vector with helper virus to promote viral spread in vivo. The gliomas arising in this system take longer to develop but once symptomatic are high-grade gliomas resembling glioblastoma multiformes (GBMs). Presumably additional genetic alterations, some of which are generated by retroviral insertion, contribute to the GBM phenotype in these mice. A similar strategy has been widely used to identify genetic alterations contributing to leukemia (Jonkers and Berns 1996; Liet et al. 1999) and mammary carcinoma (van Leeuwen and Nusse 1995). In this work, retroviruses were inserted into several critical genes; in some cases the insertion was in introns and in either orientation. Using a similar strategy, we identify p190RhoGAP as contributing to PDGF-induced oncogenesis.
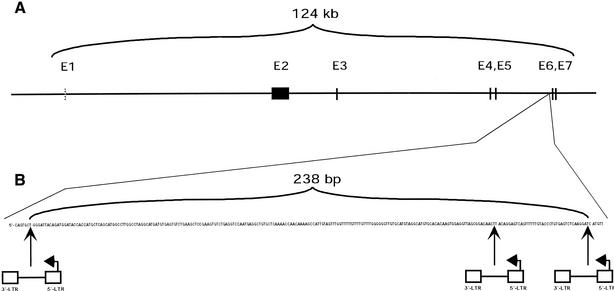
Brain tumors were induced in C57 black mice by intracerebral injection of a recombinant PDGF-B Moloney Murine Leukemia Virus (PDGF-B/MMLV) in combination with helper MMLV (Uhrbom et al. 1998). Genomic DNA was isolated from gliomas that arose, and inverse PCR (IPCR) was used to clone proviral tagged sequences (PTSs). The IPCR fragments were cloned and sequenced. The sequences were then BLAST-searched against the nonredundant database (nr), expressed sequences-tagged database (est), and high-throughput genomic sequence database (htgs). In addition, the PTSs were compared against each other in a local database, using the BLAST programs (Altschul et al. 1997). Then 20 mouse brain tumors were screened for PTSs with IPCR, and among 200 PTSs, four common integrations (loci being tagged twice or more in separate tumors) were found. One of these common PTSs, identified in three independent tumors, mapped within a segment of 238 bp that was contained within a single mouse BAC clone (AC073790; Fig. 8). Comparative analysis showed that the BAC clone mapped to human Chromosome 19q13.3. Extension of the cloned PTSs by walking along the BAC clone revealed that the proviruses tagged an intron sequence of P190 RhoGAP. The gene structure of P190 RhoGAP has not yet been identified. Similarity searches between the mouse BAC clone and the mRNA sequence of rat and human p190 RhoGAP, respectively, indicate that the mouse gene is likely to comprise seven exons, of which the first one is noncoding. This prediction was further supported by using GENSCAN (Burge and Karlin 1997), which is an exon/intron-predicting program. A combination of similarity searches and exon/intron prediction defined the mouse P190 RhoGAP to span at least 124 kb. The proviruses are integrated in opposite transcriptional direction in a large intron sequence between exons 5 and 6 and clustered within a segment as short as 238 bp. This appears to be a highly significant finding as the probability that 3 of 20 tumors would have insertions within a stretch of 238 bases is ∼1 in 1022.
Figure 8.
(A) Schematic representation of the gene structure of mouse p190 RhoGAP and positions of inserted proviral DNAs. Exon 1 is noncoding. The introns and exon 2 are proportional in size; the remaining exons are smaller than drawn. (B) Three proviruses cluster within 238 bp in the intron between exons 5 and 6 in three independent tumors.
Discussion
p190RhoGAP has been shown to be a regulator of oligodendrocyte differentiation (Wolf et al. 2001). This finding, coupled with the fact that p190RhoGAP is located on Chromosome 19q13.3, a locus that is deleted in 50%–80% of oligodendrogliomas, raised the tantalizing possibility that p190RhoGAP inhibits glioma formation by inducing cells to adopt a more differentiated phenotype. Here we show that down-regulation of phosphorylated p190 occurred in astrocytes chronically stimulated with PDGF compared with cells treated with a PDGF receptor inhibitor. Moreover, exogenous expression of p190 suppressed proliferation and altered cell shape, inducing process extension and causing the cells to adopt a more differentiated morphological phenotype. These results suggest that p190 expression promotes astrocyte differentiation.
