Abstract
The repression of the maternally inherited Igf2 allele has been proposed to depend on a methylation-sensitive chromatin insulator organized by the 11 zinc finger protein CTCF at the H19 imprinting control region (ICR). Here we document that point mutations of the nucleotides in physical contact with CTCF within the endogenous H19 ICR lead to loss of CTCF binding and Igf2 imprinting only when passaged through the female germline. This effect is accompanied by a significant loss of methylation protection of the maternally derived H19 ICR. Because CTCF interacts with other imprinting control regions, it emerges as a central factor responsible for interpreting and propagating gamete-derived epigenetic marks and for organizing epigenetically controlled expression domains.
Keywords: Genomic imprinting, Igf2, chromatin insulator, CTCF, DNA methylation
The manifestation of genomic imprinting involves the translation of gametic marks into parent of origin-dependent gene expression patterns (Bartolomei and Tilghman 1997; Horsthemke et al. 1999). The neighboring IGF2 and H19 genes emerge as paradigms of genomic imprinting, because their expression is monoallelic from opposite parental alleles and governed by shared enhancers (Bartolomei and Tilghman 1997; Horsthemke et al. 1999). The repression of the maternal IGF2 and paternal H19 alleles depends on a differentially methylated imprinting control region (ICR) in the 5′ region of the H19 gene (Olek and Walter 1997; Kaffer et al. 2000). The proposal that the complex between the H19 ICR and the 11 zinc finger protein CTCF organizes a CpG methylation-sensitive insulation of the maternal Igf2 allele (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000a,b; Bell et al. 2001; Ohlsson et al. 2001) is supported by the observations that CTCF interacts with only the maternal H19 ICR allele (Kanduri et al. 2000b) and that the chromatin insulator function is regulated by CpG methylation (Holmgren et al. 2001).
To directly prove this proposal, we mutated sequences within the H19 ICR, which we have previously shown to be in direct physical contact with CTCF. Our results document that the CTCF–H19 ICR complex is vital in the manifestation and propagation of gametic marks.
Results and Discussion
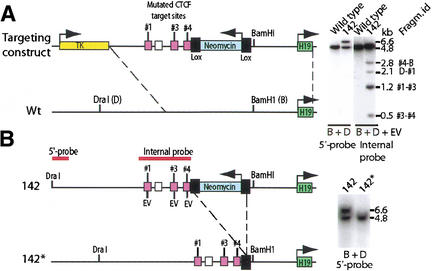
To directly demonstrate the function of the CTCF target sites within the H19 ICR in association with the establishment and manifestation of the imprinting phenomenon, we changed the sequence GTGG to ATAT in three of the four CTCF target sites within the CGCG(T/G)GGTGGCAG-core motif. This sequence change deletes essential contact points for the CTCF while preserving the CpGs responsible for the methylation-sensitive portion of the CTCF target sites (Kanduri et al. 2000b). The remaining second CTCF target site was ignored, because it does not, in contrast to the other target sites, display any marked in vivo footprint or nuclease hypersensitivity (Kanduri et al. 2000a; Szabó et al. 2000). Following electroporation of the targeting construct into ES cells and exploiting the fact that the mutations introduced an EcoRV site in each of the three mutated target sites (Kanduri et al. 2000b), we were able to identify one cell clone, #142, which had the endogenous H19 ICR replaced with all three mutated CTCF sites (Fig. 1A). The retained neomycin gene, which was flanked with loxP sites, was deleted by mating homozygous 142 mice with mice carrying a Cre transgene under the control of the β-actin promoter (Lewandoski and Martin 1997). Southern blot analysis confirmed the deletion of the neomycin gene in the offspring of these crosses to generate the 142* substrain (Fig. 1B).
Figure 1.
Schematic illustration of the knock-in strategy. (A) Recombination between targeting construct and endogenous H19 ICR as confirmed by Southern blot analysis. The recombination in the 142 clone was assessed by using a probe that is positioned 5′ of the sequence covered in the targeting construct. The properly recombined insert generated a larger size fragment due to the extra sequence information provided by the neomycin gene. All three mutated CTCF target sites (cerise boxes) replaced the endogenous sequences in the 142 clone as determined by EcoRV (EV) digestion and Southern blot analysis. The white box identifies CTCF target site #2, which was not mutated. The identities of the fragments in the Southern blot image are depicted on the far right of the image. (B) Deletion of the neomycin gene by breeding the 142 strain with mice harboring a β-actin promoter-driven Cre recombinase gene to generate the 142* substrain. The mutated allele is now indistinguishable from the wild-type allele when digested with BamHI (B) and DraI (D). See the text for additional information.
