Abstract
Although it is now well-established that boundary elements/insulators function to subdivide eukaryotic chromosomes into autonomous regulatory domains, the underlying mechanisms remain elusive. One idea is that boundaries act as barriers, preventing the processive spreading of “active” or “silenced” chromatin between domains. Another is that the partitioning into autonomous functional units is a consequence of an underlying structural subdivision of the chromosome into higher order “looped” domains. In this view, boundaries are thought to delimit structural domains by interacting with each other or with some other nuclear structure. The studies reported here provide support for the looped domain model. We show that the Drosophila scs and scs‘ boundary proteins, Zw5 and BEAF, respectively, interact with each other in vitro and in vivo. Moreover, consistent with idea that this protein:protein interaction might facilitate pairing of boundary elements, we find that that scs and scs‘ are in close proximity to each other in Drosophila nuclei.
Keywords: Insulators, boundaries, looping, domains, protein:protein interaction, pairing
The chromosomes of higher eukaryotes are subdivided into functionally autonomous domains that have distinct properties depending on whether they are transcriptionally active or silent. A good example of an “active” chromatin domain is the 35-kb β-globin locus in chicken erythrocytes. In addition to being considerably more sensitive to DNase I digestion than the flanking silenced domains (Bellard et al. 1980; Stalder et al. 1980), chromatin from the β-globin locus has a reduced ability to form pseudo-higher order structures and a two- to threefold lower level of the linker histone H5 (Verreault and Thomas 1993). There are also striking differences in the patterns of histone acetylation and methylation. Histones in the β-globin domain are hyperacetylated and have a relatively high level of methylation at Lys 4 of histone H3 (Hebbes et al. 1992; Litt et al. 2001a,b). In contrast, histones in the flanking silenced domains are hypoacetylated and are enriched in histone H3 methylated at Lys 9. The features that distinguish active and inactive domains in chicken erythrocytes are evident in other eukaryotes. For example, the silenced mating type loci of yeast are located in chromatin domains that are resistant to nuclease and restriction enzyme digestion, are highly compacted, have hypoacetylated histones, are enriched in histone H3 that is methylated on Lys 9, and have a special set of nonhistone chromosomal proteins (Grewal 2000; Huang 2002).
The subdivision of eukaryotic chromosome into domains that have a distinct chromatin organization, biochemical composition, and genetic activity requires a mechanism to separate one domain from another. Special elements called boundaries or insulators are thought to serve this purpose (Bell et al. 2001; Gerasimova and Corces 2001). Elements that function as boundaries of chromatin domains were first identified in Drosophila (Gyurkovics et al. 1990; Holdridge and Dorsett 1991; Kellum and Schedl 1991, 1992; Geyer and Corces 1992) and have subsequently been found in a diverse array of organisms including yeast, sea urchins, Xenopus, chickens, mice, and humans (Gerisamova and Corces 2001; West et al. 2002). These elements define the limits of chromosomal domains and function to establish independent units of gene activity, insulating genes or regulatory elements within a domain from the action of regulatory elements located outside in adjacent domains. Both the active chicken β-globin domain and the inactive Schizosaccharomyces pombe silent mating type domain are delimited by a pair of boundary or insulator elements (Saitoh et al. 2000; Noma et al. 2001; Thon et al. 2002). In the case of the silent S. pombe mating type locus, deletion of either of the boundary elements permits the spreading of the “silenced” mating type locus chromatin into the adjacent normally active sequences (Noma et al. 2001).
Much of our understanding of the functional properties of boundaries has come from two transgene assays. One tests a pair of boundaries for their ability to insulate reporter genes from chromosomal position effects. The other is a blocking assay in which the boundary is interposed between a reporter gene and an enhancer or silencer. Although these assays demonstrate that boundary elements are capable of establishing independent domains of gene activity outside of their normal context, they leave open the underlying mechanisms. Two general (though not mutually exclusive) models for how boundaries might function have been suggested. In the first, boundaries are thought to act as roadblocks or barriers, obstructing proteins associated with enhancers or silencers from acting on genes or regulatory elements in adjacent domains (Chung et al. 1993; Geyer 1997; Udvardy 1999). In this model, boundaries would have only an indirect role in subdividing the chromosome, defining higher domains by virtue of their ability to confine the processive spread of “active” or “silenced” chromatin to a single domain. The second model postulates that the insulator activity of boundary elements is intimately tied to the subdivision of the chromosome into discrete, physically, and functionally independent domains (Marsden and Laemmli 1979; Udvardy et al. 1985; Udvardy 1999). Boundaries would define the physical endpoints of “looped” higher order domains either by interacting with each other along the main axis of the chromosome or by interacting with some other nuclear structure.
In this paper, we have investigated whether two closely linked Drosophila boundaries, scs and scs‘, can “pair” with each other, potentially forming a looped higher order chromatin domain. scs and scs‘ flank the two divergently transcribed hsp70 genes at the 87A7 heat-shock locus and are located ∼15 kb apart. In uninduced polytene chromosomes, scs and scs‘ appear to be in close proximity; however, after heat induction, in situ hybridization experiments indicate that they are located at or near the outside edges of the domain that decondenses to give the heat-shock puff (Udvardy et al. 1985). Like other fly boundaries scs and scs‘ can protect reporter genes against chromosomal position effects and can block the action of enhancers and silencers (Kellum and Schedl 1991, 1992; Sigrist and Pirrotta 1997). Although there are promoters in close proximity to both scs and scs‘, they are separable from the sequences conferring boundary function (Glover et al. 1995; Vazquez and Schedl 1994; Zhao et al. 1995; Hogga et al. 2001).
If scs and scs‘ form a looped domain by interacting with each other, this interaction is expected to be mediated by proteins that either directly or indirectly associate with these two boundary elements. Previous studies have shown that the Zeste-white 5 (Zw5) protein interacts with scs in vitro and in vivo and that this protein has “boundary activity” in enhancer blocking assays (Gaszner et al. 1999). However, Zw5 does not bind to scs‘; instead, scs‘ has multiple target sites for the BEAF (boundary element associated factor) proteins, BEAF 32A and BEAF 32B. These BEAF binding sites are important for the boundary function of scs‘ (Zhao et al. 1995; Hart et al. 1997) and for the boundary activity of other elements containing BEAF binding sites that are located elsewhere in the fly genome (Cuvier et al. 1998, 2002). Here we show that Zw5 and BEAF interact with each other in vitro and in vivo. Consistent with the idea that this protein:protein interaction may help promote contact between scs and scs‘—forming an 87A7 looped domain— we find that Zw5 antibodies immunoprecipitate not only scs sequences but also sequences from scs‘ in chromatin immunoprecipitations (ChIPs). Finally, independent evidence that scs and scs‘ pair with each other in vivo is provided by chromosome conformation capture experiments (Dekker et al. 2002).