The above data imply that alterations in the level of RhoGAP activity, and not necessarily complete loss of activity, can have functional consequences. For example, expression of the isolated p190 GAP domain, which conferred only a threefold increase in GAP activity over control cells, was shown to inhibit Ras-induced transformation (Wang et al. 1997). It is therefore conceivable that loss of p190 on one chromosome alone, without additional loss or mutation of the remaining allele, can achieve the functional outcome of increased tumor formation. There is now increasing evidence in the literature that haploinsufficiency can promote oncogenesis, especially in the central nervous system (e.g., Nf1, p53, patched; Venkatachalam et al. 1998; Wetmore et al. 2000; McLaughlin and Jacks 2002).
Exogenously overexpressed p190 likely acts by attenuating RhoGTP signaling. Both full-length p190 and the isolated GAP domain have been shown to exhibit a strong preference for regulating Rho compared with Ras and other Rho family members (Settleman et al. 1992; Ridley et al. 1993). Given that p190 can down-regulate Rac and cdc42 in certain contexts (Coso et al. 1995), it is possible that other small G-proteins may be affected in glial cells. However, use of the Rho kinase inhibitor Y-27632 suggests that RhoGTP signaling is being inhibited, as morphological changes were seen with Rho kinase inhibitor treatment that were similar to those seen with the exogenous expression of p190.
Coinfection of differentiated astrocytes with a retrovirus that expressed PDGF and the GAP domain of p190 caused an inhibition of glial proliferation in vitro as well as a decrease in tumor burden in vivo, as compared with PDGF alone. Taken together, these data suggest that unchecked RhoGTP signaling sustains a more progenitor-like glial phenotype. In addition, it implies that the Rho regulator p190 acts to down-regulate glial proliferation and promote differentiation, thereby inhibiting tumor formation.
Role of the p190 GAP domain
The GAP domain alone was subcloned into the RCAS vector for three reasons. First, p190-induced extension of cellular processes was dependent on an active GAP domain, as the GAP inactive R1283A p190 mutant did not induce morphological changes (Fig. 2D). Second, overexpression of the GAP domain alone was able to recapitulate the morphological phenotype of the full-length protein (Fig. 2C). Third, DNA size constraints would not allow use of the entire p190 coding region. Other studies have also used the isolated GAP domain and have found that it functions to decrease Rho activity in vitro and in vivo (Haskell et al. 2001). The limitation of this approach, however, is that the GAP domain is normally modulated by its surroundings within the native protein, and removal of the domain from its usual context can change some features of the protein. Although some of the GAP domain was cytosolic and capable of acting appropriately, it was noted that a percentage of the protein was nuclear. In the future, construction of a targeted GAP domain, by attaching a membrane-targeting motif, might be considered to more effectively target the protein to its site of action.
Oligodendrocyte differentiation and glioma formation—molecular similarities
Oligodendrocyte differentiation has been widely studied by isolating progenitor cells from neonatal rodent brains and inducing them to differentiate in vitro using well-defined media conditions (Osterhout et al. 1999). Using this cell culture system, it was previously determined that p190 expression as well as tyrosine phosphorylation (Wolf et al. 2001) increase upon differentiation. Differentiated cells were also shown to demonstrate an increase in the p190:p120 phosphotyrosine-dependent complex.
The cell culture model of gliomas used in this study also allows “progenitor” and “differentiated” cells to be analyzed biochemically. GFAP+ cells constitutively expressing PDGF achieve a progenitor-like phenotype. When PDGF signaling is inhibited using the PDGFR inhibitor PTK787, the cells become more differentiated. Indeed, the glioma system shares similar biochemical expression patterns with the primary oligodendrocyte cultures. In both systems, p190 expression and phosphorylation, as well as p190:120 complex formation, increase in the “differentiated” cells.
Interestingly, PDGF is both a mitogen capable of maintaining healthy oligodendrocytes in the precursor state (McKinnon et al. 1990; Tang et al. 2000), as well as an oncogene capable of initiating tumorigenesis when expressed aberrantly in fully differentiated glia. This parallel, as well as the parallels in biochemical expression patterns, support the hypothesis that glioma cells and glial progenitors are similar entities and that gliomagenesis results from differentiation gone awry.
