Abstract
In human cells infected with herpes simplex virus 1 the double-stranded RNA-dependent protein kinase (PKR) is activated but phosphorylation of the α subunit of eukaryotic translation initiation factor 2 (eIF-2) and total shutoff of protein synthesis is observed only in cells infected with γ1z34.5− mutants. The carboxyl-terminal 64 aa of γ134.5 protein are homologous to the corresponding domain of MyD116, the murine growth arrest and DNA damage gene 34 (GADD34) protein and the two domains are functionally interchangeable in infected cells. This report shows that (i) the carboxyl terminus of MyD116 interacts with protein phosphatase 1α in yeast, and both MyD116 and γ134.5 interact with protein phosphatase 1α in vitro; (ii) protein synthesis in infected cells is strongly inhibited by okadaic acid, a phosphatase 1 inhibitor; and (iii) the α subunit in purified eIF-2 phosphorylated in vitro is specifically dephosphorylated by S10 fractions of wild-type infected cells at a rate 3000 times that of mock-infected cells, whereas the eIF-2α-P phosphatase activity of γ134.5− virus infected cells is lower than that of mock-infected cells. The eIF-2α-P phosphatase activities are sensitive to inhibitor 2. In contrast to eIF-2α-P phosphatase activity, extracts of mock-infected cells exhibit a 2-fold higher phosphatase activity on [32P]phosphorylase than extracts of infected cells. These results indicate that in infected cells, γ134.5 interacts with and redirects phosphatase to dephosphorylate eIF-2α to enable continued protein synthesis despite the presence of activated PKR. The GADD34 protein may have a similar function in eukaryotic cells. The proposed mechanism for maintenance of protein synthesis in the face of double-stranded RNA accumulation is different from that described for viruses examined to date.
Keywords: growth arrest and DNA damage gene 34
The γ134.5 gene of herpes simplex virus 1 (HSV-1) encodes a 263-aa protein consisting of three domains: a 160-aa amino-terminal domain, 10 repeats of 3 aa (Ala-Thr-Pro), and a 73-aa carboxyl-terminal domain. This protein performs at least two functions (1–6). Thus mutants lacking this gene are highly attenuated in experimental rodent animal systems and exhibit a premature shutoff of protein synthesis after initiation of viral DNA synthesis in infected human cells (4–6). Whereas preliminary data indicate that the capacity to replicate in experimental animal systems requires the entire gene, the function that precludes the shutoff of protein synthesis maps in the carboxyl-terminal domain of γ134.5 (ref. 7; unpublished data). The carboxyl-terminal amino acids of γ134.5 are homologous to the corresponding domains of highly conserved mammalian protein known as GADD34 (growth arrest and DNA damage protein 34) (7–11). The murine GADD34 gene known as MyD116 contains 657 aa and consists of a large basic amino-terminal domain, a 38-aa sequence repeated 4.5 times, and a carboxyl terminus that can substitute for the corresponding domain of the γ134.5 to prevent the shutoff of protein synthesis (9–12).
In a recent study, it has been shown that in wild-type or mutant infected cells, the double-stranded RNA-dependent protein kinase (PKR) was activated. However, only in cells infected with the γ134.5− mutants or mutants lacking the γ134.5 domain encoding the carboxyl terminus of the protein was the α subunit of the eukaryotic initiation factor 2 (eIF-2α) phosphorylated (13). The extent of phosphorylation of eIF-2α readily accounted for the total shutoff of protein synthesis in cells infected with these mutants.
PKR is activated by double-stranded RNA (14). Because most viruses either synthesize double-stranded RNA during their replication or form double-stranded RNA structures, they encode genes that block activation of PKR. The observation that in wild-type infected cells PKR was activated but eIF-2α was not phosphorylated raised the possibility that the γ134.5 protein acts to preclude the shutoff of protein synthesis by a mechanism different from that reported for other viruses. In this report we show that this is in fact the case.
MATERIALS AND METHODS
Cells and Viruses.