Results
Zw5 localizes to scs but not scs‘ in polytene chromosomes
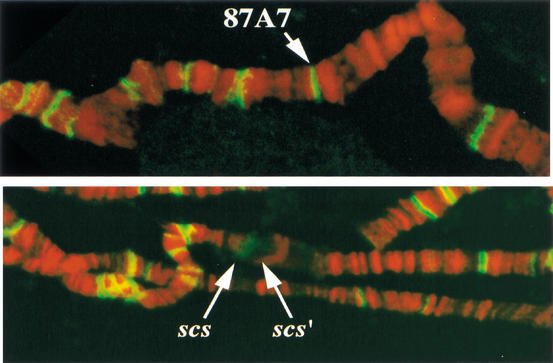
The ∼90 kD Zw5 protein has eight evenly spaced C2H2 zinc fingers in its C-terminal half and recognizes a 15-bp sequence. When this binding sequence is multimerized, the resulting multimer is capable of partially recapitulating the boundary activity of scs in an enhancer blocking assay (Gaszner et al. 1999). The boundary function of the multimer depends on Zw5. There is single copy of the Zw5 motif in scs, and in gel shift and footprinting experiments, the Zw5 protein only binds to DNA fragments from scs that contain this sequence. The motif is not present elsewhere in 87A7 or in scs‘, and the Zw5 protein does not bind to fragments derived from scs‘ in vitro (Gaszner et al. 1999; data not shown). As illustrated in Figure 1A, antibody staining of salivary gland polytene chromosomes reveals that Zw5 protein localizes to many sites (>100). As expected, there is a single anti-Zw5 band at the 87A7 locus in polytene chromosomes prepared from non-heat-shocked larvae. In polytenes from heat-shocked larvae (Fig. 1B), Zw5-specific staining is localized to the proximal edge or scs side of the puff. Note that Zw5 is not observed on the distal side of the puff (scs‘) or at the neighboring 87C heat-shock locus. Nor are there any other Zw5 containing bands in immediate proximity to 87A7. This is consistent with database searches of the fly genome for the Zw5 binding motif.
Figure 1.
Zw5 localizes to scs in polytene chromosomes. A Zw5 polyclonal antibody was used to probe fixed salivary gland polytene chromosomes prepared from control larvae grown at 18°C, and larvae heat-shocked for 30 min at 37°C. The DNA was counterstained with propidium iodide, and the preparations were then visualized by confocal microscopy. Zw5 is in green, DNA is in red. More than 100 different sites are labeled with the Zw5 antibody in polytene chromosomes from non-heat-shocked larvae. (Top) One of these sites on the third chromosome is the condensed 87A7 heat-shock locus. In polytene chromosomes from heat-shock larvae, the two 87A7 hsp70 genes are induced, and the locus forms a large puff. (Bottom) scs is on the proximal side of the 87A7 puff; scs‘ is on the distal side. Zw5 staining is seen on the proximal, scs, side of the puff. No staining is observed on the distal, scs‘, side of the puff.
Mapping of Zw5 in the 87A7 locus by ChIP
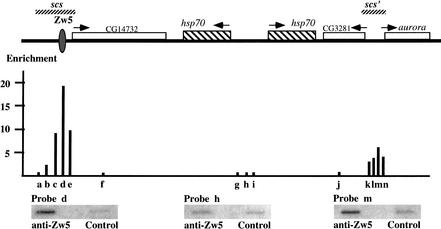
We used ChIP to confirm that the Zw5 protein seen at 87A7 in polytene chromosomes localizes exclusively to scs. We first focused our attention on a 1.2-kb region containing the scs element. As illustrated in the diagram in Figure 2, probe d, which spans the single Zw5 binding site, shows the highest degree of association with the Zw5 protein and is enriched 18-fold in the Zw5 immunoprecipitate. Probes c and e (Fig. 2), which are located to either side of the Zw5 binding side, are also enriched in the Zw5 immunoprecipitate; however, in each case the enrichment is only about one half that of the probe containing the Zw5 binding site. Presumably this drop off is owing to the relatively short length of the DNA fragments. Indeed, sequences on the proximal side of scs that are father away from the Zw5 binding site show only a very modest (Fig. 2, probe b) or little, if any, (Fig. 2, probe a) enrichment in the Zw5 immunoprecipitate. These two probes are more distant from the Zw5 binding site, and DNA fragments containing complementary sequences would not typically extend as far as the scs Zw5 binding site.
Figure 2.
Distribution of Zw5 across the 87A7 heat-shock locus. The chromatin immunoprecipitation (ChIP) procedure of Orlando and Paro (1993; see Materials and Methods) was used to examine the distribution of Zw5 across the 87A7 locus. Affinity-purified rabbit anti-Zw5 antibody (Gaszner et al. 1999) or preimmune rabbit serum was used to immunoprecipitate the CsCl-purified cross-linked chromatin. DNA recovered from each pellet was amplified by LM-PCR (Zhao et al. 1995), slot blotted, and then probed with short fragments (a–n) derived from different sequences in the 87A7 locus. The approximate position of each fragment used as a probe is indicated below the map of 87A7 in the diagram and is given in base pairs in the Materials and Methods section. The degree of enrichment in the Zw5 immunoprecipitate was calculated relative to preimmune serum control and is plotted in the diagram. The signal for probes a, f, and g–j in the Zw5 immunoprecipitate was less than that of the preimmune serum, while all other probes gave signals in the Zw5 immunoprecipitate that were greater than the preimmune serum. The highest degree of enrichment in this experiment is for probe d, which is nearly 20-fold greater in the Zw5 immunoprecipitate than in the immunoprecipitate from the control preimmune serum. Probe d spans the Zw5 binding site. The degree of enrichment for probes around scs‘ was between three- and sixfold. Probes k and m contain BEAF binding sites.
We next asked whether Zw5 is associated with sequences elsewhere in 87A7. As expected probes 3′ to the proximal (Fig. 2, probe f) and distal hsp70 gene (Fig. 2, probe j), as well as several probes (Fig. 2, probes g, h, and i) in the intergenic spacer in between the two hsp70 genes, show no enrichment in the Zw5 immunoprecipitate. Although Zw5 shows no association with sequences around the two hsp70 genes, we were surprised to discover that sequences from scs‘ are enriched in the Zw5 immunoprecipitate. As illustrated in the diagram, we tested a set of four overlapping probes (Fig. 2, probes k–n) from the scs‘ element. All four of these probes are enriched in the Zw5 immunoprecipitate compared with the control. The peak association is seen with probe m (Fig. 2), which is enriched in the Zw5 immunoprecipitate more than fivefold over the control immunoprecipitate; whereas the three other probes are enriched threefold or more. Similar results were obtained in another experiment in which we immunoprecipitated cross-linked chromatin from non-heat-shocked and heat-shocked tissue culture cells. In both the control and heat-shocked samples, the probe d (Fig. 2) spanning the Zw5 binding site in scs was enriched about ninefold, whereas probe l from scs‘ was enriched ∼3.5-fold.
Why are sequences from scs‘ detected in the Zw5 immunoprecipitate? If we assume that there is a single Zw5 protein bound to the site in scs, then ∼0.3 molecules would be associated with scs‘. However, Zw5 does not localize to scs‘ in polytene chromosomes, nor does purified Zw5 bind to scs‘ fragments in vitro. One hypothesis that could potentially account for these observations is that a long-distance looping or pairing interaction between scs and scs‘ brings Zw5 into sufficiently close proximity to scs‘ that it can be cross-linked to proteins associated with this element. Because at least two cross-linking events would be required to link Zw5 to sequences from scs‘, this could explain why the yield is rather low. Interestingly, Zhao et al. (1995) reported quite similar cross-association results for BEAF. BEAF is only seen at scs‘ in polytene chromosomes, and does not bind to sequences from scs in vitro. However, in ChIP experiments, Zhao et al. (1995) found that not only was scs‘ enriched in the BEAF antibody immunoprecipitate as expected, but also there was a three- to fivefold enrichment of scs. They also suggested that pairing between scs and scs‘ might explain this cross-association.
Zw5 and BEAF interact in vitro
If scs and scs‘ are in contact with each other in vivo, this association is expected to be mediated by protein:protein interactions. Although it seems likely that many different proteins actually bind to these two elements, Zw5 and BEAF are the only proteins known both to be associated with them in vivo and to have some relevance to their boundary activity. For this reason, we decided to ask whether Zw5 and BEAF interact with each other.