Cross-talk between Rho and PDGF
The data presented in this study demonstrate that down-regulation of Rho activity via overexpression of p190RhoGAP decreases the ability of PDGF to cause tumors. It is important to note that expression of the GAP domain along with PDGF did not completely abrogate tumor development (Fig. 7B). The cellular effects of PDGF are vast and lead to signaling through a growing number of molecules, including Ras, PI3K, JAK-STAT, and PLCγ (Heldin and Westermark 1999). In addition, it is appears that a portion of the PDGF-induced tumorigenic phenotype is mediated through disregulation of Rho, and that at the same time, other signaling pathways contribute as well.
The use of retroviral insertion for the identification of genes contributing to oncogenesis provides unbiased confirmation of the importance of p190RhoGAP in this system. A striking finding was that the insertions all occurred within a short segment of the p190 gene. As we indicated in the Results section, the probability that 3 of 20 tumors would have insertions within 238 bases is ∼1 in 1022. However, even if one considers the entire p190 gene, the likelihood is extraordinarily small that insertions could be occurring by chance. The proviruses integrated in the opposite transcriptional direction within an intronic sequence. Such integrations are likely to lead to premature termination of transcription, because of the presence of a cryptic poly(A) site in the reverse 3′-LTR of the retrovirus (Jonkers and Berns 1996). The remaining allele was sequenced and found to be intact. In this screen several other genes encoding proteins in a variety of biologic processes in addition to p190RhoGAP have been identified (C. Eklöf, J. Brodd, M. Ferletta, G. Hesselager, F. Johansson, C.-F. Tiger, L. Uhrbom, and B. Westermark, in prep.). The cross-validation of these two lines of investigation presented in this manuscript suggests that other genes identified in this insertion site screen could plan critical roles in cooperating with PDGF in gliomagenesis as well.
Tumor suppressors on 19q and 1p
We have presented evidence that p190 is a putative tumor suppressor located on 19q13.3 of the human chromosome, a locus that is deleted in oligodendrogliomas. Another common chromosomal deletion in oligodendrogliomas is 1p36, which often occurs in tandem with the 19q13.3 deletion, suggesting that inactivation of genes on both loci is necessary for tumor progression (Smith and Jenkins 2000). Searches for potential tumor suppressors in 1p are ongoing at present. Both the 19q and 1p chromosomal deletions have been identified in other cancers as well (Hogarty et al. 2001; Mora et al. 2001), implying that the eventual characterization of the tumor suppressors on these loci will have ramifications for a variety of diseases.
Implications for therapy
It appears that at least a subset of gliomas is initiated by a dedifferentiation event that takes place in fully differentiated glial cells. Consequently, drugs that can induce cells to differentiate may be considered as potential therapies for gliomas. The results presented in this study indicate that Y-27632, the Rho kinase inhibitor, might be such a therapeutic agent. Treatment with Y-27632 induced process extension and caused the reduction of expression of the neuronal precursor nestin. A small molecule that is able to inhibit Rho kinase might therefore one day be a useful therapeutic strategy to induce differentiation in gliomas.
Materials and methods
Cell culture
293T cells were maintained in DMEM supplemented with 10% heat-inactivated fetal bovine serum. NIH 3T3 cells were maintained in DMEM supplemented with 10% calf serum. The DF-1 packaging cell line was maintained in DMEM supplemented with 15% calf serum. Primary brain cultures from Gtv-a neonatal mice had previously been infected with the RCAS vector expressing PDGF (Dai et al. 2001). This cell line (subsequently referred to as Gtv-a/PDGF) was maintained in DMEM supplemented with 10% calf serum. PDGF signaling of the cell line was blocked using the drug PTK787 dissolved in DMSO (Wood et al. 2000). Cells were incubated with PTK787 at a final concentration of 1 μM for 1 wk before cells were lysed for protein analysis. Cells were treated with 50 μM Rho kinase inhibitor Y-27632 (Calbiochem) dissolved in water for 2 d. Fresh primary brain cultures from Ntv-a mice were prepared as follows. Brains were removed from neonatal mice and homogenized by drawing the cellular material through a series of 18-, 21-, and 23-gauge needles. Homogenates were then plated in tissue culture dishes.