The Vero, HeLa, and SK-N-SH cell lines were obtained from American Type Culture Collection and propagated in DMEM supplemented with 5% (HeLa cells and Vero cells) or 10% (SK-N-SH cells) fetal bovine serum, respectively. HSV-1(F) is the prototype HSV-1 strain used in this laboratory (15). Recombinant virus R3616 lacks 1000 bp each from the coding domains of both copies of the HSV-1(F) γ134.5 gene (4). In recombinant R8300, the sequence encoding the carboxyl-terminal domain of the γ134.5 gene was replaced with the corresponding domain of MyD116 gene (12).
Plasmids.
pRB4891, containing a 1.37-kb cDNA encoding human protein phosphatase 1α (PP1α) was isolated from human brain cDNA library. To construct pRB4890, a BamHI–EcoRI DNA fragment encoding codons 524–657 of MyD116 was amplified by PCR from a cDNA copy of the gene described elsewhere (12) with the primers TGACTGGATGCAGAGGCGGCTCAGATTGTTC and AGCGCGCAATTAACCCTCACTAAAG, and inserted into the BamHI–EcoRI sites of pGBT9 (CLONTECH) to serve as “bait” in the two-hybrid system. The PCR conditions were 94°C for 2 min, 60°C for 3 min, and 72°C for 3 min for 25 cycles. pRB4892 containing the chimeric glutathione S-transferase (GST)–PP1α gene was constructed by ligating a 0.9-kb PCR fragment containing the entire coding sequence of PP1α except the first methionine codon into the EcoRI–SalI sites of pGEX4T-1(Pharmacia). The oligonucleotide primers were GCACTGAATTCTCCGACAGCGAGAAGCTCAAC and GCACTGTCG ACATCTGGGGCACAGGGTGGTGT. To construct pRB4893 carrying a chimeric GST carboxyl-terminal domain of γ134.5 [GST–γ134.5C], the EcoRI–SalI fragment encoding codons 146–263 of γ134.5 from pRB71 and cloned into the EcoRI–SalI sites of pGEX4T-1. The construction of pRB4873 containing an AccI–EcoRI fragment encoding the 3′ terminal 174 codons of MyD116 fused in-frame to GST [GST–MyD116C] was reported elsewhere (12). Expression of GST–MyD116C, GST–γ134.5C, and GST–PP1α fusion proteins was induced by the addition of isopropyl β-D-thiogalactoside to the medium with Escherichia coli BL21 cells transformed with either plasmid pRB4873, pRB4892, or pRB4892, followed by affinity purification of the fusion proteins from bacterial lysates on agarose beads conjugated with glutathione.
Yeast Two-Hybrid Screen.
The yeast strain Y190 (16) transformed with plasmid pRB4890, and growing on Trp− selective medium was retransformed with a human brain cDNA library (CLONTECH) and subjected to selection on Trp−/Leu−/His− medium in the presence of 25 mM 3-aminotriazole (Sigma). Moderate to fast growing colonies were restreaked on Trp−/Leu−/His− medium 5 days after plating. The library-derived plasmids were recovered by transformation of E. coli HB101 with total yeast DNA preparations, followed by selection on Leu−/ampicillin+ medium as described (16). The filter assay for β-galactosidase was done as recommended by CLONTECH.
Determination of eIF-2α-P Phosphatase Activity.
The translation initiation factor eIF-2 was purified from rabbit reticulocyte ribosomes (17). The hemin-controlled translational repressor (HCR) was partially purified to step 4 from rabbit reticulocyte lysates (18). eIF-2 (20 pmol or 2.7 μg) was reacted with step 4 HCR in 0.02 M Tris·HCl (pH 7.5), 40 mM KCl, 2.0 mM MgCl2, and 0.17 mM [γ-32P]ATP (2–10 Ci or 74–370 GBq/mmol) in a final volume of 12 μl for 25 min at 34°C to yield phosphorylated eIF-2α and eIF-2β (1.0 and 0.7 mol/mol of eIF-2, respectively). HeLa cells were harvested 15 hr after mock infection or infection with 20 plaque-forming units (pfu) of HSV-1(F), R3616, or R8300 per cell. S10 fractions were prepared from lysates of mock-infected or infected cells as described (13) and diluted to a final volume of 18 μl with 0.02 M Tris·HCl (pH 7.5), 50 mM KCl, 2 mM MgCl2, and 0.1 mM EDTA. ATP was then added to a final concentration of 0.8 mM. After 30 sec at 34°C, each sample received 1.2 μl (2 pmol) of eIF-2α-32P and was reincubated at 34°C. The rate of dephosphorylation of eIF-2α-32P was determined by placing 6.0 μl aliquots into a solution containing SDS (19) at times indicated in the figures, followed by electrophoresis on 7% denaturing polyacrylamide gels as described (19) for 21 hr at 44 V (3 V/cm), staining with Coomassie blue or silver, drying, and autoradiography. 32P remaining in eIF-2α-32P was quantified by excising this band and comparing its Cerenkov radiation (19) to that of equivalent aliquots of eIF-2α-32P that were not further incubated and were subjected to electrophoresis in parallel.