There are two different BEAF isoforms, BEAF-32A and BEAF-32B (Hart et al. 1997). Both proteins are ∼300 amino acids in length and share the same C-terminal 200 amino acids, but have different N termini. In their common C terminus, the proteins have a coiled-coil domain containing an atypical leucine zipper, and a short region found in the fly proteins Stonewall and Ravus, but there are no other known motifs elsewhere in either protein. The C terminus has been shown to mediate homotypic protein:protein interactions, whereas the two different N termini have the DNA binding activity. In vivo, BEAF is thought to exist as a trimer, with the most abundant having the isoform composition (BEAF-32A) (BEAF-32B)2. Although the two isoforms colocalize at many sites in polytene chromosomes, a subset of the sites are enriched for either the A or B isoforms, and this presumably reflects differences in the DNA binding specificity of the two proteins.
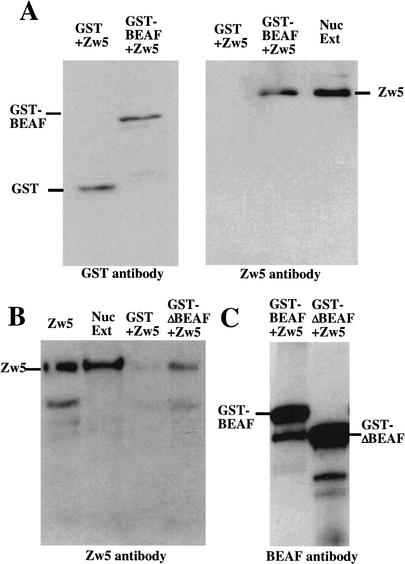
As a first test for interactions between Zw5 and BEAF we used far Western analysis. We found that biotinylated-Zw5 specifically labels recombinant BEAF-32A and BEAF-32B proteins in Western blots of bacterial extracts (data not shown). Although this finding is consistent with the idea that Zw5 and BEAF may interact, the BEAF protein in the far Western experiments has been denatured, and this may expose polypeptides that are not normally accessible. For this reason, we used a GST pull-down assay to confirm that recombinant Zw5 and BEAF can interact with each other in vitro. Equal amounts of purified GST or GST fused to full-length BEAF-32A (GST-BEAF-32A) were mixed with purified recombinant Zw5. As expected, both GST and GST-BEAF-32A are specifically bound by the glutathione beads (Fig. 3A, left). As shown in the right panel of Figure 3A, Zw5 is retained on the glutathione beads when incubated with the GST-BEAF-32A fusion protein. Because Zw5 can not be detected in the GST alone sample, the formation of the GST-BEAF-32A:Zw5 complex depends on BEAF-32A sequences.
Figure 3.
Zw5 and BEAF interact with each other in vitro. (A) Recombinant Zw5 associates with GST-BEAF-32A but not GST. Zw5 was mixed with either GST-BEAF 32A or GST alone in a 1:10 ratio. The proteins were allowed to bind for 30 min at room temperature before subsequently being added to 400 μL of PBS containing ∼25 μL of glutathione beads. After rocking overnight at 4°C, the beads were washed, and the proteins were eluted from the beads by boiling in 2× sample loading buffer. The proteins were separated by SDS-PAGE and transferred to PVDF membranes. The membrane on the left was probed with anti-GST antibody. GST+Zw5, Zw5 mixed with GST alone; GST-BEAF+Zw5, Zw5 mixed with GST-BEAF-32A. The membrane on the right was probed with Zw5 antibody. GST+Zw5, GST mixed with Zw5. Zw5 does not associate with GST. GST-BEAF+Zw5, Zw5 mixed with GST-BEAF-32A. Zw5 forms a stable complex with GST-BEAF 32A. Nuc Ext, nuclear extract used as a positive control for Zw5. (B) Recombinant Zw5 associates specifically with GST-ΔN BEAF-32A in a GST pull-down assay. Recombinant Zw5 was mixed with either GST-ΔN BEAF or GST alone in a 1:10 ratio. The proteins were allowed to interact for 30 min at room temperature before being added to 400 μL of PBS containing ∼25 μL of glutathione beads. After rocking overnight at 4°C, the beads were washed, and the proteins were eluted from the beads by boiling in 2× sample loading buffer. The proteins were separated by SDS-PAGE and transferred to a PVDF membrane. The blot was probed with anti-Zw5 antibody. Zw5, recombinant Zw5 protein; Nuc Ext, nuclear extract; GST+Zw5, Zw5 mixed with GST alone; GST-ΔN BEAF+Zw5, Zw5 mixed with GST-ΔN BEAF-32A. (C) Zw5 immunoprecipitation. GST-BEAF-32A or GST-ΔN BEAF-32A were mixed with Zw5. After incubating at room temperature, the mixture was diluted in PBS containing 25 μL of Zw5 antibody. The samples were rocked overnight at 4°C, the beads were washed, and the proteins were eluted from the beads by boiling in 2× sample loading buffer. The proteins were separated by SDS-PAGE and transferred to a PVDF membrane. The blot was probed with BEAF antibody.
As biotinylated Zw5 labeled both BEAF-32A and BEAF-32B in far Western experiments, we reasoned that the Zw5:BEAF interaction is mediated by BEAF sequences in the common C-terminal domain. To test this prediction, we generated a deletion mutant of the GST-BEAF-32A fusion protein, GST-ΔNBEAF-32A, that lacks the unique 32A N-terminal DNA binding domain. As shown in Figure 3B, the GST-ΔNBEAF-32A fusion protein, like the full-length fusion protein, interacts with Zw5. This finding indicates that the common C-terminal domain of the BEAF protein is sufficient for association with Zw5.
We also tested whether recombinant Zw5 and BEAF associate in an immunoprecipitable complex. Purified Zw5 protein was incubated with either GST-BEAF32A or GST-ΔBEAF32A and then immunoprecipitated with a Zw5 monoclonal antibody. The immunoprecipitated proteins were then analyzed by Western blotting by using either Zw5 antibody (data not shown) or antibody against BEAF (Fig. 3C). As expected, Zw5 protein was present in the Zw5 immunoprecipitate (data not shown). We also detected the GST-BEAF32A and GST-ΔBEAF32A fusion proteins (Fig. 3C). This result provides further evidence that Zw5 and BEAF can interact with each other in solution. In the reciprocal experiment, we used a BEAF monoclonal antibody instead of Zw5 for the immunoprecipitation. Unexpectedly, although we were able to immunoprecipitate GST-BEAF-32A with the BEAF monoclonal antibody, Zw5 protein was not detected (data not shown). As GST-BEAF-32A:Zw5 complexes could be recovered from the same incubations by using glutathione beads, we presume that our monoclonal antibody recognizes an epitope in the BEAF protein that either overlaps the Zw5 binding site or occludes it.
Zw5 and BEAF interact in vivo
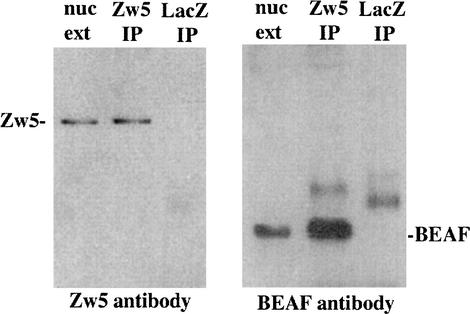
The results described in the previous sections demonstrate that recombinant Zw5 and BEAF proteins can interact with each other in vitro. We wished to establish that these two proteins also interact with each other in vivo. For this purpose, we immunoprecipitated nuclear extracts prepared from Drosophila embryos with the Zw5 monoclonal antibody or as a control with an antibody directed against bacterial β-galactosidase. The proteins recovered in the immunoprecipitate were then analyzed by gel electrophoresis and blotting.
