Abstract
RAG1 and RAG2 initiate V(D)J recombination, which is the assembly of immunoglobulin and T cell receptor genes. The N-terminal region of RAG1 can be deleted, leaving an enzymatic “core” able to catalyze the complete reaction. Here we report that the N-terminal portion of RAG1 has a distinct enzymatic role separate from the rest of the protein. It acts as an E3 ligase in the ubiquitylation of a test substrate and formation of polyubiquitin chains in vitro. This finding suggests a new way in which V(D)J recombination can be regulated and coupled to other aspects of cell physiology.
Keywords: RAG1, ubiquitin, E3 ligase, in vitro assay
V(D)J recombination is a process through which DNA segments are cleaved, shuffled, and then religated to assemble functional immunoglobulin and T cell receptor loci. The process is required for the function of the adaptive immune response and provides a large degree of diversity needed by the antigen receptors. Recombination must also be highly regulated in terms of its DNA target specificity, developmental stage, and coordination with other nuclear events such as cell cycle. Undesired consequences of aberrant recombination include translocations like those associated with several leukemias (Bassing et al. 2002; Gellert 2002).
The first enzymatic step in V(D)J recombination is the binding and then cleavage of the DNA substrate by a complex composed of the RAG1 and RAG2 proteins. These two proteins are the only known lymphoid-specific factors required for the reaction, and when expressed in nonlymphoid cells, they confer the ability to rearrange a test substrate (Oettinger et al. 1990). It came with some surprise to learn that the two proteins could be substantially truncated and yet remain functional in recombination (Sadofsky et al. 1993, 1994; Silver et al. 1993; Cuomo and Oettinger 1994; Kirch et al. 1996). These smaller “core” portions of the RAG proteins have been easier to express and purify, allowing mechanistic studies of the cleavage reaction in vitro. These core proteins can participate in the formation of higher-order DNA–protein complexes, leading to synapsis and coupled cleavage of pairs of recombination targets (Bailin et al. 1999; Landree et al. 2001; Sadofsky 2001; Jones and Gellert 2002; Mundy et al. 2002; Swanson 2002). A linear representation of the RAG1 protein is shown as Figure 1A.
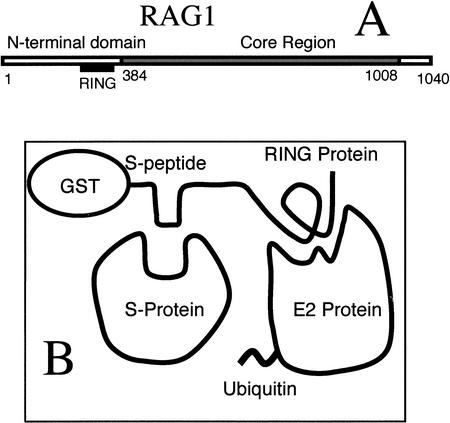
Figure 1.
(A) A linear representation of the RAG1 protein. The core region is sufficient for V(D)J recombination in cells. The N-terminal domain contains the RING motif. (B) The experimental design. The N-terminal domain is engineered into a fusion protein with GST and the S-peptide. S-protein will bind the S-peptide. If a ubiquitin-charged E2 protein interacts with the RING construct, ubiquitin will be transferred to the S-protein.
After the cleavage step, the RAG proteins participate in the joining phase of the recombination reaction. This requires at least a structural contribution to binding the four DNA ends (Landree et al. 2001; Mo et al. 2001; Tsai et al. 2002) and may also reflect interactions with the DNA repair proteins needed for processing. Thus, the core region of RAG1 is apparently sufficient for all the DNA transactions in which the enzyme participates.
Others have found that removal of the “dispensable” regions of the RAG proteins reduces the efficiency of the reaction (McMahan et al. 1997; Steen et al. 1999). In addition, the RAG proteins seem to have a role in lymphoid growth and regulation distinct from the nuclease function. Recombination of the heavy-chain locus in B cells typically involves D-to-J segment joining prior to recombination of the V segments. A mutant truncated in RAG2 is capable of catalyzing the first set of immunoglobulin rearrangements but not the later step (Kirch et al. 1998; Liang et al. 2002). It is also observed that a spontaneous frameshift mutation in RAG1 in a human patient with Omenn's syndrome leads to a more severe defect in B cell immunoglobulin rearrangement than at the T cell receptor locus (Santagata et al. 2000). These findings may reflect a role of the RAG proteins beyond their activity as a recombinase.