Determination of Phosphorylase Phosphatase Activity.
Phosphorylase b (32 μg; Sigma) was phosphorylated by incubation with 3.3 μg of phosphorylase kinase (Sigma) in 0.01 M Tris·HCl (pH 7.5), 2.0 mM MgCl2, and 0.13 mM [γ-32P]ATP (3 Ci or 111 GBq/mmol) in a final volume of 15 μl at 34°C for 10 min. Dephosphorylation was determined by adding 4.4 μg of [32P]phosphorylase to samples constituted and incubated as above in a final volume of 27 μl. At 1.5, 3, 6, and 10 min, 6.0 μl aliquots were removed and processed as described above for determining phosphatase activity.
RESULTS
The Carboxyl Terminus of the Murine GADD34 Protein (MyD116) Interacts with PP1α in the Yeast Two-Hybrid System.
We have selected the carboxyl terminus of MyD116 as bait for the two-hybrid system because (i) it and γ134.5 share amino acid sequence homology (7, 8), (ii) the carboxyl terminus of MyD116 can substitute functionally for that of the γ134.5 protein, and (iii) the codon usage in MyD116, a mammalian gene, is closer to that of yeast than that of HSV-1. Of the 106 yeast colonies screened, only one colony was positive for β-galactosidase expression. The cDNA recovered from this colony was retransformed into yeast strain Y190 with various control plasmids encoding fusion proteins (data not shown). These studies indicated that only the carboxyl terminus of MyD116 interacted with the protein encoded by the isolated cDNA. DNA sequence analysis revealed that the isolated cDNA encoded the catalytic subunit of PP1α (20).
Both MyD116 and γ134.5 Proteins Bind PP1α in Vitro.
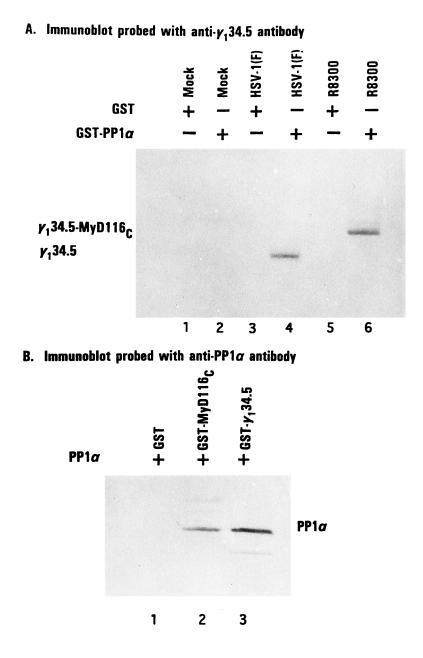
To verify and extend the observations that MyD116 and PP1α interact in yeast, a chimeric GST–PP1α gene constructed as described in Materials and Methods was expressed in E. coli. Purified GST–PP1α protein bound to beads was reacted with cell extracts prepared from HeLa cells that were either mock-infected or infected with 20 pfu of HSV-1(F) or of R8300 per cell. The proteins bound to GST–PP1α were solubilized, electrophoretically separated in denaturing gels, transferred to nitrocellulose, and reacted with antibody to the Ala-Thr-Pro repeat (1, 6) present in both γ134.5 expressed by HSV-1(F) and γ134.5–MyD116 chimeric protein expressed by R8300 recombinant virus. As shown in Fig. 1A, the GST–PP1α protein bound both γ134.5 from lysates of HSV-1(F)-infected cells and the more slowly migrating γ134.5–MyD116 chimeric protein from cells infected with R8300. GST alone did not react with either protein in the appropriate cell lysates.