As was observed with a polyclonal Zw5 antibody (Gaszner et al. 1999), our monoclonal recognizes a single protein species in embryonic nuclear extracts with an apparent molecular weight of ∼90 kD (Fig. 4). This same protein is recovered when the nuclear extract is immunoprecipitated with the Zw5 monoclonal antibody. The Zw5 protein is not, however, observed when the nuclear extract is immunoprecipitated with β-galactosidase monoclonal antibody. We next probed the immunoprecipitated samples with the BEAF monoclonal antibody. As shown in the second panel of Figure 4, our BEAF monoclonal antibody recognizes a band of the expected size (30–35 kD) in nuclear extracts. Moreover, the mobility of this protein species corresponds closely to that of recombinant BEAF-32A or BEAF-32B. The same protein species is observed in the Zw5 immunoprecipitate of nuclear extracts, but is not evident in the β-galactosidase control. These results indicate Zw5 and BEAF are in an immunoprecipitable complex in Drosophila nuclear extracts. The reciprocal experiment—immunoprecipitating BEAF from nuclear extracts and then probing for Zw5—was also tried. However, as would be expected from our results with recombinant BEAF and Zw5, we did not detect Zw5 in the immunoprecipitates though BEAF was present.
Figure 4.
Zw5 and BEAF are in an immunoprecipitable complex in Drosophila embryos. Anti-Zw5 antibody cross-linked to protein A/G beads was mixed with embryonic nuclear extract. After rocking overnight at 4°C, the beads were washed, and the proteins were eluted from the beads by boiling in 2× sample loading buffer. The proteins were separated by SDS-PAGE and transferred to PVDF membranes. The blot on the left was probed with Zw5 antibody. In the lane labeled nuc ext, the starting nuclear extract was loaded and used as the positive control for Zw5. The lane labeled Zw5 IP contains the proteins immunoprecipitated from nuclear extracts with Zw5 antibody. As evident from a comparison with the “nuc ext” lane, Zw5 protein is present in the immunoprecipitate. The lane labeled LacZ IP contains the proteins immunoprecipitated with the control β-galactosidase antibody. Note that Zw5 is not detected in the β-galactosdiase immunoprecipitate. There is, however, a weakly labeled band of ∼40 kD. The blot on the right was probed with BEAF antibody. In the lane labeled nuc ext, nuclear extract was loaded and used as the positive control for BEAF. In the Zw5 IP lane, proteins isolated from the Zw5 immunoprecipitate were loaded. As can be seen from comparison with the “nuc ext” lane, BEAF is present in the Zw5 immunoprecipitate. In the lane labeled LacZ IP, proteins isolated from the β-galactosidase immunoprecipitate were loaded. BEAF is not present; however, there is a larger protein species of ∼40 kD. As noted above, a protein species of this size was also detected when the β-galactosidase immunoprecipitate was probed with Zw5 antibody. Consequently, we suspect that it corresponds to protein that associates with the β-galactosidase and is recognized by the secondary antibody.
Zw5 interacts genetically with BEAF
To provide evidence that the protein:protein interactions between Zw5 and BEAF seen in vitro and in vivo may be functionally significant we decided to test for genetic interactions. Although mutations in zw5 have been isolated and are recessive lethal, there are no known mutations in beaf. Recently, however, Yamaguchi et al. (2001) found that overexpression of BEAF-32A in the developing eye disc by using a UAS–BEAF-32A transgene and a glass GAL4 driver causes a rough eye phenotype. This rough eye phenotype appears to be owing to the induction of apoptosis in cells just posterior to the progressing furrow. Yamaguchi et al. (2001) found that the effects of BEAF-32A overexpression on eye development can be dominantly enhanced or suppressed by mutations in other genes, and one of the enhancers they identified is su(hw), which encodes the boundary protein for the gypsy element (Gerasimova and Corces 2001). Because Zw5 and BEAF appear to physically interact with each other, we thought that changing the dose of zw5 might alter the rough phenotype induced by excess BEAF-32A protein.
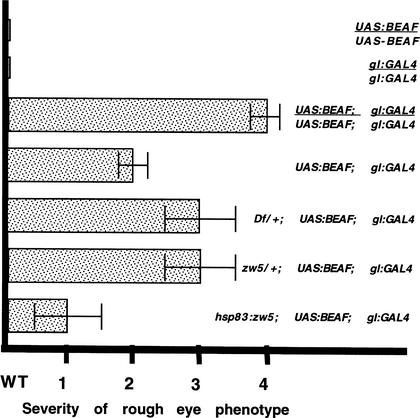
Neither the UAS–BEAF-32A transgene nor the glass GAL4 driver alone has any effect on eye development, even when the transgenes are present in two copies; however, eye development in flies carrying a single copy of each transgene is clearly abnormal and all of the BEAF/glass trans-heterozygous flies have a very similar rough eye phenotype (Fig. 5). This disruption in eye development depends on the level of excess BEAF protein, and the eye phenotype is considerably more severe in flies carrying two copies of both the BEAF expression construct and the glass driver. To test for genetic interactions, we compared the eye phenotype of female flies carrying either one or two copies of the BEAF expression construct and the glass driver with female flies carrying a single copy of each of these transgenes, in which the dose of the X-linked zw5 gene was reduced in half. In one case, we tested the effects of a small deletion that uncovers the zw5 gene, Df(1)935, and in the other case, we tested a strong loss-of-function mutation zw562j1. As can be seen in Figure 5 for both the deficiency and zw5 point mutation, a reduction in the dose of the zw5 gene enhances the rough eye phenotype. The severity of the disruption in eye development observed for these zw5/+ females is intermediate between that observed with one and two copies of the BEAF and glass GAL4 transgenes. We also tested a second, weaker zw5 mutant allele, zw590. zw590 also enhanced the rough eye phenotype induced by BEAF over expression, although not quite as strongly as the zw562j1 (data not shown).
Figure 5.
Genetic interactions between zw5 and beaf. Yamaguchi et al. (2001) showed that inducing BEAF-32A expression from a UAS–BEAF transgene using a gl:GAL4 driver has deleterious effects on eye development. We scored female flies from each cross on a scale of 0–4 based on the severity of the phenotypic effects on eye development. Wild-type females had a score of 0. The most severe rough eye phenotype was observed for females carrying two copies of both the UAS:BEAF expression construct and the gl:GAL4 driver, GMR-GAL4, and this phenotype was assigned a score of 4. An intermediate rough eye phenotype was observed for females carrying a single copy each of the UAS:BEAF and gl:GAL4 transgenes, and this phenotype was assigned a score of 2. The eye phenotype observed for each experimental cross was then compared with these three controls and assigned a score based on this comparison. Each of the experimental crosses was also scored independently by another investigator who had no knowledge of the genotypes being examined. As reported by Yamaguchi et al. (2001), animals homozygous for either the UAS:BEAF expression construct or the gl:GAL4 driver (lines 1,2) had a wild-type phenotype (#0), while animals carrying two copies of the expression construct and driver (line 3) had a severe (#4) eye phenotype. This rough eye phenotype was comparatively homogenous within the population. Females hemizygous for the expression construct and driver (line 4) were generated by mating females carrying two copies of the expression construct and the driver to w‘ males. The hemizygous females had an intermediate rough eye phenotype (#2). To obtain the female progeny heterozygous for Df(1)936 that also carry single copy of both transgenes (line 5), we crossed Df(1)935/Bal females to males hemizygous for the X-linked gl:GAL4 driver, GMR-GAL4, and homozygous for the UAS:BEAF-32A transgene. Nonbalancer females were scored. In our hands, the Df(l)935 deletion, which uncovers zw5, enhances the rough eye phenotype associated with overexpressed BEAF 32A, contrary to the findings published by Yamaguchi et al. (2001). The reason for this discrepancy is uncertain. To obtain the female progeny heterozygous for zw562j1 that also carry a single copy of both transgenes (line 6) we crossed zw562j/FM7 females to males that were hemizygous for the X-linked gl:GAL4 driver and homozygous for the UAS–BEAF-32A transgene. Female progeny lacking the FM7 balancer were examined. This strong zw5 loss-of-function allele enhances the rough eye phenotype compared with that observed for otherwise wild-type flies hemizygous for the two transgenes. The disruptions in eye development are not, however, as severe as that observed for females carrying two copies of both the expression construct and the driver. Similar results were obtained for a second independent zw5 allele, zw590. Finally, to increase the level of zw5 protein (line 7), females carrying an hsp83:zw5 cDNA transgene (line 17.1.1) over the Cyo balancer were crossed to males hemizygous for the gl:GAL4 driver, and homozygous for the UAS:BEAF-32A transgene. Females lacking the Cyo balancer were examined. They had a weaker rough eye phenotype than the single copy control. All crosses were done at 29°C.