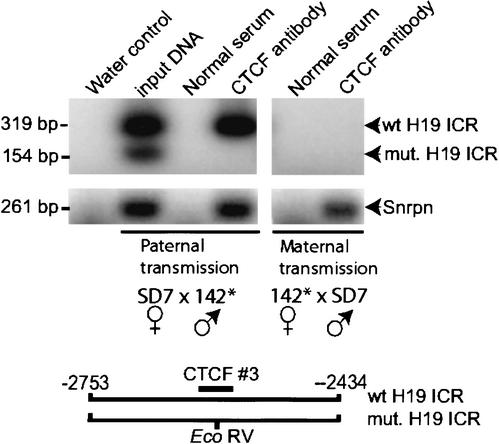
To verify that the mutations affected the association between CTCF and the maternal allele of the H19 ICR in vivo, we performed chromatin immunopurification (ChIP) analyses of formaldehyde cross-linked DNA–protein complexes from E14.5 embryos, followed by PCR amplification. As accounted for above, EcoRV restriction of the amplified fragments encompassing CTCF target site #3 allowed the discrimination between the wild-type and mutated H19 ICR alleles. Figure 2 shows that the wild-type maternal H19 ICR allele was pulled down by the CTCF antibody in agreement with a previous report (Kanduri et al. 2000b). However, the mutated H19 ICR allele could not be similarly detected upon either paternal or maternal inheritance. This difference did not depend on variation in quality between the formaldehyde-fixed protein–DNA complexes, because sequences included in the Snrpn imprinting box (Shemer et al. 2000), interacted with CTCF in both reciprocal crosses (Fig. 2). We conclude that point mutations of the H19 ICR CTCF target sites #1, #3, and #4 lead to complete failure of CTCF binding in vivo, confirming our initial anticipation that site #2 does not interact with CTCF in vivo.
Figure 2.
CTCF occupancy on wild-type and mutated H19 ICR alleles. Chromatin immunopurification analyses were performed on dispersed and formaldehyde cross-linked E14.5 embryos with the mutated H19 ICR allele inherited paternally or maternally. The PCR primers spanned CTCF target site #3, which displayed an EcoRV site only in the mutated H19 ICR allele, as indicated in the image. As an internal control, sequences of the Snrpn imprinting box were amplified from the same immunopurified DNA samples. In both instances, the sense primers were end-labeled with 32P. See the text for additional information.
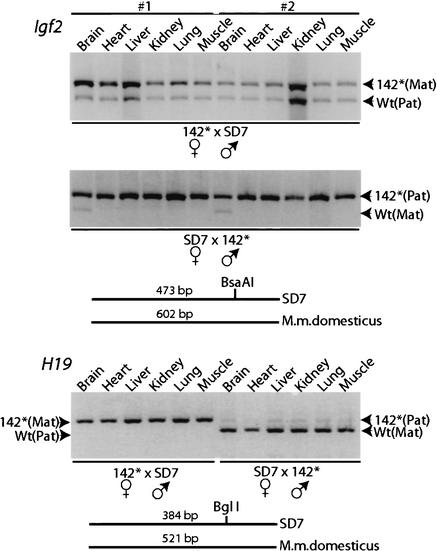
We next examined whether or not the mutations affected the imprinted state of the Igf2 gene by crossing 142* mice with SD7 mice, which were derived from a congenic strain carrying the distal end of chromosome 7 of Mus musculus spretus on a Mus musculus domesticus background (Dean et al. 1998). A polymorphic BsaAI site in the exon 4 of the Igf2 locus allowed the discrimination of allelic expression patterns in the offspring of these two strains (Dean et al. 1998). Figure 3 shows reverse transcriptase PCR (RT–PCR) analyses of a large variety of tissue types documenting that neonatal offspring from 142*×SD7 crosses (in the order female to male) robustly expressed Igf2 from both parental alleles. This loss of Igf2 imprinting was accompanied by an 11% gain of weight in neonates when compared to neonates derived from the reciprocal SD7×142* crosses (in the order female to male), which display a normal Igf2 imprinting status (Fig. 3; data not shown). We conclude that the contact points within CTCF sites #1, #3, and #4 in the H19 ICR are essential for maintaining the repressed status of the maternal Igf2 allele in most cell types of both mesodermal and endodermal lineages of the developing conceptus. In each of these instances, H19 imprinting appeared normal, although we noted a slight activation of the paternal H19 allele in offspring inheriting the mutated ICR allele paternally (Fig. 3).
