Abstract
Exonuclease 1 (Exo1) is a 5′–3′ exonuclease that interacts with MutS and MutL homologs and has been implicated in the excision step of DNA mismatch repair. To investigate the role of Exo1 in mammalian mismatch repair and assess its importance for tumorigenesis and meiosis, we generated an Exo1 mutant mouse line. Analysis of Exo1−/− cells for mismatch repair activity in vitro showed that Exo1 is required for the repair of base:base and single-base insertion/deletion mismatches in both 5′ and 3′ nick-directed repair. The repair defect in Exo1−/− cells also caused elevated microsatellite instability at a mononucleotide repeat marker and a significant increase in mutation rate at the Hprt locus. Exo1−/− animals displayed reduced survival and increased susceptibility to the development of lymphomas. In addition, Exo1−/− male and female mice were sterile because of a meiotic defect. Meiosis in Exo1−/− animals proceeded through prophase I; however, the chromosomes exhibited dynamic loss of chiasmata during metaphase I, resulting in meiotic failure and apoptosis. Our results show that mammalian Exo1 functions in mutation avoidance and is essential for male and female meiosis.
Keywords: Exonuclease 1; MMR; lymphoma, meiosis; infertility
The DNA mismatch repair (MMR) system is important for maintaining the integrity of the genome in prokaryotes and eukaryotes. It has evolved to correct mispaired bases that result from errors during DNA replication, DNA recombination, and from certain types of DNA damage. Analyses in yeast and mice also revealed an essential role of some eukaryotic MMR gene products in the control of meiotic recombination. The importance of MMR to mammals is also highlighted by the observation that germ-line mutations in several of the MMR genes are associated with hereditary nonpolyposis colorectal cancer (HNPCC), and the loss of MMR function underlies several types of sporadic cancers (Peltomaki and Vasen 1997; Peltomaki 2001).
MMR was initially characterized in bacteria (Modrich 1991; Modrich and Lahue 1996) and more recently in eukaryotic cells (Kolodner 1996). The studies of mammalian MMR focused mainly on the role of the eukaryotic MutS and MutL homologs (MSH and MLH, respectively) in the initiation of the repair reaction. These analyses showed that the early steps of MMR include the recognition of mispaired nucleotide(s) by two heterodimeric complexes: MSH2–MSH6 functions in the repair of base:base mispairs as well as a range of insertion/deletion loop mispairs (IDLs), whereas MSH2–MSH3 primarily functions in the repair of IDLs (Marsischky et al. 1996; Genschel et al. 1998; Umar et al. 1998). Subsequent to mismatch recognition, these two MutS complexes interact with MutL complexes consisting of MLH1–PMS2 or MLH1–MLH3 (Prolla et al. 1994; Li and Modrich 1995; Flores-Rozas and Kolodner 1998; Wang et al. 1999). These interactions are absolutely necessary for the activation of downstream events including the excision of the misincorporated nucleotide(s) and filling in of the resulting single-strand gap by DNA resynthesis. These later steps in MMR as well as the nature of 5′-to-3′ and 3′-to-5′ excision of mismatched bases remain poorly understood.
Studies in Escherichia coli demonstrated that the excision step of MMR occurs bidirectionally depending on the location of the initiating nick relative to the mismatch (Cooper et al. 1993; Grilley et al. 1993). It requires the participation of DNA helicase II (the uvrD/mutU product) to unwind DNA toward the mismatch (Dao and Modrich 1998; Yamaguchi et al. 1998) and the action of several functionally redundant exonucleases including RecJ or Exo VII (5′–3′) to excise mismatches starting from a 5′ nick; and Exo I, Exo VII, or Exo X (3′–5′) to remove mismatches starting from a 3′ nick (Burdett et al. 2001; Viswanathan et al. 2001).
Genetic studies in yeast and biochemical studies of MMR activity in cell extracts (Genschel et al. 1998; Umar et al. 1998; Tran et al. 1999) indicate that eukaryotes use a similar mechanism with both 3′–5′ and 5′–3′ exonuclease activities for mismatch correction. Although no 3′–5′ exonuclease specific to MMR has been identified in eukaryotes to date, the 5′–3′-specific exonuclease EXO1 has been implicated in the 5′–3′-directed excision step. EXO1 is a DNA repair nuclease of the Rad2 gene family and was originally identified as a meiotically induced 5′–3′ exonuclease in Schizosaccharomyces pombe and as a 5′–3′ exonuclease that interacts with Saccharomyces cerevisiae MSH2. In both S. pombe and S. cerevisiae it was found to play a role in mutation avoidance and mitotic recombination (Szankasi and Smith 1992, 1995; Huang and Symington 1993; Fiorentini et al. 1997; Tishkoff et al. 1997). Direct evidence for a role of yeast EXO1 in MMR was provided by the observation that it physically interacts with MSH2 and MLH1 (Tishkoff et al. 1997; Tran et al. 2001). The functional interaction between EXO1 and other MMR proteins was demonstrated by the synergistic potentiation of mutation rates in exo1 mutant strains that also carried weak mutator mutations in MLH1, PMS1, and MSH2. In addition to a role in the excision step of MMR, these studies also suggested a structural role for EXO1 in stabilizing multiprotein complexes containing MMR proteins (Amin et al. 2001).
Besides its function in somatic cells, EXO1 is required for meiotic recombination, as exo1 mutant yeast strains exhibit increased meiosis I nondisjunction and decreased meiotic intergenic recombination (Khazanehdari and Borts 2000; Kirkpatrick et al. 2000). Further studies in S. cerevisiae revealed additional EXO1 functions in various aspects of DNA metabolism, including the processing of DNA double-strand breaks, in which EXO1 appears to be able to substitute for the Mre11–Rad50–Xrs2 complex (Tsubouchi and Ogawa 2000), the repair of UV damage to DNA (Qiu et al. 1998), and the generation of single-stranded DNA at telomeres (Maringele and Lydall 2002).
The human EXO1 gene was mapped to Chromosome 1q42–43 (Schmutte et al. 1998; Tishkoff et al. 1998) and is expressed in a wide variety of tissues with elevated expression being observed in testis, thymus, colon, and placenta (Tishkoff et al. 1998). Initial biochemical analyses confirmed earlier findings in yeast showing that human EXO1 exhibits a number of nucleolytic activities, including 5′–3′ exonuclease and flap endonuclease activities (Lee and Wilson 1999; Lee et al. 2002). In addition, human EXO1 was implicated in MMR through its interaction with MSH2, MSH3, and MLH1 (Tishkoff et al. 1998; Schmutte et al. 2001). More recent biochemical studies suggested that human EXO1 is required for both 5′- and 3′-nick-directed DNA mismatch repair (Genschel et al. 2002).
The role of human EXO1 in cancer predisposition remains unclear. Recently, germ-line variations in EXO1 including a protein-truncating change and a number of missense changes were found in patients with HNPCC and atypical HNPCC (Wu et al. 2001). However, because the tumors from these patients showed loss of heterozygosity (LOH) of the EXO1 mutant allele but not the wild-type allele, it was unclear whether loss of normal EXO1 function was causative for the cancer development in these cases. A subsequent study showed that some of the initially reported EXO1 variants including the protein-truncating change were polymorphisms that were also found in normal individuals (Jagmohan-Changur et al. 2003). The data presented in these two studies indicate that germ-line mutations in EXO1 are at best a very rare cause of HNPCC, but the investigators did not address whether EXO1 mutations might cause a later onset or less penetrant form of cancer susceptibility.