Figure 1.
Association of PP1α with γ134.5 and MyD116. (A) Replicate HeLa cell cultures were harvested in lysis buffer containing 10 mM Hepes (pH 7.6), 250 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride, and 2 mM benzamidine 18 hr after mock infection or infection with 10 pfu of HSV-1(F) or R8300 per cell. After 30 min on wet ice and low speed centrifugation to remove nuclei, the supernatant fluids were precleared with GST beads and then reacted with GST phosphatase 1 bound beads at 4°C overnight. After extensive rinsing, the proteins bound to beads were solubilized by boiling in disruption buffer containing 50 mM Tris·HCl (pH 7.0), 5% 2-mercaptoethanol, 2% SDS, and 2.75% sucrose, electrophoretically separated on denaturing 12% polyacrylamide gels, and transferred to a nitrocellulose sheet, and the blot was probed with anti-γ134.5 serum (1). The positions of γ134.5 protein and of the chimeric protein γ134.5–MyD116 are shown. (B) An aliquot of GST–PP1α fusion protein bound to beads was reacted with 25 units of thrombin (Sigma) in PBS at room temperature. After 8 hr, the mixture was spun in a table top centrifuge and the supernatant fluid containing PP1α was then dialyzed against lysis buffer, reacted with GST, GST–MyD116C, or GST–γ134.5C bound to beads and processed as described in A. PP1 was detected with antiphosphatase 1α antibody (Upstate Biotechnology, Lake Placid, NY).
In the second experiment, purified PP1α was mixed with beads carrying GST or chimeric proteins consisting of GST fused to the amino acids 146–263 of γ134.5 or to amino acids 485–657 of MyD116. The proteins bound to GST or to the chimeric proteins was solubilized, subjected to electrophoresis on a denaturing gel, and reacted with anti PP1α antibody. As shown in Fig. 1B, both chimeric proteins but not GST brought down a protein with an apparent Mr of 38,000 (molecular weight markers not shown) and which reacted with the anti-PP1α antibody.
Phosphatase Activity Is Essential for Protein Synthesis in Infected Cells.
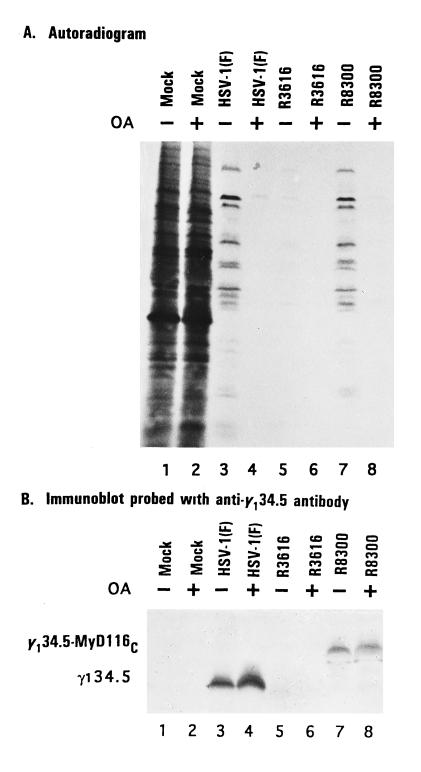
To test whether phosphatase activity is essential for sustained protein synthesis in infected cells, replicate monolayer cultures of SK-N-SH cells were either mock-infected or infected with 20 pfu of HSV-1(F), R3616, or R8300 per cell. At 2 hr after infection, replicate, mock-infected, or infected cultures were exposed to okadaic acid (25 ng/ml). After an additional 12 hr of incubation at 37°C, the cells were labeled for 1 hr with [35S]methionine and then harvested, solubilized in disruption buffer, and subjected to electrophoresis in denaturing polyacrylamide gels and autoradiography. The results (Fig. 2A) show okadaic acid totally inhibited protein synthesis in cells infected with wild-type HSV-1(F), the γ134.5− virus R3616, or in cells infected with the R8300 recombinant expressing the γ134.5–MyD116 chimeric protein. In contrast, okadaic acid had no apparent effect on protein synthesis in mock-infected cells.