Figure 3.
RT–PCR analyses of Igf2 and H19 expression patterns in various organs dissected from different neonatal offspring (labeled #1 and #2) of reciprocal crosses between 142* and SD7 mice. These assays exploited species-specific BsaAI and BglI polymorphisms in exon 4 of Igf2 (Dean et al. 1998) and exon 5 of H19 (Sasaki et al. 1995), respectively, as schematically indicated in the figure. The crosses are presented in the order female to male. See the text for additional information.
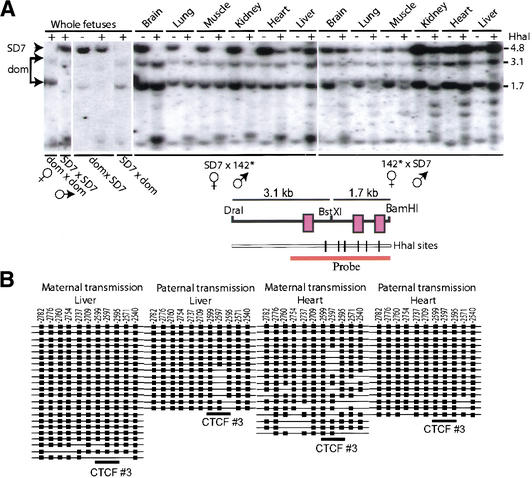
Our strategy to keep the crucial CpGs intact in the mutated CTCF target sites allowed us to directly assess if the methylation privilege status was lost as a consequence of the inability to interact with CTCF. CpG methylation analyses of the parental alleles, which were discriminated by using a polymorphic BstX1 site that restricts 142* H19 ICR, but not the SD7 allele (Fig. 4A), was initially performed by restricting the DNA with the methylation-sensitive enzyme Hha I followed by Southern blot analysis. Although it is well-established that the maternal H19 ICR allele is normally unmethylated (Olek and Walter 1997), Figure 4A shows that the mutation of the contact sequences for the CTCF sites resulted in significant de novo methylation, when maternally inherited. This observation was confirmed by bisulphite methylation analyses of the CTCF target site #3 of the mutated H19 ICR allele in heart and liver. Figure 4B shows that the normally maintained unmethylated status at this CTCF target site when maternally inherited (Olek and Walter 1997; Liang et al. 2000) is practically lost when passaged through the female germline. Interestingly, paternal transmission of the mutated H19 ICR also led to partial methylation of the maternally inherited wild-type H19 ICR allele in some tissues, such as heart and liver (Fig. 4A). Although this pattern of de novo methylation must be limited due to absence of expression of the maternal Igf2 allele in the very same tissue specimens (Fig. 3) and shows variation among different conceptuses (data not shown), it suggests the existence of a trans-sensing mechanism. This deduction converges with earlier proposals of trans-sensing within the Igf2/H19 imprinting domain (LaSalle and Lalande 1996; Forne et al. 1997).
Figure 4.
CpG methylation analyses. (A) Southern blot analysis of genomic DNA extracted from various organs dissected from neonatal offspring of reciprocal crosses between 142* and SD7 mice. The parental alleles were discriminated by exploiting a polymorphic BstXI site, as indicated in A. (B) Bisulphite sequencing analyses of the mutated H19 ICR allele between −2836 to −2479 bp from the H19 transcriptional start site, covering the mutated CTCF target site #3. The wild-type and mutant H19 ICR alleles were discriminated on the basis of the introduced point mutations within CTCF target site #3. The tissue type and the sex of the parent transmitting the mutant allele is indicated above each scheme. Filled boxes indicate methylated CpGs and no box indicates no methylation at the positions indicated above each row.
Our results document the dual feature of the CTCF–methylation link. In addition to the ability of CTCF to interpret the methylation status of its target sites within the H19 ICR, CTCF also maintains a methylation-free domain at and around its target sites. Although it is likely that this latter feature involves protection by the CTCF-H19 ICR complex towards the de novo methylation machinery, it is probable that many other neighboring protein–DNA complexes play a similar role. This deduction is supported by our observation that the mutated, maternal H19 ICR allele remains methylation-free in the unfloxed 142 strain containing the transcriptionally active neomycin gene just 3′ of the H19 ICR (data not shown). Given that the de novo methylation of the mutated H19 ICR in the floxed 142* strain is not complete, it is likely that additional factors interact with other linker regions between the positioned nucleosomes (Kanduri et al. 2002) to jointly maintain the methylation-free domain of the maternally inherited H19 ICR allele with CTCF. One such candidate factor is Oct-1, which has been reported to interact with the H19 ICR between CTCF target sites #2 and #3 to provide partial protection against de novo methylation (Hori et al. 2002).