To examine the biological roles of mammalian Exo1 and to clarify the role of Exo1 in cancer predisposition, we generated a mouse line with an inactivation mutation in the Exo1 gene by gene targeting. The analysis of this mouse line revealed that inactivation of Exo1 causes a distinct DNA repair defect that is similar to the repair defect observed in Msh6 mutant mice and increases the risk to develop cancers. Our results also demonstrate that Exo1 function is essential for meiotic progression through metaphase I and that Exo1 inactivation results in male and female sterility.
Results
Generation of Exo1 mutant mice
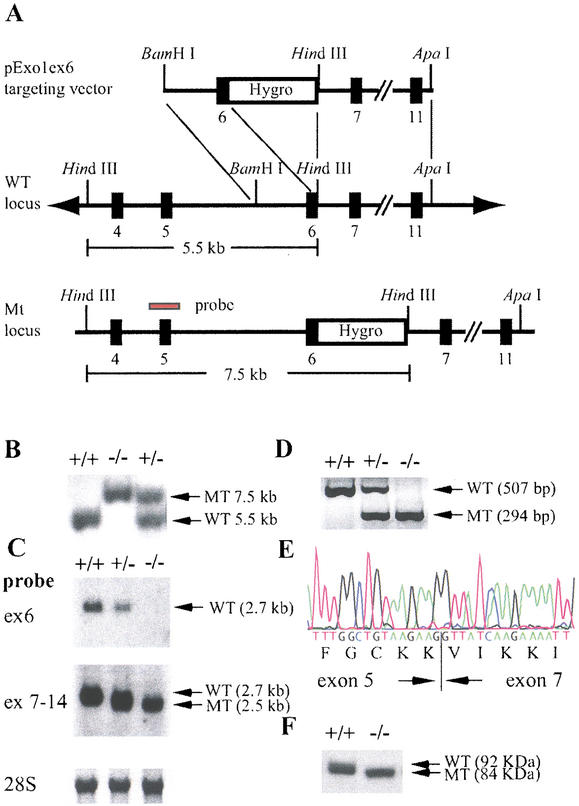
To generate Exo1 mutant mice, a gene-targeting vector was constructed by inserting a PGKhygromycin resistance cassette into the EcoRV restriction site in exon 6 corresponding to codons 249 and 250 of the Exo1 cDNA. The targeting vector was designed to disrupt the I-nuclease domain of Exo1, which is essential for the exonuclease function of the protein (Fig. 1A; Szankasi and Smith 1995; Lieber 1997). The linearized targeting vector was electroporated into ES (embryonic stem) cells, and positive clones were injected into C57BL/6J blastocysts to produce chimeric animals that transmitted the targeted allele through their germ line. Genotyping of 541 F2 offspring from 78 litters obtained from 20 heterozygous F1 mating pairs identified 135 wild-type, 274 heterozygous, and 132 homozygous mutant mice (Fig. 1B). These results indicate that the mutation is transmitted in a Mendelian fashion and does not interfere with normal embryonic development.
Figure 1.
Disruption of the mouse Exo1 gene by homologous recombination. (A) Map of the targeting vector, the Exo1 wild-type locus, and the modified locus. The probe used for Southern blot analysis is indicated (red bar). (B) Southern blot analysis of HindIII-digested genomic DNA from F2 animals; +/+, wild-type; +/−, heterozygote; −/−, homozygote. (C) Northern blot analysis of poly(A) RNA from Exo1 ES cell lines using exon 6 (ex 6) and exons 7–14 (ex 7–14) probes. (D) RT–PCR analysis of total RNA from Exo1 ES cell lines using primers located in exons 5 and 7. (E) DNA sequence of the mutant RT–PCR product in Exo1−/− mice shown in D. Note that exon 5 is spliced to exon 7 in frame, confirming the deletion of exon 6. (F) Western blot analysis of protein extracts from wild-type and Exo1−/− ES cells using an anti-Exo1 antibody. Note the slightly faster migrating protein species in Exo1−/− cells.
The Exo1 mutation was further studied on the RNA and protein level. Northern blot analysis of poly(A) RNA revealed that in Exo1−/− mice, exon 6 was absent in the Exo1 message as a result of alternate splicing (Fig. 1C). The deletion of exon 6 in the mutant Exo1 mRNA was confirmed by reverse transcriptase PCR (RT–PCR) analysis in Exo1+/− and Exo1−/− ES cell lines and in the tissues from Exo1+/− and Exo1−/− mice (brain, heart, lung, thymus, spleen, liver, kidney, pancreas, small and large intestines, skin, ovary, and testis were analyzed; Fig. 1D; data not shown). Sequencing of the mutant RT–PCR products showed that in the modified Exo1 mRNA, exon 5 was fused in frame to exon 7, resulting in the deletion of codons 181–252, which are encoded by exon 6 (Fig. 1E). Western blot analysis revealed a protein of 92 kD in wild-type cells and a slightly faster migrating protein of 84 kD in the Exo1−/− cells, confirming the deletion of exon 6 on the protein level (Fig. 1F). These results indicate that we generated a mouse line carrying a deletion mutation in the I-nuclease domain.
Loss of function in yeast strains carrying the equivalent I-domain deletion
To further investigate the functional consequences of this Exo1 mutation, we generated an equivalent deletion in the yeast EXO1 gene (deletion of codons 182–255). In contrast to the wild-type EXO1 gene, when the mutant exo1 gene was expressed under control of its native promoter on an ARS CEN LEU2 vector (pRDK1020), it did not complement the mutator phenotype of an exo1Δ strain or an exo1Δ strain containing the exo1Δ enhancer mutation pms1-A130V (data not shown). High-level expression of either the wild-type or mutant gene on an LEU2 2μ plasmid in a wild-type strain did not cause an increase in mutation rate. These results suggest that the mouse Exo1 mutation studied here is a loss-of-function mutation.
Inactivation of Exo1 and defective MMR in Exo1−/− ES cells
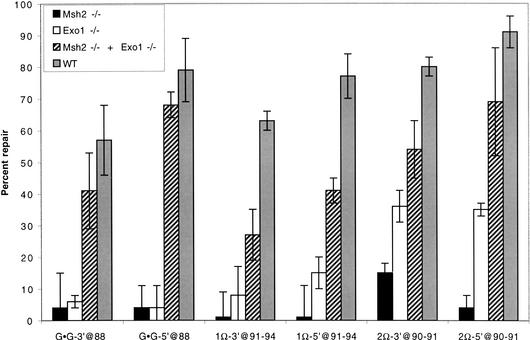
To verify the functional inactivation of Exo1 in the homozygous mutant mice and to study the role of Exo1 in mammalian MMR, we established Exo1−/− and Msh2−/− ES cell lines from 3.5-day-old blastocysts. Extracts from Exo1−/−, Msh2−/−, and wild-type cells were tested for their ability to promote repair of a G–G mispair substrate that contained a single-stranded nick located either 5′ or 3′ to the mismatched base. No mismatch repair activity was detected in the Exo1−/− or Msh2−/− extracts, whereas robust repair was seen in the wild-type extracts (Fig. 2). Complementation by combining Exo1−/− and Msh2−/− extracts restored the repair activity to wild-type levels (Fig. 2), indicating that the lack of repair activity observed in either Exo1−/− or Msh2−/− extract is not caused by preparation of generally inactive cell extracts but, rather, is caused by the specific absence of Exo1 or Msh2 activity, respectively. Similarly, no repair was seen with substrates that contained single-base insertion–deletion loops (IDL; Fig. 2). In contrast, considerable repair was detected when dinucleotide IDL substrates were tested (Fig. 2). These results confirmed that deletion of the I-domain amino-acid residues encoded by exon 6 results in the functional inactivation of Exo1 and suggest that Exo1 plays a particularly important role in the repair of base:base and single-base IDL mispairs. Extracts from Exo1+/− cells repaired all substrates tested, similar to what was observed with extracts of wild-type cells (data not shown), indicating that as in yeast, the existence of the mutant protein in the Exo1 heterozygotes does not interfere with the function of the wild-type gene product.