Immunoprecipitation and immunoblotting
Control and PTK787-treated Gtv-a/PDGF cells were lysed in NP-40 lysis buffer (1% NP-40, 0.5% deoxycholate, 1 mM sodium vanadate, 10 mM Tris at pH 7.5, 150 mM NaCl, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 250 μg/mL PefaBloc). Lysates were incubated on ice for 20 min followed by centrifugation at 14,000 rpm at 4°C for 10 min. The supernatants were then subjected to the Pierce Micro BCA protein concentration assay. Lysates containing equal amounts of protein were immunoprecipitated and subjected to Western blot analysis using a variety of antibodies. The monoclonal anti-p120 and anti-p190 used in immunoprecipitations were generous gifts from Sarah Parsons (University of Virginia, Charlottesville, VA). Monoclonal anti-p190 (UBI), monoclonal anti-nestin (Chemicon), monoclonal anti-phosphotyrosine (PY99, Santa Cruz), and polyclonal anti-p120 (Santa Cruz) were used as primary antibodies in Western blotting.
Constructs, transfections, and fluorescence microscopy
Gtv-a/PDGF cells were transfected with a variety of HA-tagged molecules in the pKH3 vector using Lipofectamine 2000 transfection reagent (Invitrogen). The following pKH3 vectors were generous gifts from Ian Macara (University of Virginia, Charlottesville, VA): wild-type p190, GAP defective R1283A p190, constitutively active Rho G14V, and dominant-negative Rho S19N.
The morphologies of the transfected cells were studied using immunofluorescence to detect the HA epitope. Briefly, cells were washed three times with PBS, fixed with 4% paraformaldehyde for 15 min, and permeabilized with 0.1% Triton X-100 in PBS for 5 min. Cells were then incubated with 5% donkey serum for 1 h, followed by incubation with the F7 anti-HA monoclonal antibody (Santa Cruz) at a 1:200 dilution for 2 h. Cells were washed three times before incubation with an anti-mouse secondary antibody conjugated to the Alexa 488 (green) or 594 (red) fluorophore at a concentration of 1:1000. Cells were then mounted for visualization using ProLong mounting media (Molecular Probes). In addition, wild-type and mutant GAP domains fused to GFP were constructed as follows. PCR primers were designed to amplify the cDNA corresponding to amino acids 1280–1513 of p190 RhoGAP. Wild-type p190 and GAP defective R1283A p190 in the pKH3 vector were used as templates. PCR products and the EGFP-C1 vector (Clontech) were digested with EcoRI and BamHI and ligated together. GFP fusion constructs as well as GFP alone were transfected into the Gtv-a/PDGF cells and visualized using the GFP filter on an epifluorescence microscope.
BRDU detection
Gtv-a/PDGF cells, plated on coverslips, were transfected with either pValo, a β-galactosidase-expressing plasmid, or HA-tagged wild-type or R1283A p190 in the pKH3 vector as described above. The media on the cells was changed ∼12 h after transfection to minimize the toxicity of the transfection reagent. Then 48 h after transfection, BRDU labeling reagent (Roche) was added to the media at a concentration of 1:1000 according to the manufacturer's instructions. Cells were incubated at 37°C for 7 h, and processed for immunofluorescence as follows. Cells were washed three times in PBS and then incubated with a fixation solution (70% ethanol, 50 mM glycine) at −20°C for 20 min. Cells were washed and incubated with a monoclonal anti-BRDU antibody (Roche) at a concentration of 1:10, along with either the Y11 polyclonal anti-HA antibody (1:200, Santa Cruz) or a polyclonal anti-β-galactosidase antibody (1:250, Molecular Probes). Antibodies were added to the incubation buffer provided by the manufacturer (Roche). After incubation with primary antibodies at 37°C for 30 min, cells were washed with PBS and incubated with anti-mouse Alexa-594 and anti-rabbit Alexa-488 secondary antibodies at ratios of 1:1000 at 37°C for 30 min. Cells were then washed a final time and mounted on coverslips using ProLong mounting media (Molecular Probes).
RCAS vector construction
Human PDGF-B/c-sis was amplified by PCR from the RCAS-PDGF-IRES-GFP vector (Dai et al. 2001) flanked by NotI and EcoRI sites. The GAP domain of p190RhoGAP, flanked by SmaI and ClaI sites, was amplified by PCR from both the p190wt/PKH3 and the p190R1283A/PKH3 vectors, kind gifts from Ian Macara (University of Virginia, Charlottesville, VA). Both PDGF and the GAP domain were inserted into the RCAS vector via the YIG shuttle vector, with PDGF upstream and GAP downstream of an IRES sequence, creating the RCAS-PDGF-IRES-GAP vector.