Sequence comparison of alignments of RAG1 from diverse species indicates that the core is significantly more conserved than the N-terminal 383 amino acid residues (Sadofsky 2001), leading us to speculate that this region may not be constrained by the same evolutionary pressures as the core. The most striking sequence feature in the N-terminal region is the absolute conservation among species of a cluster of cysteine and histidine residues. Almost all other positions show variation in sequence identity and even insertions (with respect to the mouse sequence) outside of this cluster. These conserved residues suggested a function as a zinc binding domain, and crystallization of the region, as an isolated 116-residue peptide, confirmed this and revealed dimerization behavior as well (Rodgers et al. 1996). Subsequent sequence comparison placed this domain within a subgroup of zinc binding motifs termed a “RING finger” (Aravind and Koonin 2000). This is a significant refinement, because it is increasingly apparent that the RING motif defines a growing subset of ubiquitin ligases, whose number may reach into the hundreds in the human genome (Jackson et al. 2000; Weissman 2001). It has been suggested, and remains to be disproved, that all RING-containing proteins may function in this capacity (Lorick et al. 1999; Fang et al. 2003).
Ubiquitin is a small (76-residue) protein that is highly conserved and universally expressed among all eukaryotes. It is used as a protein modifier, attached singly or in chains, to other proteins. The first function identified for ubiquitylation was as a proteosomal degradation signal. Ubiquitylation was subsequently found to influence, directly or indirectly, most aspects of cell biology including cell-cycle control, differentiation, and DNA repair (see Weissman 2001 and references therein). Ubiquitylation requires a cascade of enzymatic steps. First, free ubiquitin is bound through a thiol linkage to a ubiquitin-activating enzyme called E1. This step requires the hydrolysis of ATP. Next, the ubiquitin is transferred to a carrier protein, called an E2, using another thiol linkage. There are perhaps dozens of E2 family members. Finally, the ubiquitin is covalently joined to its target through the formation of an isopeptide bond at a free amino group. This last step requires a ubiquitin ligase, called an E3. There are several families of E3 enzymes, amounting to hundreds of proteins, of which the RING family is one. Specificity for the target protein is provided by the E3 or the combination of E2 and E3 proteins.
In this report we demonstrate that the N-terminal portion of RAG1, containing the RING motif, is capable of serving as an E3 ligase in vitro.
Results and Discussion
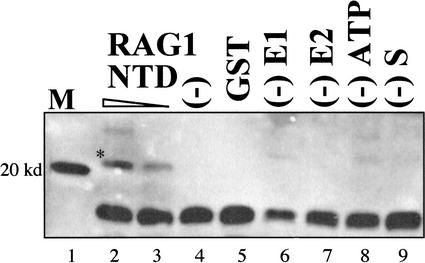
E3 ligases of the RING variety tether a substrate to the ubiquitin-charged E2 protein. The RING structure appears to both bind the E2 and activate the transfer of the ubiquitin. Demonstrating the putative E3 ligase activity for an uncharacterized protein is more challenging when a natural substrate is unknown. To demonstrate an enzymatic activity, we therefore adopted a strategy recently used to demonstrate the E3 ligase activity of two yeast RING-containing proteins, Hrd1p (Bays et al. 2001) and Doa10 (Swanson et al. 2001). The experimental design is presented in Figure 1B. A synthetic protein is assembled containing three components: GST (glutathione S-transferase), the 15-residue S-peptide, and the RING-containing protein. A separate protein, the S-protein, binds tightly to the S-peptide and is thereby targeted for ubiquitylation, provided that the RING interacts with an E2. The reaction requires the E1 enzyme, an E2, the RING-containing fusion protein, ATP, the S-protein target, and free ubiquitin. Modification of the S-protein is subsequently detected by immunoblotting using anti-ubiquitin antibody. A blot reflecting the transfer of ubiquitin to the S-protein is presented in Figure 2, as follows: Lane 1 is a 20-kD marker, which runs slightly faster than the ubiquitylated S-protein. Lane 2 is the complete reaction using 1 μg of the RAG1 N-terminal domain in the fusion construct. The asterisk highlights the ubiquitylated S-protein. Lane 3 represents the complete reaction, albeit containing only 0.3 μg of the fusion protein. Each of the remaining lanes is missing one of the necessary factors. There is no RING-containing fusion protein in lane 4, and GST alone in place of the fusion is shown in lane 5. Lane 6 lacks the E1 protein, lane 7 lacks the E2, lane 8 lacks ATP, and the S-protein was not included in the otherwise complete reaction in lane 9. This experiment demonstrates that the RAG1 N-terminal segment was capable of stimulating the transfer of ubiquitin in a dose-dependent manner. We note that this experiment, which is not presented in historical order, used the E2 protein UbcH10. The intensely positive band at the bottom of Figure 2 (see also Figs. 3, 4) is a ubiquitin dimer that is present in the commercial ubiquitin preparation. The more abundant ubiquitin monomer is not shown here.