This report documents that the CTCF target sites manifest not only the repressed status of the maternally inherited Igf2 allele, but also the methylation-free domain of the maternal H19 ICR allele in somatic cells. This latter issue is of particular interest with respect to the acquisition of the methylation mark during male germline development, because it implies that CTCF must be down-regulated and removed from the maternal H19 ICR allele in adult testis prior to de novo methylation. Although CTCF is indeed down-regulated during differentiation of spermatogonia into spermatocytes (J. Whitehead, L. Liu, C. Kanduri, V. Pant, M. Lezcano, W.-Q. Yu, A. Kerjean, M. Parvinen, A. Paldi, E. Klenova, V. Lobanenkov, and R. Ohlsson, unpubl.), this would seemingly be at odds with the vital role for CTCF in cell survival and development (G.N. Filippova, J. Whitehead, S. Fagerlie, S. Vatolin, K. Foley, D. Loukinov, E.M. Pugacheva, J.E. Ulmer, J.M. Moore, Y.J. Hu, E.M. Klenova, C. Kemp, S.J. Collins, P.E. Neiman, R. Ohlsson, and V.V. Lobanenkov, unpubl.). We have earlier shown that a paralog of CTCF, termed BORIS for ”Brother of Regulator of Imprinted Sites,” is expressed exclusively in CTCF-negative spermatocytes in adult mouse testis (Loukinov et al. 2002). Because our preliminary observation shows that BORIS replaces CTCF on H19 ICR target sites prior to imprint acquisition of the maternally derived H19 ICR allele (J. Whitehead, L. Liu, C. Kanduri, V. Pant, M. Lezcano, W.-Q. Yu, A. Kerjean, M. Parvinen, A. Paldi, E. Klenova, V. Lobanenkov, and R. Ohlsson, unpubl.), we speculate that the CTCF → BORIS switch is essential for the elimination of the methylation-protective properties of the CTCF–H19 ICR complex in pachytene spermatocytes.
Materials and methods
Knock-in strategy
Three of the four CTCF target sites (Fig. 1) were mutated to change the core sequence from GTGG to ATAT, as has been described (Kanduri et al. 2000b). The targeting vector was assembled in the pGEMT vector, as indicated in Figure 1. The herpes simplex virus thymidine kinase expression cassette is followed by 4.3 kb of genomic 5′-sequence (long genomic arm), the PGK-neobpA expression cassette flanked by loxP sites (“floxed”), ∼2.6 kb of genomic 3′-sequence (including the H19 transcriptional start site) and the pGEMT vector backbone. The vector was linearised at the singular Nde1 site, electroporated into R1 embryonic stem cells (Nagy et al. 1993) and colonies were selected with G418 and Gancyclovir. Homologous integration events were screened by Southern blotting (see below). The neomycin gene was deleted by mating homozygous mutant mice with a transgenic mouse strain carrying the Cre-recombinase gene under the control of the β-actin promoter (Lewandoski and Martin 1997).
Southern analysis for confirming recombination
Genomic DNA was isolated using Wizard genomic DNA isolation kit (Promega). DNA was digested with respective enzymes, analyzed by electrophoresis on a 1% agarose gel blotted onto a Hybond-N+ membrane (Amersham Pharmacia). A 1.3-kb PCR-generated fragment (−6750 to −5406 bp with respect to the transcription start site of the mouse H19 gene) outside the targeting vector sequence was used to test for correct genomic integration of the targeting construct. To assess the presence of all three mutated CTCF target sites, DNA was digested with Dra1, BamH1, and EcoRV and probed with an internal probe spanning the sequence −4296 to −1759 bp upstream of the H19 transcription start site, covering all four CTCF sites in the H19 ICR region. For screening of the recombinant clones, genomic DNA was digested with Dra1 and BamH1 and probed with a 5′ external probe. The presence of the neomycin gene allowed the discrimination of the mutated and wild-type alleles.