Expression of both the PDGF and GAP domain transcripts was tested by transfecting the RCAS vector into DF-1 cells using Fugene (Roche). Cells were lysed after 36 h, and expression of the GAP domain was tested via immunoblotting, using both an anti-HA antibody and an anti-p190 antibody (Transduction Laboratories). Expression of PDGF was tested by transfecting 293T cells with the RCAS vectors using the Lipofectamine 2000 transfection reagent. The media from the transfected 293T cells was then incubated with serum-starved NIH 3T3 cells. 3T3 lysates were then subjected to Western blotting with the anti-phosphotyrosine antibody PY99 (Santa Cruz) to detect phosphorylation of the PDGF receptor.
Infection of primary brain cultures
RCAS-PDGF-IRES-GFP, RCAS-PDGF-IRES-wtGAP, and RCASPDGF-IRES-GAP/R1283A were transfected into the DF-1 packaging cell line using Fugene. Primary brain cultures from Ntv-a neonatal mice were prepared as described above. Supernatants of confluent transfected DF-1 cells were filtered through a 0.45-μm filter to remove cellular debris, and were then used to infect primary brain cultures of Gtv-a or Ntv-a neonatal mice. Primary brain cultures were subjected to three overnight infections each with fresh DF-1 supernatant. After infection, cells were allowed to recover for 2 d. Subsequently, cells were trypsinized, and 1 × 105 cells of each construct were plated. Cells were allowed to grow for designated amounts of time, and were trypsinized and recounted to generate growth curves.
In vivo infection of tv-a transgenic mice
A confluent plate of transfected DF-1 cells (1 × 106) was trypsinized and pelleted by centrifugation. Then 10% of the cells were lysed and tested for expression levels of wild-type and mutant GAP domain. Cells were resuspended in 10 μL of media and placed on ice before injection. Finally, 1 μL of the cell suspension was injected into the forebrain of neonatal Gtv-a or Ntv-a mice, using a 10-μL gas-tight Hamilton syringe.
Tumor analysis
Mice were killed 12 wk after injection, or earlier if the mice showed hydrocephalus or paralysis. Brains were removed from the skull, fixed in 10% formalin for a minimum of 3 d with shaking, and embedded in paraffin. Then 5-μm sections were cut with a Leica microtome and stained with hematoxylin and eosin (H&E). Brains in which tumors were detected were stained with a polyclonal anti-HA antibody (Santa Cruz) or a monoclonal anti-p190 antibody (Transduction Laboratories) by immunohistochemistry to determine the extent of spread of the HA-tagged GAP domain.
In brief, immunohistochemistry was performed as follows: brain sections were deparaffinized overnight and incubated with an antigen unmasking reagent (Vector Lab) at a dilution of 1:100 under steam heating for 30 min. Sections were incubated in 1% hydrogen peroxide in methanol for 30 min to eliminate endogenous peroxidase activity. Sections were then blocked with 5% donkey serum diluted in PBS/0.5% Tween (PBS-T) for 1 h, followed by incubation with primary antibody (1:200 in PBS-T). Sections were washed and incubated with a biotinylated secondary antibody (Vector Labs) diluted 1:200 in PBS-T for 1 h. Avidin conjugated peroxidase (ABC kit from Vector Labs) was added at room temperature for 1 h, followed by DAB (Vector Labs). Sections were counterstained with hematoxylin and mounted. Negative controls were subjected to the same procedure without exposure to primary antibody.
Tumor induction by MMLV/PDGF-B and genomic DNA preparation
The generation of brain tumors using MMLV/PDGF-B and preparation of genomic DNA have been previously described (Uhrbom et al. 1998).
Inverse polymerase chain reaction (IPCR)
Provirally tagged sequences (PTSs) were cloned with inverse PCR (IPCR). One microgram of genomic DNA from tumor material was cut with Pst1 and Rsa1, respectively. After phenol extraction and ethanol precipitation, DNA was ligated to a circular construct. The ligase was heat-inactivated, followed by precipitation of DNA in ammonium acetate and ethanol. The DNA was dissolved in ddH2O and used as template in the first PCR. In the subsequent nested PCR, 1/50 of the first reaction served as template. PCR reactions were performed in a total volume of 50 μL containing 1× PCR buffer (Applied Biosystems), 50 μM of each dNTP (Amersham Pharmacia Biotech Inc.), 2.5 units of Taq polymerase (Applied Biosystems), 0.4 μM of each primer, and 1% DMSO. Amplifications were made in a Perkin Elmer GeneAmp PCR System 2400 machine for 40 cycles with melting at 94°C for 30 sec, annealing at 60°C (primary PCR)/65°C (nested PCR) for 30 sec, and elongation at 72°C for 1 min. PCR products were visualized on a 1.6% agarose gel stained with ethidium bromide.