If a twofold reduction in the amount of Zw5 protein is sufficient to enhance the deleterious effects of BEAF overexpression, one might expect that increasing the level of Zw5 protein above that in wild-type females would have the opposite effect. This is the case. Introduction of an hsp83:zw5 cDNA transgene into the BEAF/glass GAL4 trans-heterozygous female flies reduces the severity of the rough eye phenotype (Fig. 5).
Linkage of scs and scs‘ in vivo
The physical and genetic interactions between Zw5 and BEAF documented above would be compatible with a model in which Zw5 bound to scs interacts with BEAF bound to scs‘. This protein:protein association would help bring the scs and scs‘ insulators into close proximity to each other, forming a looped higher order chromatin domain. In this case the presence of scs‘ sequences in the Zw5 ChIP would be explained by the cross-linking of Zw5 protein bound to scs to the BEAF protein associated with scs‘. On the other hand, because Zw5 and BEAF can interact with each other in the absence of DNA, there is an alternative and equally plausible explanation for the unexpected cross-association observed in the ChIP experiments. In this scenario, Zw5 would also be cross-linked indirectly to scs‘ sequences by virtue of its interaction with scs‘-bound BEAF; however, this Zw5 protein would not be bound to scs but, instead, would be “free” protein not associated with any chromosomal DNA sequence.
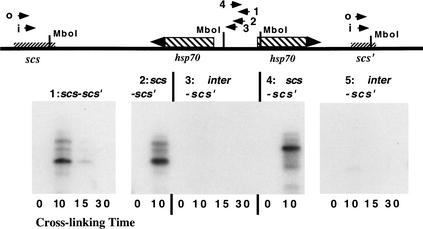
To distinguish between these two possibilities we used the chromosome conformation capture procedure recently described by Dekker et al. (2002). Drosophila embryonic nuclei were cross-linked with formaldehyde for increasing lengths of time and, after quenching and washing, restricted with MboI. The restriction enzyme was heat inactivated, and the nuclei were diluted into ligation buffer and ligated overnight. The samples were then deproteinized and PCR-amplified by using primers from scs‘ combined with primers from either scs, or the region around the 5′ ends of the two hsp70 genes and the intergenic spacer. After amplification, the reaction products were analyzed by gel electrophoresis and visualized directly either by ethidium staining (data not shown) or by blotting to filters and probing with a 32P-labeled fragment complementary to scs‘ (Fig. 6).
Figure 6.
scs and scs‘ are paired in Drosophila embryos. The chromosome conformation capture procedure of Dekker et al. (2002) was used to determine whether scs and scs‘ are in close proximity to each other in vivo. Formaldehyde cross-linked Drosophila embryonic nuclei were restricted with MboI and then ligated. After ligation, the cross-linking was reversed and the DNA was PCR-amplified with primer combinations derived from scs‘ and either scs or the 5′ ends of the two 87A7 hsp70 genes. The PCR products were then detected with either ethidium bromide staining or by hybridization with a probe derived from scs‘. The primer combinations for each experiment were as follows. (1) scs-scs‘: samples in each lane were cross-linked for the times indicated and then PCR-amplified for 40 cycles using scs-i and scs‘-i as primers. (2) scs-scs‘: samples were cross-linked for the times indicated and then PCR-amplified in two steps. In the first step, the samples were PCR-amplified for 20 cycles with scs-o and scs‘-o as primers. In the second step, an aliquot of from the first step was PCR-amplified for 20 cycles with scs-i and scs‘-i as primers. (3) scs‘-inter: samples were cross-linked for the times indicated and then PCR-amplified in two steps. In the first step, the samples were amplified for 20 cycles with inter-1 and scs‘-o as primers. In the second step, an aliquot from the first step was PCR-amplified for 20 cycles with inter-2 and scs‘-i as primers. (4) scs-scs‘: samples were cross-linked for the times indicated and PCR-amplified in two steps. In the first step, the samples were amplified for 20 cycles with scs-o and scs‘-o as primers. In the second step, an aliquot from the first step was PCR-amplified for 20 cycles with scs-o and scs‘-i as primers. (5) scs‘-inter: samples were cross-linked for the times indicated and PCR-amplified in two steps. In the first step, the samples were amplified for 20 cycles with inter-3 and scs‘-o as primers. In the second step, an aliquot from the first step was PCR-amplified for 20 cycles with inter-3 and scs‘-i as primers. As a positive control, we also ligated a mixture of Sau3a-digested plasmids containing scs, scs‘, and the 87A7 intergenic spacer. We then PCR-amplified using scs-scs‘ or scs‘-inter primer pairs and detected the mixed ligation products by ethidium bromide staining or by probing filters with the scs‘. The expected hybrid scs‘-scs and scs‘-inter amplification products were observed.
The scs primer, scs-i, used in the experiment shown in the first panel of Figure 6 is ∼550 bp from the nearest downstream MboI restriction site in scs, whereas the scs‘ primer, scs‘-i, is ∼200 bp from the nearest MboI site in scs‘. If these two MboI restriction fragments are ligated to each other in the correct orientation, we expect to generate a PCR product of ∼750 bp. As can be seen in Figure 6, #1, a band of the predicted size is strongly labeled in the 10-min cross-linked sample. It is also present in the 15-min sample, but in much lower yield. In addition to the very prominent 750-bp band, several less strongly labeled larger fragments are observed in the 10-min cross-linked sample (and, in longer exposures, in the 15-min sample). Similar experiments with PCR primers and probes from regions elsewhere in the fly genome indicate these larger molecular fragments are most likely derived from the ligation of incompletely digested restriction fragments that extend to MboI sites beyond the first downstream scs or scs‘ MboI restriction site. Although PCR products of the expected size are present in the 10- and 15-min cross-linked samples, they are not observed in the uncross-linked sample or in the sample cross-linked for 30 min. Thus, cross-linking is required to generate the hybrid scs-scs‘ ligation product. Moreover, it would appear that the ligation of the scs and scs‘ fragments can be inhibited by excessive cross-linking.
Even though scs and scs‘ are separated by ∼15 kb, it seemed possible that these two elements might be brought into sufficiently close proximity so that they could be cross-linked to each other at a relatively high frequency simply because the intervening 87A7 DNA is packaged into chromatin. If this were the case, cross-linking of scs or scs‘ to sequences located within 87A7 should occur at the same or higher frequency. To investigate this possibility, we decided to test the sequences around the 5′ ends of the two hsp70. This region is located midway between scs and scs‘, and like the other sequences from within the 87A7 locus, probes from the intergenic spacer region showed essentially the same background level of enrichment in the Zw5 ChIPs (Fig. 2). For the first experiment, the primer combination was inter-4 from the intergenic spacer and scs‘-o (Fig. 6 and legend). With these two primers we expected to observe a fragment of 1,650 bp if the intergenic MboI fragment ligated to the MboI fragment from scs‘. However, unlike scs, no amplification products were detected with the inter-4:scs‘-o primer combination (data not shown, see also below).