Chromatin immunopurification analyses
Dispersed embryonic cells were formaldehyde cross-linked (in 1% formaldehyde for 10 min at 37°C), and the DNA–protein complexes were immunopurified using anti-CTCF antibody and protein A 4 Fast Flow Sepharose beads (Pharmacia-Upjohn), as has been described (Kuo and Allis 1999; Kanduri et al. 2000b). To examine an association between H19 ICR and CTCF, the immunopurified DNA was PCR amplified with primers spanning the mutated CTCF target site #3: sense primer 5′-CTCAGTGGTCGATATATGGTTT-3′; antisense primer 5′-TGAGT CAAGTTCTCTTGGTTC-3′. PCR conditions were 95°C for 3 min, 28× (94°C for 40 sec, 54°C for 40 sec, 72°C for 40 sec). To compare the quality of the formaldehyde cross-linked samples, sequences included in the Snrpn imprinting box were amplified from the same samples, as follows: sense primer 5′-ATCCTGGATGCAAGAGCTGT-3′; antisense primer 5′-GCCGCACGTACAGTTACA-3′. PCR conditions were: 95°C for 3 min, 28× (94°C for 40 sec, 58°C for 45 sec, 72°C for 50 sec). The amplified sequence is positioned in the first exon of Snrpn (nucleotides 3901–4162 in GenBank file AF130843). In both instances, the sense primers were endlabeled with 32P-γ-ATP and polynucleotide kinase, as has been described (Kanduri et al. 2000b).
Allelic RT–PCR analyses
To determine the imprinting status of the Igf2 and H19 genes, homozygous 142* mice were mated with SD7 mice (a congenic strain carrying the distal end of chromosome 7 of Mus musculus spretus on a Mus musculus domesticus background). RNA was isolated from individual organs of 2-day-old neonatals or foetuses at E14.5 using TriPure DNA/RNA isolation reagent (Boehringer Mannheim). RNA was treated with RQ1 DNase I (Promega) to remove any contaminating genomic DNA. RT–PCR analyses were done using Qiagen one-step RT–PCR kit as per guidelines. Primers and PCR conditions were the same as previously published protocols for Igf2 (Dean et al. 1998) and H19 (Sasaki et al. 1995). The PCR product was purified and digested with BsaA1 to determine the allelic origin of the Igf2 transcripts and with Bgl1 for the H19 transcripts. Fragments were separated on 2% agarose gel and stained with ethidium bromide for visualization.
Methylation analyses
For Southern blot analysis, genomic DNA was isolated from tissues of 2-day-old neonatal mice. Fifteen micrograms of DNA were digested with Dra1, BamH1, and BstX1 and with/without the methylation-sensitive restriction enzyme Hha1. Bstx1 has a unique site present only in the Dom allele but not in the SD7 allele at −3537 bp of the H19 transcription start site. Bisulfite treatment of DNA was carried out using an established protocol (Olek et al. 1996), with the following adaptations: DNA was restricted with an excess amount of Dra1 and BamH1 to generate suitably small fragments containing the target sequence. One microgram of digested DNA was denatured with 0.3 M NaOH at 37°C for 15 min, then mixed with 2 volumes of 2% low-melting agarose dissolved in water. The mixture was pipetted into prechilled mineral oil to form the DNA–agarose beads. The prepared beads were then incubated with 1.2 mL 5.0 M NaHSO3/20 mM Hydroquinone solution covered by mineral oil and incubated at 51°C for 6 h. Treated DNA beads were equilibrated with TE (1 mM EDTA, 10 mM Tris-HCl at pH 8.0), 6 × 15 min. Following desulphonation with 0.2 M NaOH, 2 × 15 min and equilibration with MilliQ water, 2 × 15 min at RT, the DNA beads were subjected to PCR amplification reactions, using the following published primers and conditions (Tremblay et al. 1997): Bhha5t2, BHha5t, and BHha5t3 (the final PCR product covers bases −2836 to −2479 relative to the H19 transcriptional start site). Amplified fragments were cloned into the pGEMT-Easy vector (Promega), and subsequently sequenced with the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems).
Acknowledgments
We are grateful to Dr. Wolf Reik for generously providing both SD7 mice and sequence information of the Mus musculus spretus H19 ICR, to Dr. Gail Martin for providing the Cre-transgenic mouse line, and to Dr. Ragnar Mattsson at the Lund University transgenic core facility for help with blastocyst injections. This work was supported by the Swedish Science Research Council (VR), the Swedish Cancer Research Foundation (CF), the Swedish Pediatric Cancer Foundation (BCF), and the Lundberg Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Rolf.Ohlsson@ebc.uu.se; FAX 46-18-4712683.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.254903.