Cloning and sequencing of IPCR fragments
IPCR products were cloned into pGEM-T Easy vectors (Promega). They were sequenced from both directions with universal M13-21 and SP6 primers, and the sequencing was performed on an ABI Model 377 DNA Sequencer (Applied Biosystems) with the Big Dye Terminator Cycle Sequencing Ready Reaction Kit protocol (Applied Biosystems).
Sequence analysis
To investigate if the PTSs mapped to already known oncogenes, we BLAST-searched them against the nonredundant, expressed sequence-tagged (est), and high-throughput genomic sequence databases (htgs). Sequences were determined as matching using a cutoff value for the probability value of 10−25 or less, and after visual inspection of the homology. Local databases were created using the BLAST programs (Altschul et al. 1997) for finding overlapping sequences among the cloned PTSs.
Prediction of the mouse P190 RhoGAP structure was done by doing pairwise alignment of the mRNA sequence for rat (M94721) and human (AF159851) P190RhoGAP, respectively, with the mouse BAC clone (AC073790). Exon/intron prediction was done using GENSCAN (http://genes.mit.edu/GENSCAN.html).
Acknowledgments
We thank Dr. Josefin Brodd for sequencing the p190 alleles; Sarah Parsons and Ian Macara for helpful reagents; Katia Manova, Kevin Curran, and Eric Suh of the MSKCC Molecular Cytology Core Facility; Eddie Nerio for the histology work; Raisa Louft-Nisenbaum for valuable technical assistance; and Debra Alston for help in preparation of the manuscript. This work was supported by NIH MSTP grant GM07739 (R.M.W.); the Robert Wood Johnson Jr. Charitable Trust (R.M.W.); NIH grants GM57966 and CA96582 (M.D.R.); CA94842 and CA894314-1 (E.C.H.); the Tow, Bressler, and Searle Scholar's Foundations (E.C.H.); and The Swedish Cancer Foundation and Children's Cancer Foundation (B.W.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL m-resh@ski.mskcc.org; FAX (212) 717-3317.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1040003.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur WT, Burridge K. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol Biol Cell. 2001;12:2711–2720. doi: 10.1091/mbc.12.9.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur WT, Petch LA, Burridge K. Integrin engagement suppresses RhoA activity via a c-Src-dependent mechanism. Curr Biol. 2000;10:719–722. doi: 10.1016/s0960-9822(00)00537-6. [DOI] [PubMed] [Google Scholar]
- Bello MJ, Vaquero J, de Campos JM, Kusak ME, Sarasa JL, Saez-Castresana J, Pestana A, Rey JA. Molecular analysis of chromosome 1 abnormalities in human gliomas reveals frequent loss of 1p in oligodendroglial tumors. Intl J Cancer. 1994;57:172–175. doi: 10.1002/ijc.2910570207. [DOI] [PubMed] [Google Scholar]
- Boerner JL, Danielsen A, McManus MJ, Maihle NJ. Activation of Rho is required for ligand-independent oncogenic signaling by a mutant epidermal growth factor receptor. J Biol Chem. 2001;276:3691–3695. doi: 10.1074/jbc.M003801200. [DOI] [PubMed] [Google Scholar]
- Bogler O, Wren D, Barnett SC, Land H, Noble M. Cooperation between two growth factors promotes extended self-renewal and inhibits differentiation of oligodendrocyte-type-2 astrocyte (O-2A) progenitor cells. Proc Natl Acad Sci. 1990;87:6368–6372. doi: 10.1073/pnas.87.16.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- Chang JH, Gill S, Settleman J, Parsons SJ. c-Src regulates the simultaneous rearrangement of actin cytoskeleton, p190RhoGAP, and p120RasGAP following epidermal growth factor stimulation. J Cell Biol. 1995;130:355–368. doi: 10.1083/jcb.130.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coso OA, Chiariello M, Yu JC, Teramoto H, Crespo P, Xu N, Miki T, Gutkind JS. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes & Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danen EH, Sonneveld P, Sonnenberg A, Yamada KM. Dual stimulation of Ras/mitogen-activated protein kinase and RhoA by cell adhesion to fibronectin supports growth factor-stimulated cell cycle progression. J Cell Biol. 2000;151:1413–1422. doi: 10.1083/jcb.151.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoyelle C, Vasse M, Korner M, Mishal Z, Ganne F, Vannier JP, Soria J, Soria C. Cerivastatin, an inhibitor of HMG-CoA reductase, inhibits the signaling pathways involved in the invasiveness and metastatic properties of highly invasive breast cancer cell lines: An in vitro study. Carcinogenesis. 2001;22:1139–1148. doi: 10.1093/carcin/22.8.1139. [DOI] [PubMed] [Google Scholar]
- Dupont H, Blancq M. Formation of complexes involving RasGAP and p190 RhoGAP during morphogenetic events of the gastrulation in Xenopus. Eur J Biochem. 1999;265:530–538. doi: 10.1046/j.1432-1327.1999.00689.x. [DOI] [PubMed] [Google Scholar]
- Guha A, Dashner K, Black PM, Wagner JA, Stiles CD. Expression of PDGF and PDGF receptors in human astrocytoma operation specimens supports the existence of an autocrine loop. Intl J Cancer. 1995;60:168–173. doi: 10.1002/ijc.2910600206. [DOI] [PubMed] [Google Scholar]
- Haskell MD, Nickles AL, Agati JM, Su L, Dukes BD, Parsons SJ. Phosphorylation of p190 on Tyr1105 by c-Src is necessary but not sufficient for EGF-induced actin disassembly in C3H10T1/2 fibroblasts. J Cell Sci. 2001;114:1699–1708. doi: 10.1242/jcs.114.9.1699. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- Hogarty MD, Maris JM, White PS, Guo C, Brodeur GM. Analysis of genomic imprinting at 1p35–36 in neuroblastoma. Med Pediatr Oncol. 2001;36:52–55. doi: 10.1002/1096-911X(20010101)36:1<52::AID-MPO1014>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Holland EC. Glioblastoma multiforme: The terminator. Proc Natl Acad Sci. 2000;97:6242–6244. doi: 10.1073/pnas.97.12.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Gliomagenesis: Genetic alterations and mouse models. Nat Rev Genet. 2001;2:120–129. doi: 10.1038/35052535. [DOI] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes & Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- Jonkers J, Berns A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim Biophys Acta. 1996;1287:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- Kulkarni SV, Gish G, van der Geer P, Henkemeyer M, Pawson T. Role of p120 Ras-GAP in directed cell movement. J Cell Biol. 2000;149:457–470. doi: 10.1083/jcb.149.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liet JM, Piloquet H, Marchini JS, Maugere P, Bobin C, Roze JC, Darmaun D. Leucine metabolism in preterm infants receiving parenteral nutrition with medium-chain compared with long-chain triacylglycerol emulsions. Am J Clin Nutr. 1999;69:539–543. doi: 10.1093/ajcn/69.3.539. [DOI] [PubMed] [Google Scholar]
- McKinnon RD, Matsui T, Dubois-Dalcq M, Aaronson SA. FGF modulates the PDGF-driven pathway of oligodendrocyte development. Neuron. 1990;5:603–614. doi: 10.1016/0896-6273(90)90215-2. [DOI] [PubMed] [Google Scholar]
- McLaughlin ME, Jacks T. Thinking beyond the tumor cell: Nf1 haploinsufficiency in the tumor environment. Cancer Cell. 2002;1:408–410. doi: 10.1016/s1535-6108(02)00078-8. [DOI] [PubMed] [Google Scholar]
- Mora J, Cheung NK, Chen L, Qin J, Gerald W. Loss of heterozygosity at 19q13.3 is associated with locally aggressive neuroblastoma. Clin Cancer Res. 2001;7:1358–1361. [PubMed] [Google Scholar]
- Olson MF, Paterson HF, Marshall CJ. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature. 1998;394:295–299. doi: 10.1038/28425. [DOI] [PubMed] [Google Scholar]