Figure 2.
Immunoblot with anti-ubiquitin antibody detects ubiquitylated S-protein. Two concentrations of the RAG1 NTD fusion protein (1.0 and 0.3 μg per reaction) gave a graded degree of ubiquitylated product in lanes 2 and 3 (*). No product is detected in lanes 4 and 5 lacking any fusion protein or substituted with GST protein. The reaction also depends on each of the components that are individually left out of lanes 6–9.
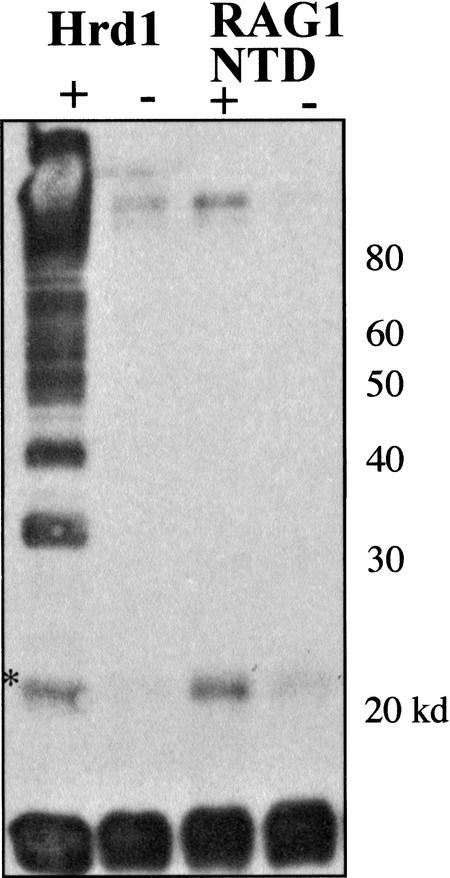
Figure 3.
Comparison of the RAG1 NTD E3 ligase with the yeast Hrd1p. The yeast protein in lane 1 yields a ladder of polyubiquitylated products, whereas the RAG1 NTD protein in lane 3 seems to produce only the monoubiquitylated form (*). The immunoblot is developed against ubiquitin.
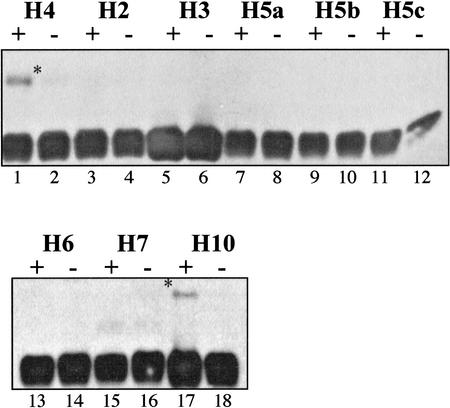
Figure 4.
The RAG1 E3 ligase was assayed with a series of E2 proteins. For each E2 protein, a pair of lanes either contained (+) or lacked (−) the RAG1 NTD fusion protein. A ubiquitylated product was detected only with UbcH4 and UbcH10 as E2 partner. The immunoblot was developed with anti-ubiquitin antibody, and a ubiquitylated product was detected. Lanes 1 and 2 are the same data presented in Figure 3.