References
- Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- Dean W, Bowden L, Aitchison A, Klose J, Meneses J, Reik W, Feil R. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: Association with abberant phenotypes. Development. 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- Forne T, Oswald J, Dean W, Saam J, Bailleul B, Dandolo L, Tilghman S, Walter J, Reik W. Loss of the maternal H19 gene induces changes in Igf2 methylation in both cis and trans. Proc Natl Acad Sci. 1997;94:10243–10248. doi: 10.1073/pnas.94.19.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Holmgren C, Kanduri K, Dell G, Ward A, Mukhopadhya R, Kanduri M, Lobanenkov V, Ohlsson R. CpG methylation regulates the Igf2/H19 insulator. Curr Biol. 2001;11:1128–1130. doi: 10.1016/s0960-9822(01)00314-1. [DOI] [PubMed] [Google Scholar]
- Hori N, Nakano H, Takeuchi T, Kato H, Hamaguchi S, Oshimura M, Sato K. A dyad Oct-binding sequence functions as a maintenance sequence for the unmethylated state within the H19/Igf2-imprinted control region. J Biol Chem. 2002;277:27960–27967. doi: 10.1074/jbc.M202280200. [DOI] [PubMed] [Google Scholar]
- Horsthemke B, Surani MA, James TC, Ohlsson R. The mechanisms of genomic imprinting. In: Ohlsson R, editor. Genomic imprinting: An interdisciplinary approach. Berlin: Springer-Verlag; 1999. pp. 91–118. [DOI] [PubMed] [Google Scholar]
- Kaffer CR, Srivastava M, Park KY, Ives E, Hsieh S, Batlle J, Grinberg A, Huang SP, Pfeifer K. A transcriptional insulator at the imprinted H19/Igf2 locus. Genes & Dev. 2000;14:1908–1919. [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, Holmgren C, Franklin G, Pilartz M, Ullerås E, Kanduri M, Liu L, Ginjala V, Ulleras E, Mattsson R, et al. The 5′-flank of the murine H19 gene in an unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr Biol. 2000a;10:449–457. doi: 10.1016/s0960-9822(00)00442-5. [DOI] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi C-F, Wolffe A, Ohlsson R, Lobanenkov A. Functional interaction of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000b;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Kanduri M, Kanduri C, Mariano P, Vostrov A, Quitsche W, Lobanenkov V, Ohlsson R. Multiple nucleosome positioning sites regulate the CTCF-mediated insulator function of the H19 imprinting control region. Mol Cell Biol. 2002;22:3339–3344. doi: 10.1128/MCB.22.10.3339-3344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M, Allis C. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Martin G. Cre-mediated chromosome loss in mice. Nat Genet. 1997;17:223–225. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- Liang L, Kanduri C, Pilartz M, Svensson K, Song J, Wentzel P, Eriksson U, Ohlsson R. Dynamic readjustment of parental methylation patterns of the 5′-flank of the mouse H19 gene during in vitro organogenesis. Int J Dev Biol. 2000;44:785–790. [PubMed] [Google Scholar]
- Loukinov D, Pugacheva E, Vatolin S, Pack S, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov A, et al. BORIS, a novel male germline-specific protein associated with epigenetic reprogramming events, shares the same 11 Zn-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J. Derivation of completely culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Olek A, Walter J. The pre-implantation ontogeny of the H19 methylation imprint. Nat Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- Olek A, Oswald J, Walter J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 1996;24:5064–5066. doi: 10.1093/nar/24.24.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Ferguson-Smith A, Shum A, Barton S, Surani M. Temporal and spatial regulation of H19 imprinting in normal and uniparental mouse embryos. Development. 1995;121:4195–4202. doi: 10.1242/dev.121.12.4195. [DOI] [PubMed] [Google Scholar]
- Shemer R, Hershko A, Perk J, Mostoslavsky R, Tsubert B-Z, Cedar H, Buiting K, Razin A. The imprinting box of the Prader-Will/Angelman syndrome domain. Nat Genet. 2000;26:440–443. doi: 10.1038/82571. [DOI] [PubMed] [Google Scholar]
- Szabó P, Tang S-H, Rentsendorj A, Pfeifer G, Mann J. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- Tremblay K, Duran K, Bartolomei MS. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]