Figure 2.
(A) Autoradiographic images of electrophoretically separated, [35S]methionine-labeled proteins from cell lysates infected with the indicated viruses in the presence or absence of okadaic acid (OA). SK-N-SH cells were either mock-infected or infected with 20 pfu of HSV-1(F), R3616, or R8300 per cell. At 2 hr after exposure to the viruses, the cells were overlaid with medium 199V (1) supplemented with or without okadaic acid (25 ng/ml). At 14 hr after infection, the cells were overlaid for 1 hr with 1 ml of medium 199V lacking methionine but supplemented with 50 μCi of [35S]methionine (specific activity, >1000 Ci/mmol; Amersham) for 1 hr, then harvested, solubilized, subjected to electrophoresis in denaturing 12% polyacrylamide gels, transferred to a nitrocellulose sheet, and subjected to autoradiography as described (6). (B) Immunoblot of the nitrocellulose sheet in A probed with anti-γ134.5 antibody (1).
To test a trivial hypothesis that okadaic acid blocked in some fashion the synthesis of γ134.5 or the γ134.5-MyD116 proteins in infected cells, the lysates of cells infected with wild-type and recombinant viruses were solubilized, subjected to electrophoresis in denaturing gels, electrically transferred to a nitrocellulose sheet, and reacted with antibody to the γ134.5 protein. As shown in Fig. 2B, the lysates of cells infected with HSV-1(F) or with R8300-expressed γ134.5 or γ134.5–MyD116 protein, respectively. The antibody did not react with lysates of mock-infected cells or cells infected with the γ134.5− virus.
The eIF-2α-32P Phosphatase Activity Is Highly Abundant in Wild-Type or R8300 Recombinant Virus-Infected Cells and Significantly Decreased in γ134.5− Virus-Infected Cells.
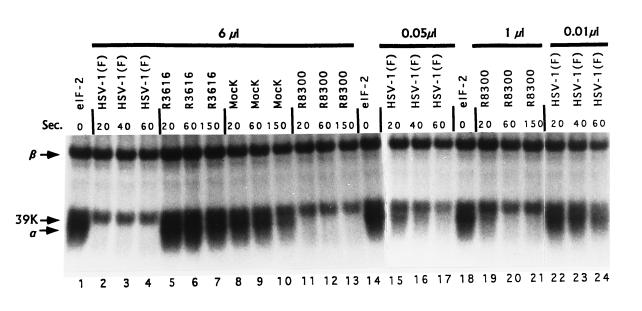
We tested whether the shutoff of protein synthesis in cells infected with γ134.5− virus R3616 was determined at the level of eIF-2α phosphatase activity. S10 fractions were prepared from lysates of HeLa cells either infected or mock-infected and tested for their ability to dephosphorylate eIF-2α-32P. Unreacted eIF-2α-32P (Fig. 3, lanes 1, 14, and 18) showed three labeled bands representing eIF-2α, eIF-2β, and a small amount of a Mr 39,000 protein that is a contaminant distinct from eIF-2α (17). Reaction of this eIF-2 with the S10 fraction from mock-infected cells showed a moderate rate of loss of radioactivity from eIF2α-32P (Figs. 3 and 4A). This eIF-2α-32P phosphatase activity was reduced ≈3-fold in cells infected with γ134.5− virus. In sharp contrast, the S10 fraction of cells infected with the wild-type virus or with the R8300 mutant virus exhibited so marked a level of eIF-2α-32P phosphatase activity that virtually all added eIF-2α-32P radioactivity was removed within the first 20 sec of reaction (Fig. 3, lanes 2 and 11). To verify that this phosphatase activity does not continue after the reaction mixture is mixed with the SDS denaturing solution, we tested and found that eIF-2α-32P is completely stable following addition to a mixture of SDS and S10 fraction from cells infected with either wild-type or R8300 recombinant virus (data not shown).