We observed that the ubiquitylated S-protein product runs with a mobility consistent with a singly ubiquitylated species. Frequently, E3 ligases are capable of performing multiple rounds of ubiquitylation to form a high-molecular-weight chain of adducts. Indeed, polyubiquitylation can be obtained in this assay. We duplicated the previously published observation (Bays et al. 2001) that the RING domain of yeast Hrd1p is capable of stimulating such polyubiquitin additions (Fig. 3). Here we compared the E3 activity of Hrd1p with the RAG1 N-terminal domain. Both proteins were prepared as similar fusion constructs. Complete reactions, as in Figure 2, were assembled, but no E3 protein was added in lanes marked (−). Clearly, Hrd1p was able to create a large number of ubiquitylated species ascending to the top of the blot. In contrast, the RAG1 construct appears to only yield the singly modified S-protein. The band near the top of the gel may represent ubiquitylation of one of the other proteins in the reaction, and is not always dependent on exogenous E3. These reactions were performed with the human E2 protein UbcH4, expressed in Escherichia coli. In this experiment we only witnessed monoubiquitylation of the artificial substrate. We explore the issue of mono- versus polyubiquitylation later in this report.
The two E2 proteins used in the experiments presented above (UbcH4 and UbcH10) are the only two we have found capable of cooperating with the RAG1 E3 in the synthetic assay. We present a survey of nine E2 proteins in Figure 4 as follows: Each pair of lanes contains an E2 protein (named at the top) with (+) or without (−) the RAG1 N-terminal domain fusion construct. Only the two proteins already described, UbcH4 and UbcH10, yielded the ubiquitylated S-protein (asterisk) in an E3-dependent manner. Similar results were obtained with shorter constructs containing the minimal RAG1 RING (data not shown).
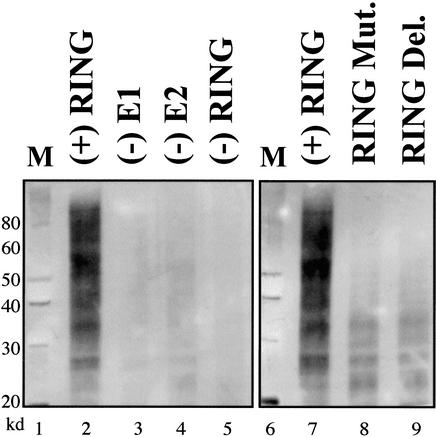
Given the artificial nature of the ubiquitylation assay used here, it is not clear whether the monoubiquitin modification we observed on the test substrate represents the natural behavior of RAG1 acting as an E3 ligase. For example, the BRCA1/BARD1 RING proteins add single ubiquitin adducts to exogenous substrates such as histones in vitro, but are also capable of the polyubiquitylation of the RING protein itself (Chen et al. 2002; Mallery et al. 2002). The biologic significance of these observations is still under evaluation. We also found differing behavior under other experimental conditions. When tested without the S-protein artificial substrate, we also detected a smear on the immunoblot representing polyubiquitylation (Fig. 5). In Figure 5, lanes 2 and 5, show the formation of high-molecular-weight protein when complete reactions were assembled containing the isolated RAG1 RING (residues 250–350 as a GST fusion) with the E2 protein UbcH5b. Lanes 3–5 show the expected dependence upon the three enzymatic components, when each is individually excluded. Lane 8 shows the effect of mutating conserved residues in the RING. In this lane we specifically targeted two conserved residues, replacing the cysteines at positions 290 and 293 with alanines. The high-molecular forms are almost entirely eliminated. Identical results were obtained with the deletion of half of the RING, as presented in lane 9. The protein used here includes the RAG1 sequence from residues 250 through 307, truncating 43 residues from the RING. Additional point mutations (H307A and C310A in one construct, and the four positions 290, 293, 307, and 310 simultaneously) similarly eliminated the E3 ligase activity (data not shown). These results are fully consistent with the dependence of the RING family of proteins upon zinc binding through contact with the conserved amino acid residues. Similar results were also obtained using the related E2 protein UbcH5c.
Figure 5.
The minimal RAG1 RING can stimulate formation of polyubiquitin chains. This anti-ubiquitin immunoblot shows high-molecular-weight proteins in a complete reaction including the E2 protein UbcH5b (lanes 2,7). The reaction is dependent on the presence of the E1, E2, and RING proteins (lanes 3–5, respectively). Mutation of the RING by two point mutations, C290A and C293A (lane 8), or deletion of half of the RING (lane 9) strongly reduces activity to a similar degree.