To confirm that sequences from scs are joined to scs‘ in formaldehyde cross-linked Drosophila nuclei we used a two-step PCR amplification procedure. In the first step, we PCR-amplified the ligation mixture with an “outside” primer combination of scs-o and scs‘-o. As indicated in Figure 6, these primers are located upstream, respectively of the scs-i and scs‘-i and are expected to give a hybrid scs-scs‘ amplification product of 1,000 bp. In the second step, an aliquot of the reaction mix was PCR-amplified with the “inside” primer combination of scs-i and scs‘-i. The only PCR products from the first reaction that should be amplified in the second are appropriately oriented hybrid scs-scs‘ DNA fragments that contain sequences complementary to both the scs and scs‘ internal primers. As shown in Figure 6, #2, the expected 750-bp hybrid fragment (plus the same larger fragments seen in reaction #1) is observed in the sample cross-linked for 10 min, but can not be detected in the non-cross-linked control. Further evidence that scs and scs‘ are joined comes from the two-step PCR reaction #4. In this case, the ligation products were first PCR-amplified by using the same scs-o:scs‘-o primer combination that was used for the initial amplification in reaction #2. However, in the second step, we combined scs-o with scs‘-i instead of scs-i. Because scs-o is ∼750 bp from the downstream scs MboI restriction site, the hybrid scs-scs‘ amplification product generated from the ligated scs:scs‘ restriction fragment should be ∼200 bp larger than the scs-i:scs‘-i amplification product, or ∼950 bp. As can be seen in Figure 6, #4, a strongly labeled band of this size is observed in the cross-linked sample but not in the non-cross-linked control.
Although we did not observe any PCR amplification products corresponding to hybrids between the hsp70 intergenic spacer and scs‘ using the inter-4:scs‘-o primer combination, it seemed possible that the ability to ligate cross-linked fragments might depend on their relative orientation. For this reason, we retested for joined MboI scs‘:hsp70 intergenic spacer fragments using primers from the intergenic spacer that were in the opposite orientation. In reaction #3 (Fig. 6), we first PCR-amplified with the outside inter-1 and scs‘-o primer combination (which should give a ∼700-bp fragment) and then, in the second step, amplified with the inside primers combination inter-2 and scs-i. We expected to observe a ∼500 bp fragment; however, this fragment was not detected in any of the cross-linked samples (Fig. 6, #3). In reaction #5, we first PCR-amplified with the inter-3:scs‘-o combination and then, in the second step, amplified with the inter-3:scs-i combination. If the intergenic spacer region is joined to scs‘, we should observe an inter-2:scs-i hybrid DNA fragment of ∼300 bp. Although a fragment of approximately this size can be detected in the 10-min cross-linked sample, it is present in only very low yield compared with the PCR products derived from scs-scs‘ hybrid DNA fragments.
These findings indicate that scs and scs‘ are in close proximity in vivo. In this case, the unexpected cross-association seen in the ChIP experiments could be explained by the model in which Zw5 bound to scs can interact with and be cross-linked to BEAF protein bound to scs‘.
Discussion
Boundary elements or insulators subdivide eukaryotic chromosomes into functionally and structurally autonomous domains. Two models have been proposed to account for these activities. In one model, boundary elements act simply as barriers, blocking the spread of “active” or “silenced” chromatin from one domain to the next. Because active and silenced chromatin differ so extensively in the nature and extent of histone protein modification, in nonhistone protein composition and in their degree of compaction, the barrier activity of the boundary or insulator would in itself be sufficient to establish structurally distinct domains. In the other model, boundary elements serve to couple the functional and structural subdivision of the chromosome. They determine the limits of higher-order “looped” chromatin domains either by interacting with each other or with some other nuclear structure. In the most extreme version of this model, the insulator activity of the boundary would be absolutely dependent on this interaction, whereas in less extreme versions, the ability to establish independent units of gene activity might only be partially dependent on this interaction.
Evidence supporting one or the other model can be found in the literature. For example, in their studies on the silenced mating type domain of S. pombe, Noma et al. (2001) found that deletion of one of the boundary elements leads to the unidirectional spread of silenced chromatin into the normally active region immediately adjacent to the deleted boundary. This observations is consistent with the expectations of the barrier model. In contrast, a bidirectional spread of silenced chromatin would be expected if interactions between the two mating type boundary elements are essential for establishing the silenced mating domain. Conversely, recent work on the su(Hw) insulator indicates that boundaries might interact with each other. The first hint of interactions between su(Hw) insulators came from studies on pairing sensitive silencing of mini-white by the Ubx Polycomb Response Element (PRE). Sigrist and Pirrotta (1997) found that the addition of a su(Hw) insulator to the PRE-mini-white construct enabled transgenes inserted at very distant chromosomal sites and even on different chromosomes to pair with each other and repress mini-white expression. Consistent with the idea that pairing between su(Hw) insulators is responsible for this long distance silencing, cytological studies by Gerasimova et al. (2000) revealed that su(Hw) insulators coalesce into a small number of insulator “bodies” located at the periphery of the nucleus, and can drag nearby DNA sequences to these bodies. That pairing may, in fact, be relevant to boundary activity is suggested by the unexpected finding that enhancer blocking activity is inhibited when two su(Hw) insulators, instead of one, are placed between an enhancer and a promoter (Cai and Shen 2001; Muravyova et al. 2001). It is thought that the two su(Hw) insulators pair with each other, and that this pairing loops out a mini-domain, allowing the upstream enhancer to activate the downstream promoter.
The results presented here are consistent with idea that the scs and scs‘ boundary elements interact with each other in vivo, in this case forming a ∼15-kb looped higher order domain that includes the two 87A7 hsp70 genes. That scs and scs‘ might contact each other was first suggested by ChIP experiments using Zw5 and BEAF antibodies. Although the Zw5 protein does not bind to sequences from scs‘, we found that scs‘ is enriched in Zw5 immunoprecipitates. Conversely, though BEAF does not bind to sequences from scs, Zhao et al. (1995) reported that scs is slightly enriched in BEAF immunoprecipitates. For Zw5, the degree of enrichment is less than that of a single bona fide target sequence (Zw5:scs); nevertheless, it is clearly above background. Moreover, this cross-association can not be explained simply by the close proximity of the two boundary elements as sequences within 87A7 that are located much nearer to scs than scs‘ show no evidence of enrichment in Zw5 immunoprecipitates.
If scs and scs‘ pair with each other, this coupling is likely to be mediated by proteins associated with each boundary element. Consistent with this expectation, we found that Zw5, which binds to scs in vivo, can interact with BEAF, which binds to scs‘. Experiments with recombinant proteins indicate that this interaction is direct and involves the C-terminal domain of the BEAF protein, which is shared by the BEAF-32A and BEAF-32B isoforms. Two lines of evidence suggest that the Zw5:BEAF interactions seen in vitro with recombinant proteins are recapitulated in vivo. First, BEAF protein is in an immunoprecipitable complex with Zw5 in nuclear extracts from Drosophila embryos. Second, zw5 and beaf interact genetically. Reducing zw5 activity enhances the deleterious effects of BEAF-32A overexpression in the eye, whereas increasing zw5 activity suppresses these effects. This genetic interaction would be compatible with some version of a simple titration mechanism in which Zw5 mitigates the disruptions in development arising from BEAF overexpression by associating with the excess BEAF protein.