Figure 2.
Mismatch repair is impaired in Exo1−/− ES cells. DNA MMR activity was assayed in Exo1−/−, Msh2−/−, Exo1−/− + Msh2−/−, and wild-type (WT) cell extracts as described (Thomas et al. 1995). Substrates designated with Ω contain the number of extra nucleotides that accompany the symbol. Substrates are designated with a 3′ when the nick is in the (−) strand at the AvaII site (position −264), or with a 5′ when the nick is in the (−) strand at the Bsu36I site (position +276). The nucleotide position of the mismatch or unpaired bases in the lacZ complementation gene is indicated after the @, where position +1 is the first transcribed base of the lacZα complementation gene. The (−) sign designates the strand containing the extra nucleotide(s). WT, wild-type mouse ES cells. Cell extracts were prepared as described (Thomas et al. 1995) from ES cells. The results are averages based on counting >500 plaques per variable in three independent experiments. Error bars represent the standard deviations. Blue/white ratios of plaque color (not shown) demonstrated that, when observed, MMR was directed to the nicked strand.
Microsatellite instability in Exo1−/− mutant mice
Microsatellite instability (MSI), a type of replication error occurring at simple sequence repeats, is commonly associated with inactivation of MMR genes such as Msh2 and Mlh1. To study whether Exo1 inactivation is associated with MSI, we performed a quantitative analysis at microsatellite loci in genomic DNA from mouse tails (Table 1). At the mononucleotide marker U12235, Exo1−/− mice displayed 14% unstable alleles, a significant increase in MSI as compared with wild-type mice (3%, p = 0.0007). This level of MSI was similar to that observed in MMR-deficient Mlh1−/− mice (16%; Table 1). We also analyzed MSI at two dinucleotide markers: D7Mit91 and D17Mit123. In contrast to the mononucleotide repeat marker, we did not detect significant instability at either of these loci (Table 1). On the other hand, Mlh1−/− mice showed high levels of instability at both markers as compared with wild-type mice (15% at D7Mit91, p = 0.003; 28% at D17Mit123, p < 0.0001; Table 1). These results are in good agreement with the results obtained in the cell extracts described above (Fig. 2). They further support the idea that Exo1 plays an important role in the repair of single-base IDL mismatches and is less important in the repair of two-base IDL mismatches.
Table 1.
Microsatellite instability
| Marker
|
Wild type
|
Exol−/−
|
Mlhl−/−
|
|---|---|---|---|
| U12235 (A)n | 3% (7/244)a | 14% (11/79)b | 16% (12/73) |
| D7Mit91 (CA)n | 2% (3/124) | 0% (0/72)c | 15% (19/123) |
| D17Mit123 (CA)n | 6% (8/134) | 3% (2/68)d | 28% (19/69) |
Mutation frequency as determined by the number of unstable alleles divided by the total number of alleles scored.
Highly significant, p = 0.0007 versus wild type for marker U12235.
Not significant, p = 0.2992 versus wild type for marker D7Mit91.
Not significant, p = 0.5002 versus wild type for marker D17Mit123.
Mutator phenotype in Exo1−/− cells
To investigate the effect of Exo1 inactivation on mutagenesis, we determined the mutation rate at the Hprt locus in Exo1−/−, Msh2−/−, and wild-type ES cells by fluctuation analysis (Wierdl et al. 1996; Abuin et al. 2000). Exo1−/− cells displayed a 30-fold increase in mutation rate compared with wild-type cells. However, this mutation rate increase is fivefold lower than in Msh2−/− cells, in which we observed a 150-fold increase compared with wild-type cells (Table 2). The 95% confidence intervals for the measurements show that the fivefold difference is significant. These results indicate that Exo1−/− cells still retain some MMR capacity, which may be attributable to other exonuclease activities involved in the process.
Table 2.
Mutation rates at the Hprt locusa
| Genotype
|
Mutation rateb (×10−6)
|
Confidence limitsc (×10−6)
|
Fold increase
|
|---|---|---|---|
| WT | 0.02 | 0.0005–0.1 | 1 |
| Exol−/− | 0.6 | 0.3–0.9 | 30 |
| Msh2−/− | 3.0 | 2.4–3.7 | 150 |
Fluctuation analysis was performed as recently described (Abuin et al. 2000).
Mutation rates were determined according to calculations derived by Wierdl et al. 1996).
95% confidence interval for mutation rate (Dixon and Massey 1968).
Reduced survival and increased susceptibility to lymphoma in Exo1−/− mutant mice
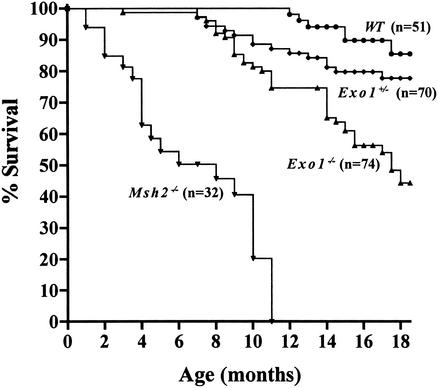
To study the long-term effect of Exo1 inactivation on survival and cancer susceptibility, we followed a large cohort of Exo1−/− mice (n = 74), Exo1+/− mice (n = 70), wild-type mice (n = 51), and Msh2−/− mice (n = 32) over a period of 18 mo. We found that Exo1−/− mice had a significantly reduced survival when compared with age-matched Exo1+/− and wild-type mice, with only 50% of Exo1−/− mice being alive at 17 mo (Fig. 3). In contrast, 80% of Exo1+/− mice and 90% of wild-type mice were still alive at this age. The reduction in lifespan observed in Exo1−/− mice was not as severe as that of Msh2−/− mice, where 50% of mice died by 8 mo of age and all animals were dead by 12 mo of age (Fig. 3).
Figure 3.
Survival of Exo1 mutant mice. The survival curve was generated using the Prism (GraphPad Prism 2.0) software package. The differences between the Exo1−/− versus Exo1+/+ and Exo1+/− curves are significant (p < 0.0001 and p = 0.0012, respectively). The difference between the Exo1+/− and Exo1+/+ curves is not significant (p = 0.2937). For comparison, the survival of Msh2−/− mice on similar genetic background is shown (Smits et al. 2000). Statistical analysis is according to the log rank test.