Figure 3.
Autoradiographic images of purified, electrophoretically separated eIF-2 after reaction with various amounts of S10 fractions of HeLa cells mock-infected or infected with HSV-1(F), R3616, or R8300. The assays were carried out as described. The amount of S10 fractions used are indicated on the top. Arrows marked α and β indicate the position of the α and β subunits of eIF-2; the arrow marked 39K indicates the position of a trace 39-kDa protein unrelated to eIF-2α.
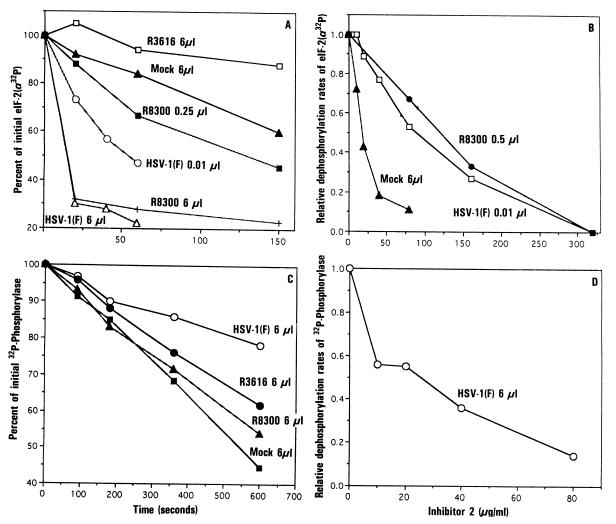
Figure 4.
Phosphatase activity in S10 fractions of HeLa cells mock-infected or infected with 20 pfu of HSV-1(F), R3616, or R8300 per cell. The procedures were carried out as described. (A) Fraction of residual radioactivity in eIF-2α-32P after reaction with variable amounts of S10 fractions from mock-infected or infected cells. (B) Effect of inhibitor 2 on the rate of dephosphorylation of eIF-2α-32P by the phosphatase activity in the S10 fractions. The results are expressed as the relative rate of activity seen in duplicate samples containing no added inhibitor 2. (C) [32P]phosphorylase phosphatase activity. The procedure was the same as in A except that the dephosphorylation of [32P]phosphorylase by the S10 fractions was assayed. The results are an average of two determinations and are expressed as the percentage of [32P]phosphorylase remaining, when compared with zero time samples. (D) Effect of inhibitor 2 on the relative rate of [32P]phosphorylase phosphatase activity. The assay was done as described in B.
To quantify the increase in eIF-2α-32P phosphatase activity associated with wild-type or R3800 recombinant virus infection of HeLa cells, the S10 fractions from these cells were progressively diluted until the rate of dephosphorylation of eIF-2α-32P could be measured accurately. Only when the S10 fraction of wild-type infected cells was diluted 600-fold could the eIF-2α-32P phosphatase activity be measured accurately, and it was still 5-fold greater than that of mock-infected cells (Figs. 3 and 4A). These findings indicate that the eIF-2α-32P phosphatase activity of wild-type infected cells increased ≈3000-fold. Similar analyses indicated that in cells infected with the R8300 recombinant virus this activity increased ≈50-fold (Figs. 3 and 4A). The removal of radioactivity in these samples represented dephosphorylation and not proteolysis inasmuch as it was not associated with loss of eIF-2α protein as measured by staining. In addition, the marked increase in eIF-2α-P phosphatase activity seen in S10 fractions of wild-type virus-infected cells may be specific for eIF-2α inasmuch as neither the Mr 39,000 phosphoprotein nor eIF-2β-32P was significantly affected under the same conditions (Fig. 3). Finally, the results shown in Figs. 3 and 4 were reproducible and obtained in two determinations with each of three S10 fraction from mock-infected or infected HeLa cells.
The eIF-2α-32P Phosphatase Activity Is Sensitive to PP1 Inhibitor 2.