The presence of a second enzymatic activity in the N-terminal region of RAG1 may resolve some questions. It has been puzzling that more than 35% of RAG1 can be removed from the protein with little real consequence on the DNA binding, cleavage, and recombination aspects of V(D)J enzymology. Why should such a large region of the protein be maintained? Furthermore, sequence comparison among the many species for which sequence data are now available shows a distinctly lower degree of conservation within the N-terminal region compared to the almost absolute conservation of the core region. Our finding that the N-terminal region can function as a ubiquitin ligase suggests that this part of the protein may have a distinct role, independent of the nuclease activity. Since the core region is sufficient to catalyze the full recombination reaction, it seems that this E3 activity is not an obligate part of the DNA recombination mechanism. Perhaps its role is more indirect. There are important levels of regulation of V(D)J recombination that remain ill-defined. One important issue is the relationship between chromatin accessibility and the targeting of recombination (for review, see Bassing et al. 2002; Gellert 2002). We can speculate that the RAG1 E3 ligase could play a direct role in chromatin modification, because histones are themselves ubiquitylated (Jason et al. 2002), or RAG1 E3 ligase may play an indirect role through other known functions of ubiquitylation in DNA repair, cell-cycle regulation, and differentiation (see Weissman 2001 and references therein). It was recently shown that RAG2 is regulated by ubiquitin-dependent degradation, which raises additional possibilities for a role of an E3 ligase (Mizuta et al. 2002).
Materials and methods
Plasmid construction and production of GST fusion proteins
The RAG1 N-terminal domain (DNA encoding residues 1–383 of the mouse sequence) and minimal RAG1 RING (DNA encoding residues 250–350) were amplified by PCR and cloned into the pET42b(+) vector (Novagen) downstream of the GST-S peptide using the NcoI and XhoI sites. Point mutations and deletions within the RING, as described in the text, were also prepared by PCR. All inserts were verified by DNA sequencing.
GST-S, GST-S-Hrd1p (Bays et al. 2001), GST-S-RAG1 NTD, and RAG1-derived RING proteins were expressed in E. coli BL21(DE3) pLysS (Novagen), purified on Ni-NTA agarose beads (QIAGEN), and confirmed by immunoblotting using anti-His-Tag (Novagen) or anti-GST (Santa Cruz Biotechnology) antibodies.
In vitro analysis of RAG1-mediated protein ubiquitylation
In vitro ubiquitylation reactions were performed as described (Bays et al. 2001; Swanson et al. 2001) with modifications. In brief, 0.5 μg yeast Uba1 (Boston Biochem), 0.5 μg E2 enzyme (see below), 1.5 μg of the S-protein (Biozyme Laboratories), and 1 μg GST-S-RAG1 proteins were incubated in 250 μM bovine ubiquitin (Sigma), 6.6 mM ATP, 50 mM Tris-HCl at pH 7.5, 2.5 mM MgCl2, and 0.5 mM DTT for 4 h at 30°C. Reactions were stopped by addition of SDS loading buffer and boiled for 3 min. S-protein ubiquitylation was analyzed by 15% SDS-PAGE and visualized by anti-ubiquitin immunoblotting (P4G7 monoclonal anti-ubiquitin antibodies, Covance). Experiments evaluating polyubiquitin chains were similarly analyzed using 10% SDS-PAGE.
E2 enzymes were obtained from Boston Biochem (UbcH2, UbcH3, UbcH5a, UbcH5b, UbcH5c, UbcH6, UbcH7, and UbcH10) or purified on Ni-NTA agarose from expression plasmids for UbcH4 and UbcH6 (kindly provided by Dr. M. Scheffner, Köln University), and UbcH10 (kindly provided by Dr. R. Basavappa, University of Rochester). All commercial and home-made enzymes behaved similarly in our assays.
Acknowledgments
Dr. M. Hochstrasser and M. Locher (Yale) were exceptionally generous with their time and encouragement in helping us establish the assay. We are also grateful to Dr. R. Basavappa, and Dr. M. Scheffner for sharing their reagents with us. These results were made possible by the support of the NIH (AI41711) and institutional funds from the Albert Einstein College of Medicine. M.S. is a scholar of the Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sadofsky@aecom.yu.edu; FAX (718) 430-8541.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1058103.