- Osterhout DJ, Wolven A, Wolf RM, Resh MD, Chao MV. Morphological differentiation of oligodendrocytes requires activation of Fyn tyrosine kinase. J Cell Biol. 1999;145:1209–1218. doi: 10.1083/jcb.145.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994;145:1175–1190. [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Self AJ, Kasmi F, Paterson HF, Hall A, Marshall CJ, Ellis C. Rho family GTPase activating proteins p190, bcr and rho GAP show distinct specificities in vitro and in vivo. EMBO J. 1993;12:5151–5160. doi: 10.1002/j.1460-2075.1993.tb06210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Olson MF, Marshall CJ. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 2001;20:755–766. doi: 10.1093/emboj/20.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settleman J, Albright CF, Foster LC, Weinberg RA. Association between GTPase activators for Rho and Ras families. Nature. 1992;359:153–154. doi: 10.1038/359153a0. [DOI] [PubMed] [Google Scholar]
- Sharma SV. Rapid recruitment of p120RasGAP and its associated protein, p190RhoGAP, to the cytoskeleton during integrin mediated cell–substrate interaction. Oncogene. 1998;17:271–281. doi: 10.1038/sj.onc.1201921. [DOI] [PubMed] [Google Scholar]
- Smith JS, Jenkins RB. Genetic alterations in adult diffuse glioma: Occurrence, significance, and prognostic implications. Front Biosci. 2000;5:D213–D231. doi: 10.2741/smith. [DOI] [PubMed] [Google Scholar]
- Tang DG, Tokumoto YM, Raff MC. Long-term culture of purified postnatal oligodendrocyte precursor cells. Evidence for an intrinsic maturation program that plays out over months. J Cell Biol. 2000;148:971–984. doi: 10.1083/jcb.148.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikoo A, Czekay S, Viars C, White S, Heath JK, Arden K, Maruta H. p190-A, a human tumor suppressor gene, maps to the chromosomal region 19q13.3 that is reportedly deleted in some gliomas. Gene. 2000;257:23–31. doi: 10.1016/s0378-1119(00)00387-5. [DOI] [PubMed] [Google Scholar]
- Uhrbom L, Hesselager G, Nister M, Westermark B. Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus. Cancer Res. 1998;58:5275–5279. [PubMed] [Google Scholar]
- van Golen KL, Wu ZF, Qiao XT, Bao LW, Merajver SD. RhoC GTPase, a novel transforming oncogene for human mammary epithelial cells that partially recapitulates the inflammatory breast cancer phenotype. Cancer Res. 2000;60:5832–5838. [PubMed] [Google Scholar]
- van Leeuwen F, Nusse R. Oncogene activation and oncogene cooperation in MMTV-induced mouse mammary cancer. Semin Cancer Biol. 1995;6:127–133. doi: 10.1006/scbi.1995.0018. [DOI] [PubMed] [Google Scholar]
- Venkatachalam S, Shi YP, Jones SN, Vogel H, Bradley A, Pinkel D, Donehower LA. Retention of wild-type p53 in tumors from p53 heterozygous mice: Reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DZ, Nur EKMS, Tikoo A, Montague W, Maruta H. The GTPase and Rho GAP domains of p190, a tumor suppressor protein that binds the M(r) 120,000 Ras GAP, independently function as anti-Ras tumor suppressors. Cancer Res. 1997;57:2478–2484. [PubMed] [Google Scholar]
- Wetmore C, Eberhart DE, Curran T. The normal patched allele is expressed in medulloblastomas from mice with heterozygous germ-line mutation of patched. Cancer Res. 2000;60:2239–2246. [PubMed] [Google Scholar]
- Wolf RM, Wilkes JJ, Chao MV, Resh MD. Tyrosine phosphorylation of p190 RHOGAP by Fyn regulates oligodendrocyte differentiation. J Neurobiol. 2001;49:62–78. doi: 10.1002/neu.1066. [DOI] [PubMed] [Google Scholar]
- Wood JM, Bold G, Buchdunger E, Cozens R, Ferrari S, Frei J, Hofmann F, Mestan J, Mett H, O'Reilly T, et al. PTK787/ZK 222584, a novel and potent inhibitor of vascular endothelial growth factor receptor tyrosine kinases, impairs vascular endothelial growth factor-induced responses and tumor growth after oral administration. Cancer Res. 2000;60:2178–2189. [PubMed] [Google Scholar]
- Zrihan-Licht S, Fu Y, Settleman J, Schinkmann K, Shaw L, Keydar I, Avraham S, Avraham H. RAFTK/Pyk2 tyrosine kinase mediates the association of p190 RhoGAP with RasGAP and is involved in breast cancer cell invasion. Oncogene. 2000;19:1318–1328. doi: 10.1038/sj.onc.1203422. [DOI] [PubMed] [Google Scholar]