To investigate whether the reduced survival of Exo1−/− mice was caused by increased tumor development, we killed 26 Exo1−/− mice, 27 Exo1+/− mice, and 24 wild-type mice between the ages of 16 and 18 mo. We found that 80.7% (21/26 mice) of the Exo1−/− mice that were still alive had developed tumors by this age. In contrast, only 37% (10/27 mice) of the Exo1+/− mice and 37.5% (9/24 mice) of the wild-type mice developed tumors at a similar age (p = 0.0020, Exo1−/− vs. Exo1+/− mice; p = 0.0034, Exo1−/− vs. wild-type mice). There was no difference in tumor incidence between the Exo1+/− and wild-type mice (p = 1.0000). It should be noted that this analysis likely underestimates the tumor incidence in the Exo1−/− mice as our analysis did not include the Exo1−/− mice that had died in the first 16 mo of the survival study because of the development of tumors. Histopathological examination of the tumors showed that the majority of Exo1−/− mice developed lymphoma (69.2%, 18 out of 26 Exo1−/− mice). This incidence in lymphoma in Exo1−/− mice was significantly higher than those observed in Exo1+/− mice (29.6%, 8/27 mice; p = 0.0060, Exo1−/− vs. Exo1+/−) or wild-type mice (20.8%, 5/24 mice; p = 0.0008, Exo1−/− vs. wild-type). In addition, Exo1−/− mice also developed other tumors at lower frequency, including gastrointestinal adenoma (5/26 mice), bronchioalveolar adenoma (5/26 mice), uterine adenocarcinoma (1/26 mice), ovarian adenocarcinoma (1/26 mice), coagulating gland adenoma (1/26 mice), hepatic adenoma (1/26 mice), hemangiosarcoma (1/26 mice), and schwannoma (1/26 mice). However, similar tumor types were also observed at similar incidences in Exo1+/− and wild-type mice. For example, the incidence of gastrointestinal tumors did not differ between Exo1−/− and wild-type mice (p = 0.1919). A significant number of Exo1−/− mice (42.3%, 11/26 mice) also developed two or more different types of tumor at the same time, whereas only 11% (3/27) of Exo1+/− mice and none (0/24) of the wild-type mice developed more than one different tumor type (p = 0.0135, Exo1−/− vs. Exo1+/−; p = 0.0003, Exo1−/− vs. wild-type). These data suggest that Exo1 inactivation predisposes mice to the development of tumors late in life and specifically increases the risk of lymphoma.
Meiotic defects in mutant males
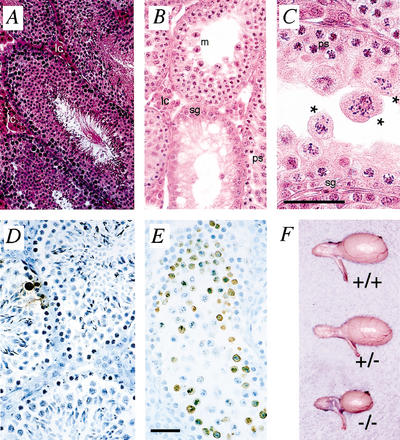
Exo1−/− male and female mice were infertile when mated with wild-type mates. Paraffin-embedded 5-μm sections of adult testis revealed an abnormal seminiferous epithelium in Exo1−/− males compared with Exo1+/+ littermates. In wild-type males, the seminiferous epithelium displayed a range of spermatogenic cells beginning at the basement membrane with the primary spermatogonia and extending in toward the tubular lumen as spermatogenesis proceeded (Fig. 4A). Within the lumen, tails of mature spermatozoa (s) were evident, indicating full completion of spermatogenesis within these tubules (Fig. 4A). In contrast, the seminiferous tubules of Exo1−/− males are severely depleted in spermatogenic cells (Fig. 4B,C). Spermatogonia and spermatocytes are visible proximal to the basement membrane within the tubules of Exo1−/− testes (dark pink cells, sg; punctate staining cells, ps), and appear to progress through pachynema in a normal fashion, as demonstrated by the appearance of pachytene spermatocytes (Fig. 4C, ps) and metaphase cells (Fig. 4B, m). By metaphase, however, spermatocytes from Exo1−/− males display abnormal spindle structures, with chromosomes misaligned across the spindles (Fig. 4C, asterisks). As a result, most of the Exo1−/− spermatocytes become apoptotic at metaphase, as evidenced by significantly increased numbers of TUNEL-positive cells within the lumen of the seminiferous tubules in Exo1−/− testes compared with what is seen in Exo1+/+ males (Fig. 4D,E). Despite the high numbers of apoptotic and abnormal metaphase cells, some cells from Exo1−/− males do progress through to meiosis II as indicated by a very small number of nonmotile spermatozoa that can be retrieved from the epididymides of Exo1−/− adult males. Epididymal sperm counts revealed that Exo1−/− adult males had ∼50 spermatozoa per male compared with 8 × 107 spermatozoa per wild-type male. The testis size in Exo1−/− adult males is therefore only about 30%–50% of those in wild-type and heterozygote littermates (Fig. 4F).
Figure 4.
Testis morphology in males. (A–C) Hematoxylin and eosin staining of testis sections from Exo1+/+ (A) and Exo1−/− (B,C) adult males. Note the unusual spindle structures within the seminiferous tubules of Exo1−/− males (C, asterisks). (D,E) TUNEL staining to detect apoptotic cells (brown precipitate) in Exo1+/+ (D) and Exo1−/− (E) adult males. (E) Note the increased level of apoptosis in the testes of Exo1−/− males. (F) Comparison of testis size in males of different Exo1 genotypes. lc, leydig cells; m, metaphase I cells; ps, pachytene spermatocytes; sg, spermatogonia; s, spermatozoa. Bars, 50 μm.
Normal meiotic progression from leptotene through pachytene in Exo1 mutant males
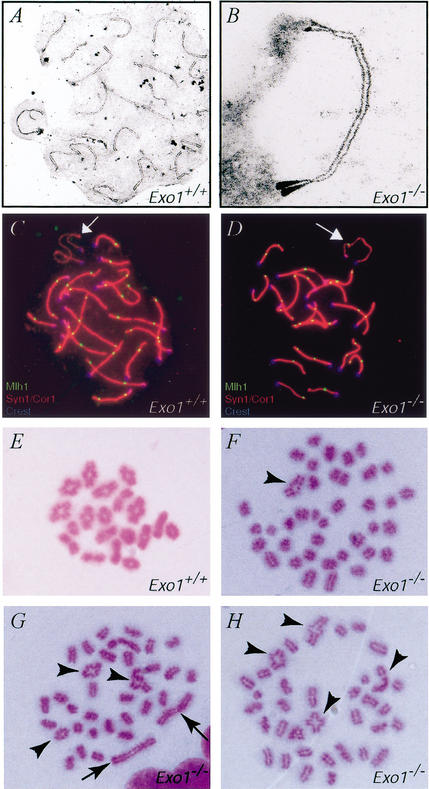
We monitored meiotic progression in adult Exo1+/+ and Exo1−/− males using various markers that are expressed specifically at different time points of meiotic prophase I. Rad51 was used as a positive marker for leptotene and zygotene cells as well as a negative marker for pachytene cells. We found that Rad51 localized normally to unpaired chromosomes in leptotene and zygotene spermatocytes from Exo1−/− mice and that Rad51 foci were lost at pachynema in a normal fashion in these animals (data not shown). Progression through prophase I up until pachynema was also assessed by accumulation of the synaptonemal complex proteins Syn1/Scp1 and Cor1/Scp3 (data not shown). To confirm these findings, we used electron microscopy to examine silver-stained chromosomal spreads and found that spermatocytes from Exo1−/− mice showed fully synapsed bivalents at pachynema (Fig. 5B), a normal pachytene chromosomal configuration. In addition, the localization of Mlh1, another midpachytene marker that represents sites of crossing over, showed that in spermatocyte spreads from both Exo1+/+ and Exo1−/− males, Mlh1 was found at a frequency and localization pattern that mirrored the predicted sites/frequency of crossovers in wild-type mice, with ∼26–29 foci per nucleus (Fig. 5C,D). Moreover, Mlh1 foci were consistently localized to the pseudoautosomal region (PAR) of the XY bivalent, an obligate crossover site (Fig. 5C,D, arrows). The localization of multiple meiotic markers such as Cor1/Scp3, Syn1/Scp1, Rad51, and Mlh1 at specific times throughout prophase I and the complete synapsis observed at the electron microscopic level indicate that meiosis proceeds normally until pachynema in Exo1−/− males.