The Zw5:BEAF protein:protein interactions documented here would clearly account for the cross-association of scs‘ and scs sequences seen in the respective ChIPs. Thus, the association of Zw5 with BEAF bound to scs‘ would explain why scs‘ sequences are detected in the Zw5 immunoprecipitates. Conversely, the interaction of BEAF with Zw5 bound to scs would explain why scs sequences might be enriched in the BEAF immunoprecipitate. However, what is not clear from these experiments is whether Zw5 bound to scs is actually complexed with BEAF bound to scs‘, thereby bringing these two boundary elements together. It is equally possible that the cross-association in both cases is due solely to “free” protein. Although we can not rule out this later explanation, chromosome conformation capture experiments provide independent evidence that scs pairs with scs‘ in vivo. Using this procedure, we can readily detect cross-linking between sequences in scs and scs‘. As was the case in the ChIP experiments, the linking of scs to scs‘ does not seem to be owing simply to the close proximity of these two boundary elements in the chromosome. Thus, we detect only little cross-linking of scs‘ to the 5′ ends of the two 87A7 hsp70 genes and the intergenic spacer, even though these DNA sequences are located much closer to scs‘ than to scs.
While our results are compatible with a model in which scs pairs with scs‘, establishing a looped higher order chromatin domain spanning the 87A7 heat-shock locus, several important questions remain open. First, is the pairing between scs and scs‘ mediated solely by Zw5:BEAF complexes or are other interactions involved? Though a definitive answer to this question must await the identification of the other proteins associated with these two elements, it is interesting to note that Cuvier et al. (2002) have recently reported that BEAF interacts with the nonhistone chromosomal protein D1. Because we have found that D1 binds to the AT-rich sequences in scs in vitro (A. Udvardy and P. Schedl, unpubl.), it is conceivable that D1:BEAF interactions might also facilitate the pairing of scs and scs‘ . Second, is pairing relevant to the genetically defined insulator activities of these two boundaries and of other boundaries? Answering this question will require assays that specifically measure the effects of pairing on insulator activity; however, as was observed for the su(Hw) insulator and the Ubx PRE, we found that the scs element strongly enhanced long distance silencing of mini-white by the Mcp PRE (Muller et al. 1999). Finally, on a more general note, if pairing between boundary elements serves to subdivide the chromosome into a series of higher order domains, what mechanisms generate the appropriate specificity in boundary:boundary interactions? In the absence of specificity mechanisms that define some type of interaction hierarchy, the organization of higher domains would be completely different from one copy of the chromosome to the next.
Materials and methods
Far Western
Recombinant Zw5 was expressed and purified (from pQE-Zw5) as described in Gaszner et al. (1999). This Zw5 protein was then biotinylated by using Pierce’s EZ-Link Sulfo-NHS-LC-Biotin. Both BEAF 32A and BEAF 32B were expressed from pET3b(NSEB) bacteria (Zhao et al. 1995).
GST-BEAF fusion/purification
PCR was used to engineer a SalI restriction site just upstream of the transcription start site of the BEAF 32A cDNA, and a NotI site after the transcriptional stop. The cDNA was inserted into the SalI/NotI sites in pGEX-4T-3, creating a N-terminal GST-BEAF 32A fusion, GST-BEAF 32A. The plasmid was transformed into Bl-21 Escherichia coli. Expression of the fusion protein was performed as detailed in Amersham Pharmacia Biotech's GST Purification Modules manual with the following exceptions: IPTG was added at a final concentration of 0.1 mM. Induction was carried out at room temperature for 3 h. Induction was checked by Coomassie staining after the cells were lysed in PBS by cell disruption. Purification was performed by column method as detailed in Amersham Pharmacia Biotech's GST Purification Modules manual. The fusion protein was eluted with 25 mM glutathione plus 1.0% Triton X-100. For the GST-ΔN BEAF construct, primers were used to generate a PCR fragment with a EcoRI site on the 5′ side of the common domain between BEAF 32A and BEAF 32B. There was a NotI site on the 3′ side in the same position as it was in GST-BEAF 32A. This fragment of the cDNA was inserted into the EcoRI/NotI sites in pGEX-4T-3, creating a N-terminal GST fusion to the common domain of BEAF 32A and BEAF 32B. Expression and purification was the same as for GST-BEAF 32A.
GST-BEAF pull-down assays
The GST-BEAF 32A fusion protein was mixed in a 10:1 ratio with recombinant pQE-Zw5 and 25 μg BSA. Recombinant GST (equal in amount to the GST-BEAF 32A fusion protein) alone was also mixed in a 10:1 ratio with recombinant zw5 and 25 μg BSA in a separate reaction. The binding was allowed to proceed at room temperature for 30 min in a 96-well microtiter plate. The two binding reactions were then added to equal volumes of glutathione beads (∼25 μL) in 400 μL of PBS. The bead mixtures were rocked overnight at 4°C. The beads were washed six times with PBS plus 1.0% Triton X-100. The protein was eluted from the beads by boiling just prior to being loaded onto an SDS gel. The same protocol was also used for the pull-down of pQE-Zw5 by GST-ΔN BEAF and for the pull-down of pQE9-Zw5Zn by both GST-BEAF 32A and GST-ΔN BEAF.
Preparation of embryonic nuclei and nuclear extracts
Embryos 0–12 h old were collected, dechorionated for 90 sec with 0.5× bleach, and then rinsed with 500 mL 0.7% NaCl/0.04% Triton X-100, followed by 500 mL 0.7% NaCl. After drying, the embryos were suspended in 5 mL/g of buffer I (15 mM Hepes-KOH at pH 7.6, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 0.35 M sucrose, 1 mM DTT, 1 mM Na2S2O5, 0.5–1 μg/mL antipain, leupeptin), homogenized. After filtering, the nuclei were harvested by centrifuging at 2,000 xg for 10 min, resuspended in 5 mL/g buffer I, and the suspension was overlaid on an equal volume of buffer II (buffer I + 0.8 M sucrose). The suspension was centrifuged at 2,000 xg for 10 min, and the nuclear pellet was resuspended in low-salt buffer (100 mM KCl, 20 mM Hepes, 1 mM EDTA, 5 mM MgCl2, 20% glycerol, 1 mM DTT). High-salt buffer (same as low salt but with 2 M KCl) was added to make the final concentration of salt 0.8 M. The mixture was placed on a shaker for at least 30 min at 4°C and then spun for 1 h at 40,000 xg. After the lipid layer was removed, the supernatant was aliquoted into tubes and frozen in liquid nitrogen.
Coimmunoprecipitations
Zw5-5F monoclonal antibody and a commercially available β-gal monoclonal antibody were cross-linked to a mixture of protein A/G agarose beads. Approximately 25 μL of the cross-linked beads were added to 100 μL of 0–12-h embryonic nuclear extract in 400 μL of Co-IP buffer [20 mM Hepes at pH 7.5, 150 mM NaCl, 2.5 mM MgCl2 × 6H2O, 250 mM sucrose, 0.05% (w/v) Tergitol NP-40, 0.5% (v/v) Triton X-100]. The beads containing nuclear extracts were rocked overnight at 4°C. The beads were washed six times with Co-IP buffer lacking magnesium; 50 μL of 2× sample loading buffer was added to the beads, and the mixture was boiled. The extracts were then separated by SDS-PAGE. The proteins were transferred to PVDF membranes, which were probed with BEAF 70 antibody. Anti-mouse HRP was used to detect the BEAF associated with the Zw5 that had been immunoprecipitated.
Antibody
The anti-Zw5 polyclonal antibody has previously been described (Gaszner et al. 1999). Both anti-Zw5 and anti-BEAF monoclonal antibodies were generated by the Core Facilities of Princeton University. The BEAF antibody recognizes both BEAF 32A and BEAF 32B recombinant proteins on a Western (data not shown).