References
- Aravind L, Koonin EV. The U box is a modified RING finger—A common domain in ubiquitination. Curr Biol. 2000;10:132–134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- Bailin T, Mo X, Sadofsky MJ. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol Cell Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109:45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Chen A, Kleiman FE, Manley JL, Ouchi T, Pan ZQ. Auto-ubiquitination of the BRCA1/BARD1 RING ubiquitin ligase. J Biol Chem. 2002;277:22085–22092. doi: 10.1074/jbc.M201252200. [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Oettinger MA. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Lorick KL, Jensen JP, Weissman AM. RING finger ubiquitin protein ligases: Implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin Cancer Biol. 2003;13:5–14. doi: 10.1016/s1044-579x(02)00095-0. [DOI] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD. The lore of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- Jason LJ, Moore SC, Lewis JD, Lindsey G, Ausio J. Histone ubiquitination: A tagging tail unfolds? Bioessays. 2002;24:166–174. doi: 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- Jones JM, Gellert M. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J. 2002;21:4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch SA, Sudarsanam P, Oettinger MA. Regions of RAG1 protein critical for V(D)J recombination. Eur J Immunol. 1996;26:886–891. doi: 10.1002/eji.1830260425. [DOI] [PubMed] [Google Scholar]
- Kirch SA, Rathbun GA, Oettinger MA. Dual role of RAG2 in V(D)J recombination: Catalysis and regulation of ordered Ig gene assembly. EMBO J. 1998;17:4881–4886. doi: 10.1093/emboj/17.16.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landree MA, Kale SB, Roth DB. Functional organization of single and paired V(D)J cleavage complexes. Mol Cell Biol. 2001;21:4256–4264. doi: 10.1128/MCB.21.13.4256-4264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HE, Hsu LY, Cado D, Cowell LG, Kelsoe G, Schlissel MS. The ‘dispensable’ portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallery DL, Vandenberg CJ, Hiom K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002;21:6755–6762. doi: 10.1093/emboj/cdf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan CJ, Difilippantonio MJ, Rao N, Spanopoulou E, Schatz DG. A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol Cell Biol. 1997;17:4544–4552. doi: 10.1128/mcb.17.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta R, Mizuta M, Araki S, Kitamura D. RAG2 is down-regulated by cytoplasmic sequestration and ubiquitin-dependent degradation. J Biol Chem. 2002;277:41423–41427. doi: 10.1074/jbc.M206605200. [DOI] [PubMed] [Google Scholar]
- Mo X, Bailin T, Sadofsky MJ. A C-terminal region of RAG1 contacts the coding DNA during V(D)J recombination. Mol Cell Biol. 2001;21:2038–2047. doi: 10.1128/MCB.21.6.2038-2047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy CL, Patenge N, Matthews AG, Oettinger MA. Assembly of the RAG1/RAG2 synaptic complex. Mol Cell Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- Rodgers KK, Bu Z, Fleming KG, Schatz DG, Engelman DM, Coleman JE. A zinc-binding domain involved in the dimerization of RAG1. J Mol Biol. 1996;260:70–84. doi: 10.1006/jmbi.1996.0382. [DOI] [PubMed] [Google Scholar]
- Sadofsky MJ. The RAG proteins in V(D)J recombination: More than just a nuclease. Nucleic Acids Res. 2001;29:1399–1409. doi: 10.1093/nar/29.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky MJ, Hesse JE, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Gomez CA, Sobacchi C, Bozzi F, Abinun M, Pasic S, Cortes P, Vezzoni P, Villa A. N-terminal RAG1 frameshift mutations in Omenn's syndrome: Internal methionine usage leads to partial V(D)J recombination activity and reveals a fundamental role in vivo for the N-terminal domains. Proc Natl Acad Sci. 2000;97:14572–14577. doi: 10.1073/pnas.97.26.14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Spanopoulou E, Mulligan RC, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen SB, Han JO, Mundy C, Oettinger MA, Roth DB. Roles of the ‘dispensable’ portions of RAG-1 and RAG-2 in V(D)J recombination. Mol Cell Biol. 1999;19:3010–3017. doi: 10.1128/mcb.19.4.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson PC. A RAG-1/RAG-2 Tetramer Supports 12/23-Regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol Cell Biol. 2002;22:7790–7801. doi: 10.1128/MCB.22.22.7790-7801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes & Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CL, Drejer AH, Schatz DG. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes & Dev. 2002;16:1934–1949. doi: 10.1101/gad.984502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]