Figure 5.
Chromosome pairing and synapsis during meiosis I. (A,B) electron micrographs of silver-stained chromosome spreads from Exo1+/+ (A) and Exo1−/− (B) spermatocytes, showing normal pairing and synapsis at pachynema. (C,D) Correct spatial and temporal localization of Mlh1 (green, FITC signal) on meiotic chromosomes from Exo1+/+ (C) and Exo1−/− (D) mice stained with a dual antibody against Cor1 and Syn1 and TRITC/red secondary antibody. The centromere is marked with human CREST antisera (blue, Cy5 signal). (E–H) Air-dried chromosome preparations from Exo1+/+ (E) and Exo1−/− (F–H) spermatocytes, showing abnormal metaphase configurations in the absence of Exo1. Although some crossovers remain (arrowheads), the majority of chromosomes are either univalents or appear to be achiasmate bivalents (arrows).
Abnormal metaphase I in Exo1 mutant males
To further study the meiotic defect in males, we examined the meiotic events in the germ cells beyond prophase I. Air-dried giemsa-stained chromosomal preparations revealed 20 pairs of bivalent chromosomes in spermatocytes from Exo1+/+ males, with the normal appearance and frequency of chiasmata for each chromosome pair (Fig. 5E). In contrast, similar preparations from Exo1−/− mutant spermatocytes revealed mainly univalent chromosomes, with some bivalents. Quantitative analysis of metaphase I chromosome preparations from 40 spermatocyte nuclei from Exo1−/− male mice revealed that, among 1600 chromosomes scored, >75% were univalents (Fig. 5F–H), instead of the expected chiasmate bivalents. The remaining chromosomes were bivalents. Of these bivalents, 17% were achiasmate (completely lacking chiasmata; Fig. 5G, arrows), 15% were abnormal in appearance, and the rest appeared normal (Fig. 5F–H, arrowheads). The extensive occurrence of these chromosomal configurations, together with the abnormal metaphase figures observed at the histological level (Fig. 4C), indicate that these achiasmate and abnormal bivalents undergo premature separation before aligning at the metaphase plate. The heterogeneity of chromosomal configurations observed within and between each nucleus suggests a dynamic loss of chiasmata specifically at metaphase I that results in checkpoint activation and apoptosis (Fig. 4E).
Meiotic phenotypes in Exo1 mutant females
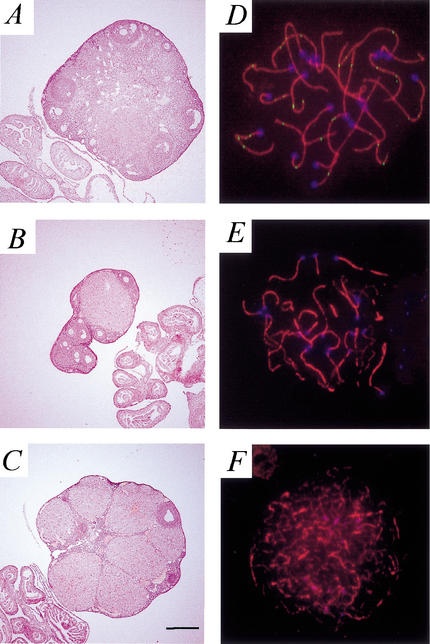
To examine the ovarian phenotype of Exo1−/− females, ovaries were prepared for routine histology. Ovaries of Exo1−/− adult females contained follicles of various sizes, ranging from primordial to antral follicles and postovulatory corpora lutea (Fig. 6B,C). At 10 wk of age, ovaries from Exo1+/+ and Exo1−/− females are similar in appearance and content (data not shown). In older Exo1−/− females (at 7 mo of age), however, the ovarian size is often smaller than that of age-matched wild-type littermates (Fig. 6A–C). At these older ages, there is considerable heterogeneity in both the size and morphology of the ovaries from Exo1−/− females, both within the same animal (Fig. 6B,C are a pair of ovaries from one animal) and between age-matched females.
Figure 6.
Ovarian morphology and meiotic progression in Exo1−/− females. (A–C) Gross morphology of the ovaries from Exo1+/+ (A) and Exo1−/− (B,C) female mice at 7 mo of age. Note the loss of oocytes and the smaller size of the ovary from Exo1−/− females. Oviductal and uterine structures look normal in the absence of Exo1. Magnification: A–C, 16×. (D–F) Chromosome spreads from oocytes taken at day 2 pp from Exo1−/− pups. Anti-Cor1 antibody is shown in red (TRITC), anti-Dmc1/Rad51 antibody is in green (FITC), and CREST autoimmune serum, indicating the centromeres, is in blue (CY5). Oocytes from Exo1−/− females progress through early pachynema, as demonstrated by complete synapsis of chromosomes and residual Dmc1/Rad51 foci (D), and into early diplotene (E), as demonstrated by the single cores with large regions of chromosomes remaining synapsed and the absence of Dmc1/Rad51 foci. (F) These oocytes then enter dictyate arrest, as demonstrated by the gradual degradation of chromosomal cores, but persistence of Cor1 and the clustering of centromeres.
Oocyte progression through meiotic prophase I was assessed by immunofluorescence of chromosomal spreads from Exo1−/− female pups at day 2 postpartum (pp), at which time oocytes should have reached the late prophase I to dictyate stages. As in male germ cells, chromosomes appear to synapse normally to achieve a complete pachytene configuration, as demonstrated by localization of Cor1 and Dmc1 (a meiosis-specific RecA homolog that colocalizes with Rad51; Fig. 6D). Oocytes progress normally through diplonema in the absence of functional Exo1 (Fig. 6E) and were found a few days after birth to be in dictyate arrest (Fig. 6F). These oocytes arrive in dictyate in the absence of any checkpoint activation, thus allowing them to progress through early follicular development stages, a situation similar to that seen in Mlh1−/− females.
Discussion
We have generated a mouse line with an inactivation mutation in Exonuclease 1. This mouse line carries a mutation that results in the deletion of exon 6 in the Exo1 mRNA, thereby eliminating the majority of amino-acid residues that encode the I-nuclease domain. As a result, extracts from Exo1−/− ES cells failed to support either 5′- or 3′-nick-directed MMR, presumably because of loss of the exonuclease activity. Similarly, the genetic analysis of yeast strains carrying an equivalent EXO1 deletion also demonstrates that this mutation results in loss of function. The functional importance of this nuclease domain has also been demonstrated in studies in which mutations altering conserved amino acid residues within this domain were tested for their effect on either exonuclease activity or biological function of Exo1 (Sokolsky and Alani 2000; Lee et al. 2002; Lewis et al. 2002). Therefore, the disruption of this critical domain in the Exo1 mutant mice is likely to cause inactivation of its exonuclease activity.