ChIP
Formaldehyde cross-linking and ChIP were performed as described by Orlando and Paro (1993). Schneider 2 tissue culture cells were grown at 25°C under standard conditions. Heat-shocked chromatin was prepared from cells that were rapidly warmed to and incubated at 37°C prior to formaldehyde treatment. Twenty-five microliters of affinity-purified rabbit anti-Zw5 antibody (Gaszner et al. 1999) or preimmune rabbit serum was used for immunoprecipitation. DNA recovered from the immune pellet was amplified by LM-PCR (Zhao et al. 1995), applied to nylon membranes, and hybridized with radioactively labeled probes. DNA for probes a–e was purified as subfragments of a larger genomic clone; DNA for probes e–n was isolated from clones of corresponding PCR fragments. Individual probes are described below. Slot blot hybridization signals were measured by using PhosphorImager (Molecular Dynamics). Following is a list of probes, with length and GenBank accession no.:position: probe a, 292 bp, X63731:511–803; probe b, 185 bp, X63731:803–988); probe c, 393 bp, X63731:988–1381; probe d, 123 bp, X63731:1381–1504; probe e, 291 bp, X63731:1504–1795; probe f, 194 bp, AC007889.8:47,118–47,292; probe g, 206 bp, J01103:372–578; probe h, 229 bp, J01103:543–772; probe i, 230 bp, J01103:737–967; probe j, 217 bp, AC007889.8:57,030–57,237; probe k, 199 bp, X63732:31–230; probe l, 191 bp, X63732:169–360; probe m, 216 bp, X63732:337–553; and probe n, 222 bp, X63732:535–757.
Chromosome conformation capture
Approximately 0.4 g of nuclei were used for each cross-linking reaction. Cross-linking was performed as described in Dekker et al. (2002) with the following exception: After cross-linking, the nuclei were resuspended in 1 mL of cross-linking buffer without formaldehyde. One hundred microliters from each cross-linking reaction was digested to completion with MboI in a final 200 μL volume. The enzyme was inactivated by incubation for 30 min at 65°C. One-tenth of each digestion was incubated with T4 ligase and ATP overnight at room temperature in a final volume of 250 μL for the cross-linked nuclei and 23 μL for the uncross-linked nuclei. The cross-links were reversed by overnight incubation at 65°C in the presence of proteinase K. The DNA was purified by phenol-chloroform extraction and was ethanol precipitated. The purified DNA was resuspended in 20 μL of water, and the entire volume was used for a single PCR. Touchdown PCR was used. The annealing temperature was dropped from 72°C to 55°C in 3°C increments. With the exception of the two 72°C cycles, there were three cycles at each temperature. PCR products were separated on a 1% agarose gel and transferred to Zetaprobe (Bio-Rad) by using standard Southern blotting techniques. The blots were probed with an ∼150-bp PCR product generated from scs‘ sequence.
Primers
Primers were as follows: scs-o, GTAGTTTGACTTTCTCTGT TAATCAC; scs-i, GGTGGCAAATGAACTGC; inter-1, AAAG CATATGCAAAAACCGT; inter-2, TGGTTACAACTCAAAG GGGCG; inter-3, CTCATTGACTGGAGCTATCCG; inter-4, CATATGTTGCTGATGCGGATAGC; scs‘-o, GCTGTTCTG GTTCACAGTCACCAG; and scs‘-i, GTGCGACGAATTAA CATATTTTCA.
Acknowledgments
We acknowledge R. Mishra, G. Desphande, G. Shanower, D. Gohl, and K. Carroll for helpful discussions, and T. Graham for comments on the manuscript. We also thank K. Carroll and D. Gohl for help with experiments, J. Goodhouse for assistance with the confocal microscope, and G. Grey for providing fly food. BEAF cDNAs were obtained from C. Hart, and the BEAF overexpression flies were from Dr. M. Yamaguchi. This work was supported by a grant from NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pschedl@molbio.princeton.edu; FAX (609) 258-1028.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1052003.
References
- Bell AC, West AG, Felsenfeld G. Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- Bellard M, Kuo MT, Dretzen G, Chambon P. Differential nuclease sensitivity of the ovalbumin and β-globin chromatin regions in erythrocytes and oviduct cells of laying hen. Nucleic Acids Res. 1980;8:2737–2750. doi: 10.1093/nar/8.12.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai HN, Shen P. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science. 2001;291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Cuvier O, Hart CM, Laemmli UK. Identification of a class of chromatin boundary elements. Mol Cell Biol. 1998;18:7478–7486. doi: 10.1128/mcb.18.12.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier O, Hart CM, Kas E, Laemmli UK. Identification of a multicopy chromatin boundary element at the borders of silenced chromosomal domains. Chromosoma. 2002;110:519–531. doi: 10.1007/s00412-001-0181-1. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer–promoter interaction. Genes & Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG. Chromatin insulators and boundaries: Effects on transcription and nuclear organization. Ann Rev Genet. 2001;35:193–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- Geyer PK. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- Geyer P, Corces V. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes & Dev. 1992;6:1865–1973. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Grewal SI. Transcriptional silencing in fission yeast. J Cell Physiol. 2000;184:311–318. doi: 10.1002/1097-4652(200009)184:3<311::AID-JCP4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Gyurkovics H, Gausz J, Kummer J, Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990;9:2579–2585. doi: 10.1002/j.1460-2075.1990.tb07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart CM, Zhao K, Laemmli UK. The scs‘ boundary element: Characterization of boundary element-associated factors. Mol Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes TR, Clayton AL, Thorne A, Crane-Robinson C. Core histone hyperacetylatiopn co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 1992;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogga I, Mihaly J, Barges S, Karch F. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol Cell. 2001;8:1145–1151. doi: 10.1016/s1097-2765(01)00377-x. [DOI] [PubMed] [Google Scholar]
- Holdridge C, Dorsett D. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol Cell Biol. 1991;11:1894–1900. doi: 10.1128/mcb.11.4.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 2002;30:1465–1482. doi: 10.1093/nar/30.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Schedl P. A position effect assay for boundaries of higher order chromatin domains. Cell. 1991;64:941–951. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- ————— A group of scs elements function as domain boundaries in an enhancer-blocking assay. MCB. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science. 2001a;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001b;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden MP, Laemmli UK. Metaphase chromosome structure: Evidence for a radial loop model. Cell. 1979;17:849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. The Mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics. 1999;153:1333–1356. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Bell AC, Recillas-Targa F, West AG, Simpson M, Pikaart M, Felsenfeld G. Structural and functional conservation at the boundaries of the chicken β-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila Polycomb Response Element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder J, Larsen A, Engel JD, Dolan M, Groudine M, Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980;20:451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Thon G, Bjerling P, Bunner CM, Verhein-Hansen J. Expression-state boundaries in the mating-type region of fission yeast. Genetics. 2002;161:611–622. doi: 10.1093/genetics/161.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy A. Dividing the empire: Boundary chromatin elements delimit the territory of enhancers. EMBO J. 1999;18:1–8. doi: 10.1093/emboj/18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy A, Maine E, Schedl P. The 87A7 chromomere: Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;l85:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- Vazquez J, Schedl P. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 1994;13:5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Thomas JO. Chromatin structure of the β-globin chromosomal domain in adult chicken erythrocytes. Cold Spring Harb Symp Quant Biol. 1993;58:15–24. doi: 10.1101/sqb.1993.058.01.005. [DOI] [PubMed] [Google Scholar]
- West AG, Gaszner M, Felsenfeld G. Insulators: Many functions, many mechanisms. Genes & Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Yoshida H, Hirose F, Inoue YH, Hayashi Y, Yamagishi M, Nishi Y, Tamai K, Sakaguchi K, Matsukage A. Ectopic expression of BEAF32A in the Drosophila eye imaginal disc inhibits differentiation of photoreceptor cells and induces apoptosis. Chromosoma. 2001;110:313–321. doi: 10.1007/s004120100155. [DOI] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]