The role of Exo1 in 5′-nick-directed mismatch excision observed here is consistent with its known 5′–3′ double-stranded exonuclease activity. However, the role for Exo1 in 3′-nick-directed MMR is surprising. Similar findings were recently reported for human EXO1 in biochemical experiments studying mismatch-provoked excision directed by a single strand nick located either 5′ or 3′ to the mispair (Genschel et al. 2002). In that report, fractionated cell extracts were used to demonstrate that the 3′-nick-directed excision reaction required EXO1 as well as both the MutSα and MutLα complexes, whereas the 5′-nick-directed excision reaction required EXO1 and MutSα but not MutLα. The requirement of MutLα in the 3′-excision reaction combined with the demonstration that EXO1 physically interacts with MLH1 (Schmutte et al. 2001; Tran et al. 2001) and the postulated role of EXO1 in stabilizing higher-order MMR protein complexes (Amin et al. 2001) are consistent with the idea that MutLα plays an important role in recruiting and coordinating the protein components of MMR. It is possible that Exo1 plays an MutLα-dependent structural role in the assembly of the 3′-excision complex. It may also be possible that Exo1 has 3′–5′ exonuclease activity, although such an activity has not been demonstrated. An alternative explanation for the involvement of Exo1 in 3′-directed excision is that the 3′–5′ exonuclease activities that function in 3′-directed excision might not be able to efficiently initiate excision at nicks but rather act more efficiently at gaps. In such a scenario, Exo1 could function to generate a gap that is required for these exonuclease activities.
Our in vitro biochemical studies showed that Exo1 participates in the repair of base:base mismatches or single-base IDLs, but not in the repair of dinucleotide IDLs (Fig. 2). Furthermore, inactivation of Exo1 resulted in a significant increase in MSI at a mononucleotide repeat marker, but not at dinucleotide repeat markers (Table 1). A similar although less striking requirement for Exo1 in the repair of base:base and single-base IDL mismatches was also detected in yeast in assays that detect mutations in the CAN1 gene and in those that detect frameshift mutations in homonucleotide runs located within the HOM3, LYS2, or HIS7 genes. These studies showed that inactivation of EXO1 by itself caused a significant increase in both base-substitution mutations in CAN1 and in −1 frameshift mutations in mononucleotide runs, and that loss of Exo1 greatly enhanced +1 frameshift mutations in A5, A7, and A12 homonucleotide runs in yeast mutants that were also defective in DNA polymerase ε (Tishkoff et al. 1997; Tran et al. 1999, 2001; Kirchner et al. 2000; Amin et al. 2001). Together with the observations in yeast, our results in mice indicate that Exo1 participates in the repair of single-base mismatches or single-base IDLs. They also suggest that Exo1 functions in an Msh2–Msh6-dependent repair pathway. This notion of a specific involvement of Exo1 in the Msh2–Msh6-mediated repair of base–base mismatches is supported by the direct interaction between MSH2 and EXO1 in yeast and human and the requirement for MutSα in EXO1-mediated excision repair (Genschel et al. 2002). It is now possible to test this model at the organismal level by generating mouse lines that are double mutant in Exo1 and various other MMR genes.
The MMR defect in Exo1 mutant mice caused a reduction in survival and increased cancer susceptibility. A comparison with MMR mutant mice that carry mutations in MMR genes with which Exo1 is known to interact shows that the cancer phenotype of Exo1−/− mice is less severe. Whereas the 50% survival rate of Msh2−/− or Mlh1−/− mice is typically at 6 mo of age (Reitmair et al. 1996; de Wind et al. 1998; Prolla et al. 1998; Edelmann et al. 1999b; Smits et al. 2000), it took 17 mo for half of the Exo1−/− mice to die. We specifically observed a significant increase in the development of lymphomas in Exo1−/− mice, whereas Msh2−/− or Mlh1−/− mutant mice also developed significant numbers of intestinal tumors and skin tumors in addition to lymphomas. In contrast, the phenotype of Exo1−/− mutant mice is significantly more severe than in Msh3−/− mutant mice, which do not show an increase in cancer predisposition or a reduction in survival (Edelmann et al. 2000). It is important to note that the difference in the severity in the cancer phenotype between Exo1 and Msh2 mutant mice correlates well with the differences in the strength of the mutator phenotype that we observed at the Hprt locus in these mice (Table 2).
Recent studies by Wu et al. (2001) and Jagmohan-Changur et al. (2003) identified a small number of EXO1 germ-line missense variants in patients with suspected HNPCC that were not found in normal controls. In addition, tumors from some of these patients displayed MSI. These results raise the possibility of a role for EXO1 in gastrointestinal tumorigenesis (Wu et al. 2001; Jagmohan-Changur et al. 2003). However, only a small proportion of Exo1−/− mice developed gastrointestinal adenomas and the incidence was similar to wild-type control mice. In addition, the tumors from Exo1−/− mice did not display MSI at dinucleotide repeat markers (data not shown). In contrast, mouse models for HNPCC, such as Msh2 and Mlh1 mutant mice, develop not only lymphoma but also a significant number of gastrointestinal tumors that typically display MSI (Reitmair et al. 1996; de Wind et al. 1998; Prolla et al. 1998; Edelmann et al. 1999b). Our results indicate that Exo1 does not play a significant role in the development of gastrointestinal tumors and support the view that mutations in human EXO1 might only rarely be a cause of HNPCC (Jagmohan-Changur et al. 2003).
Our study reveals an essential role of Exo1 in mammalian meiosis, as first suggested by the high expression levels of Exo1 mRNA in testis (Lee et al. 1999; data not shown). Inactivation of Exo1 results in sterility in both males and females. The meiotic defects of Exo1−/− mice are distinct from those of other MMR mutant mice. Exo1−/− mice display normal meiotic progression through pachynema, as demonstrated by the expression of various meiotic markers during the leptotene and pachytene stages and by the full synapsis of chromosomes at pachynema. In contrast, mice lacking Msh4, Msh5, or Pms2 display varying defects of synapsis, ranging from almost complete synaptic failure at zygonema (Msh4−/− and Msh5−/− mice) to incomplete or promiscuous synapsis between nonhomologous chromosomes (Pms2−/− mice; Baker et al. 1995; Edelmann et al. 1999a; Kneitz et al. 2000), suggesting that Exo1 acts at later meiotic processes than these three proteins. On the other hand, the meiotic phenotypes of Exo1−/− mice closely resemble those observed in Mlh1−/− and Mlh3−/− mice, especially with regard to the full synapsis of chromosomes at pachynema (Baker et al. 1996; Edelmann et al. 1996; Lipkin et al. 2002). Studies by Eaker et al. (2002) demonstrated that although no Mlh1 foci were present on pachytene chromosomes in Mlh1−/− mice, apoptosis was not initiated in prophase I but rather was detected in metaphase. The presence of abnormal chromosomal figures in metaphase suggests that the spermatocytes in Mlh1−/− mice are not eliminated in direct response to the initial meiotic defect, which occurs in late prophase, but at a later stage of meiosis I (Eaker et al. 2002). To date, two meiotic checkpoints have been described: a homology search checkpoint at zygonema, which is the cause of the meiotic arrest in Dmc1, Msh4, and Msh5 mutant mice (Pittman et al. 1998; Yoshida et al. 1998; Edelmann et al. 1999a; Kneitz et al. 2000), and the metaphase checkpoint that prevents cells from exiting metaphase I until all chromosome pairs have been properly oriented at the metaphase plate. The finding by Eaker et al. (2002) suggests that the loss of spermatocytes in Mlh1−/− mice is caused by the activation of a spindle assembly checkpoint that is downstream of the meiotic defect in these cells. The normal localization of Mlh1 in pachytene spermatocytes of Exo1−/− mice (Fig. 5D) suggests that the meiotic function of Exo1 is required either at a similar stage of meiosis as Mlh1 or at a later time point. It also indicates that Mlh1 localization at pachynema is not the only factor in ensuring the persistence and stability of chiasmata until metaphase. In addition, the meiotic defect in Exo1−/− spermatocytes appears to activate the same spindle assembly checkpoint leading to apoptosis, with the loss of spermatocytes occurring dynamically at the metaphase I stage. This results in a dramatic decrease in metaphase cells in the seminiferous tubules of Exo1−/− males, although some remaining abnormal cells can still be detected (Fig. 4B,C). Although studies in yeast have demonstrated that inactivation of EXO1 resulted in increased meiosis I nondisjunction, decreased spore viability, and decreased crossing-over (Khazanehdari and Borts 2000; Kirkpatrick et al. 2000), these studies did not identify the specific function for EXO1 in meiosis. Therefore, it will be important to further characterize the function of Exo1 in mammalian meiosis, and this mouse line provides a valuable tool for such studies.
Materials and methods
Generation of Exo1−/− mice
A probe corresponding to exon 6 of mouse Exo1 was used to screen the RPCI-22 129S/SvEvTac mouse genomic BAC library. Positive clones were identified and used to generate the Exo1 targeting vector. The targeting vector was constructed by inserting a PGKhygromycin cassette into the EcoRV site in exon 6 contained within a 5-kb EcoRI fragment that was previously cloned into pBluescript SK vector. Next, an 8-kb HindIII–ApaI fragment containing intron 6 and a region spanning exons 7–11 was cloned into the HindIII and ApaI sites 3′ to the PGKhygromycin resistance cassette (Fig. 1A). The targeting vector, designated pExo1ex6, was linearized at the single NotI site and electroporated into 3 × 107 WW6 ES cells according to standard protocols. After 10 d of selection in hygromycin B (200 μg/mL), DNA was isolated from resistant clones and screened by PCR. Three positive clones were identified, and the correct gene-targeting event was verified by Southern blot analysis. Two of the targeted clones were injected into C57BL/6J blastocysts to produce chimeric animals that transmitted the targeted allele through their germ lines. F1 heterozygote offspring were intercrossed to generate F2 homozygous mutant Exo1 mice.
Northern blot and RT–PCR analysis
Five micrograms of poly(A) RNA from Exo1 ES cell lines was separated in 1.0% formaldehyde agarose gels and transferred to nitrocellulose membranes. The membranes were subsequently hybridized with two probes corresponding to exon 6 and exons 7–14, respectively. To verify the Northern blot results, RT–PCR using primers located in exon 5 (forward: 5′-GCCTCGTGGC TCCGTATG-3′) and exon 7 (reverse: 5′-GCGTAAGTCAGTGT TTCGGG-3′) was performed on total RNA isolated from a variety of tissues in wild-type and Exo1 mutant animals. The mutant RT–PCR products were further analyzed by DNA sequencing.
Western blot analysis
Equal amounts of protein from ES cell extracts were separated on 10% SDS-PAGE gels and transferred onto Immobilon-P (Millipore) PVDF membranes. The membranes were blocked in 5% nonfat dry milk in TBS-T and incubated with a rabbit polyclonal antibody raised against codons 786–837 at the C-terminal region of mouse Exo1. Exo1 protein was detected by chemiluminescence after incubation with a secondary donkey anti-rabbit IgG antibody conjugated with horseradish peroxidase conjugate (Amersham).
Genetic analysis of exo1 mutant yeast strains
The S. cerevisiae strains used for complementation testing were RKY3590 MATa ura3-52 leu2Δ trp1Δ63 hom3-10 lys2∷InsE-A10 and two isogenic derivatives RKY4168 exo1∷URA3 and RKY4177 exo1∷URA3 pms1-A130V. The low-copy plasmids used were pRS315 ARS CEN LEU2, a derivative containing the cloned EXO1 gene and 2 kb of upstream sequence containing the promoter; pRDK1020 ARS CEN LEU2 EXO1; and a related derivative, pRDK1021 ARS CEN LEU2 exo1Δ6, in which EXO1 codons 182–255 were deleted. The high-copy plasmids used were pRS425 2μ LEU2 and derivatives of this vector, pRDK1022 2μ LEU2 EXO1 and pRDK1023 2μ LEU2 exo1Δ6, that contain the same EXO1 fragments as pRDK1020 and pRDK1021, respectively. All genetic methods for evaluating the effect of plasmids on the mutation rate of these strains have been described previously (Amin et al. 2001).
Cell-free extracts and mismatch repair assay
Procedures for extract and heteroduplex substrate preparation and for measuring repair activity were as described (Thomas et al. 1995). The substrates used are described in the legend to Figure 2.
Histopathological analysis of tumors
Tumors were removed from the mice and fixed in 10% neutral buffered formalin. All tumors were processed for paraffin embedding, and sections were prepared for staining with hematoxylin and eosin. Statistical analyses of tumor incidence were conducted by the Fisher's exact test.
Mutation rate analysis
Male ES cells were grown in HAT medium and subsequently plated at a density to obtain colonies initiated from single cells. Between 9 and 11 colonies were expanded sufficiently for 2 × 106 cells from each culture to be plated at a density of 105 cells per 90-mm plate. After 24 h, the medium was replaced with 6TG containing (50 μM) medium, which was replaced every 2 d. After 14 d, plates were stained, and the colonies were counted.
Mutation rates were calculated using the method of the median (Wierdl et al. 1996). In calculating mutation rates, the number of 6TG-resistant clones was not adjusted for plating efficiency because Abuin et al. (2000) reported that the plating efficiency of ES cells at low density provides an unreliable estimate of the plating efficiency at higher densities. Because we observed no gross differences between plating efficiencies at moderate-to-high densities, we performed the mutation rate analyses using the rationale of Abuin et al. (2000). Although the mutation rate will be slightly underestimated without adjusting for plating efficiency, this will not affect relative rates between different cell lines.
Microsatellite instability analysis
Tail DNAs from three Exo1−/− and wild-type mice were pooled and diluted to approximately one genome equivalent per reaction. The microsatellite loci were amplified using end-labeled primers and analyzed by electrophoresis on denaturing 6% polyacrylamide gels as previously described (Wong et al. 2002). Statistical analyses were performed according to the Fisher's exact test.
Histological analysis of reproductive organs
Ovaries and testes from adult mice were fixed in Bouin's fixative or 4% buffered formalin and processed for immunohistochemistry by routine methods. The paraffin-embedded tissues were sectioned at 5 μm. For analysis of prophase I chromosome configurations and protein localization, spermatocyte and oocyte spreads were prepared and analyzed according to previously described methods (Edelmann et al. 1999a; Kneitz et al. 2000; Lipkin et al. 2002). For these meiotic analyses, the testes were removed from adult Exo1+/+ and Exo1−/− males, and ovaries were removed from females at day 2 pp.
Acknowledgments
We are grateful to the following colleagues for generously providing antibodies and advice: Peter Moens (York University, Ontario, Canada; antisynaptonemal-complex antibodies), and Bo Jin and Marie Lia for technical assistance. This work is supported by NIH grants (CA 76329 and CA93484 to W.E., GM50006 to R.D.K.) and a Cancer Center grant to Albert Einstein College of Medicine (CA13330).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL edelmann@aecom.yu.edu; FAX (718) 430-8776.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1060603.
References
- Abuin A, Zhang H, Bradley A. Genetic analysis of mouse embryonic stem cells bearing Msh3 and Msh2 single and compound mutations. Mol Cell Biol. 2000;20:149–157. doi: 10.1128/mcb.20.1.149-157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-dependent mutator mutations: Model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21:5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Bronner CE, Zhang I, Plug A, Robatzek M, Warren G, Elliott EA, Yu J, Ashley T, Arnheim N, et al. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82:309–320. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc Natl Acad Sci. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DL, Lahue RS, Modrich P. Methyl-directed mismatch repair is bidirectional. J Biol Chem. 1993;268:11823–11829. [PubMed] [Google Scholar]
- Dao V, Modrich P. Mismatch-, MutS-, MutL-, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J Biol Chem. 1998;273:9202–9207. doi: 10.1074/jbc.273.15.9202. [DOI] [PubMed] [Google Scholar]
- de Wind N, Dekker M, van Rossum A, van der Valk M, te Riele H. Mouse models for hereditary nonpolyposis colorectal cancer. Cancer Res. 1998;58:248–255. [PubMed] [Google Scholar]
- Dixon WJ, Massey FJ. Introduction to statistical analysis. 3rd ed. New York: McGraw-Hill; 1968. pp. 439–443. [Google Scholar]
- Eaker S, Cobb J, Pyle A, Handel MA. Meiotic prophase abnormalities and metaphase cell death in MLH1-deficient mouse spermatocytes: Insights into regulation of spermatogenic progress. Dev Biol. 2002;249:85–95. doi: 10.1006/dbio.2002.0708. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kneitz B, Winand N, Lia M, Heyer J, Kolodner R, Pollard JW, Kucherlapati R. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet. 1999a;21:123–127. doi: 10.1038/5075. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Yang K, Kuraguchi M, Heyer J, Lia M, Kneitz B, Fan K, Brown AM, Lipkin M, Kucherlapati R. Tumorigenesis in Mlh1 and Mlh1/Apc1638N mutant mice. Cancer Res. 1999b;59:1301–1307. [PubMed] [Google Scholar]
- Edelmann W, Umar A, Yang K, Heyer J, Kucherlapati M, Lia M, Kneitz B, Avdievich E, Fan K, Wong E, et al. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer Res. 2000;60:803–807. [PubMed] [Google Scholar]
- Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- Grilley M, Griffith J, Modrich P. Bidirectional excision in methyl-directed mismatch repair. J Biol Chem. 1993;268:11830–11837. [PubMed] [Google Scholar]
- Huang KN, Symington LS. A 5′–3′ exonuclease from Saccharomyces cerevisiae is required for in vitro recombination between linear DNA molecules with overlapping homology. Mol Cell Biol. 1993;13:3125–3134. doi: 10.1128/mcb.13.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagmohan-Changur, S., Poikonen, T., Vilkki, S., Launonen, V., Wikman, F., Orntoft, T.F., Moller, P., Vasen, H., Tops, C., Kolodner, R.D., et al. 2003. EXO1 variants occur commonly in normal population: Evidence against a role in hereditary nonpolyposis colorectal cancer. Cancer Res. (in press). [PubMed]
- Khazanehdari KA, Borts RH. EXO1 and MSH4 differentially affect crossing-over and segregation. Chromosoma. 2000;109:94–102. doi: 10.1007/s004120050416. [DOI] [PubMed] [Google Scholar]
- Kirchner JM, Tran H, Resnick MA. A DNA polymerase ε mutant that specifically causes +1 frameshift mutations within homonucleotide runs in yeast. Genetics. 2000;155:1623–1632. doi: 10.1093/genetics/155.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DT, Ferguson JR, Petes TD, Symington LS. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics. 2000;156:1549–1557. doi: 10.1093/genetics/156.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H, Jr, Kolodner RD, Kucherlapati R, Pollard JW, Edelmann W. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes & Dev. 2000;14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes & Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- Lee B, Wilson DM., III The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- Lee B, Shannon M, Stubbs L, Wilson DM., III Expression specificity of the mouse exonuclease 1 (mExo1) gene. Nucleic Acids Res. 1999;27:4114–4120. doi: 10.1093/nar/27.20.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Nguyen LH, Barsky D, Fernandes M, Wilson DM., III Molecular interactions of human Exo1 with DNA. Nucleic Acids Res. 2002;30:942–949. doi: 10.1093/nar/30.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Karthikeyan G, Westmoreland JW, Resnick MA. Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase) Genetics. 2002;160:49–62. doi: 10.1093/genetics/160.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes & Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes & Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. Deficient DNA mismatch repair: A common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- Peltomaki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: Database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- Prolla TA, Pang Q, Alani E, Kolodner RD, Liskay RM. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- Prolla TA, Baker SM, Harris AC, Tsao JL, Yao X, Bronner CE, Zheng B, Gordon M, Reneker J, Arnheim N, et al. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet. 1998;18:276–279. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- Qiu J, Guan MX, Bailis AM, Shen B. Saccharomyces cerevisiae Exonuclease-1 plays a role in UV resistance that is distinct from nucleotide excision repair. Nucleic Acids Res. 1998;26:3077–3083. doi: 10.1093/nar/26.13.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitmair AH, Redston M, Cai JC, Chuang TC, Bjerknes M, Cheng H, Hay K, Gallinger S, Bapat B, Mak TW. Spontaneous intestinal carcinomas and skin neoplasms in Msh2-deficient mice. Cancer Res. 1996;56:3842–3849. [PubMed] [Google Scholar]
- Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58:4537–4542. [PubMed] [Google Scholar]
- Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem. 2001;276:33011–33018. doi: 10.1074/jbc.M102670200. [DOI] [PubMed] [Google Scholar]
- Smits R, Hofland N, Edelmann W, Geugien M, Jagmohan-Changur S, Albuquerque C, Breukel C, Kucherlapati R, Kielman MF, Fodde R. Somatic apc mutations are selected upon their capacity to inactivate the β-catenin downregulating activity. Genes Chromosomes Cancer. 2000;29:229–239. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1033>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Sokolsky T, Alani E. EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae. Genetics. 2000;155:589–599. doi: 10.1093/genetics/155.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- ————— A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- Thomas DC, Umar A, Kunkel TA. Measurement of heteroduplex repair in human cell extracts. In: Friedberg EC, editor. Methods: A companion to methods in enzymology. Vol. 7. San Diego, CA: Academic Press; 1995. pp. 187–197. [Google Scholar]
- Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998;58:5027–5031. [PubMed] [Google Scholar]
- Tran HT, Gordenin DA, Resnick MA. The 3′ → 5′ exonucleases of DNA polymerases δ and ε and the 5′ → 3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Simon JA, Liskay RM. Interactions of Exo1p with components of MutLα in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2001;98:9760–9765. doi: 10.1073/pnas.161175998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Risinger JI, Glaab WE, Tindall KR, Barrett JC, Kunkel TA. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics. 1998;148:1637–1646. doi: 10.1093/genetics/148.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M, Burdett V, Baitinger C, Modrich P, Lovett ST. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J Biol Chem. 2001;276:31053–31058. doi: 10.1074/jbc.M105481200. [DOI] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N. Functional specificity of MutL homologs in yeast: Evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierdl M, Greene CN, Datta A, Jinks-Robertson S, Petes TD. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics. 1996;143:713–721. doi: 10.1093/genetics/143.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, Fazzari M, Jin B, Brown AM, Lipkin M, et al. Mbd4 inactivation increases C→T transition mutations and promotes gastrointestinal tumor formation. Proc Natl Acad Sci. 2002;99:14937–14942. doi: 10.1073/pnas.232579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Berends MJ, Post JG, Mensink RG, Verlind E, Van Der Sluis T, Kempinga C, Sijmons RH, van der Zee AG, Hollema H, et al. Germline mutations of EXO1 gene in patients with hereditary nonpolyposis colorectal cancer (HNPCC) and atypical HNPCC forms. Gastroenterology. 2001;120:1580–1587. doi: 10.1053/gast.2001.25117. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Dao V, Modrich P. MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J Biol Chem. 1998;273:9197–9201. doi: 10.1074/jbc.273.15.9197. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]