To determine whether eIF-2α-32P was dephosphorylated by PP1α, we measured the rate of dephosphorylation of eIF-2α-32P as a function of the concentration of inhibitor 2, a known inhibitor of PP1. As shown in Fig. 4B, eIF-2α-32P phosphatase in the S10 fraction from wild-type virus- or R8300 virus-infected cells was similarly sensitive to inhibitor 2 with a 50% reduction in the rate of dephosphorylation attained at ≈90 and 120 μg of inhibitor 2 per ml, respectively. This suggests that activated eIF-2α-32P phosphatase is derived from PP1. However, the concentration of inhibitor 2 required for inhibition of the activated eIF-2α-P phosphatase appears to be much higher than that required to inhibit eIF-2α-32P phosphatase in mock-infected cells and also much higher than that required to inhibit PP1α in cell extracts (21). This could be due to the possible association of γ134.5 with HeLa cell PP1α, resulting in phosphatase activity directed specifically at eIF-2α-P, as suggested by the results shown in Fig. 3.
The Activated Phosphatase Activity Is Specific for eIF-2α.
As an additional test of the hypothesis that eIF-2α-32P phosphatase is specifically activated in infected cells, we measured the dephosphorylation of [32P]phosphorylase, a known substrate of serine/threonine phosphatases, by lysates of HeLa cells mock-infected or infected with wild-type virus. The results (Fig. 4C) showed that the [32P]phosphorylase phosphatase activity of HSV-1(F)-infected cells is only one-half that of mock-infected cells. The activity responsible for the dephosphorylation of [32P]phosphorylase in wild-type virus infected cells is sensitive to inhibitor 2 (Fig. 4D). The observation that phosphorylase phosphatase activity in wild-type virus-infected cells is significantly reduced relative to that of mock-infected cells suggests that the mechanism of activation of eIF-2α-P phosphatase may involve association of the γ134.5 gene product with a portion of the cellular PP1α, converting this fraction to a phosphatase activity that is specific for eIF-2α-P.
DISCUSSION
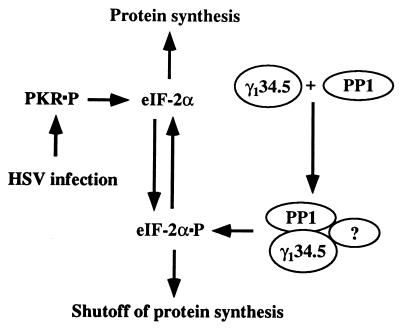
The scenario we would like to propose is as follows. In cells infected with HSV-1 accumulation of symmetric transcripts after the onset of DNA synthesis results in activation of PKR. A viral protein, γ134.5, binds PP1α, presumably through its carboxyl terminus, and redirects the activity of the enzyme to dephosphorylate eIF-2α either by itself or in conjunction with as yet unidentified proteins (Fig. 5). As a consequence, although PKR is activated, protein synthesis continues unabated. In support of this scenario are the following data.
Figure 5.
Schematic representation of the regulation of protein synthesis by γ134.5 in infected cells. As described in the text, PKR is activated after the onset of DNA synthesis in cells infected with HSV-1. In the absence of γ134.5, eIF-2α is phosphorylated and protein synthesis ceases. γ134.5 binds to PP1α and dephosphorylates eIF-2α. It is not known whether the γ134.5–PP1 complex is sufficient or requires additional factors to dephosphorylate eIF-2α. Because GADD34 substitutes for γ134.5, it is conceivable that one of the functions of GADD34 is to prevent shutoff of protein synthesis by a similar mechanism.
(i) In cells infected with both wild-type and γ134.5− mutants, PKR is phosphorylated, but only in cells infected with mutants lacking all or the 3′ domain of the γ134.5 gene is protein synthesis shut off and eIF-2α phosphorylated. The γ134.5 sequence required to block protein synthesis shut off maps in the 3′ domain of the gene and can be replaced without loss of function by the corresponding domain of MyD116, the murine GADD34 gene.
(ii) The eIF-2α-P phosphatase activities reported here may account for the differences in steady-state eIF-2α-P and protein synthesis previously noted in γ134.5− virus infected cells (13). Because PKR is not activated in mock-infected cells, the observed moderate eIF-2α-P phosphatase activity should be sufficient to prevent the degree of phosphorylation of eIF-2α required to inhibit protein synthesis. Wild-type HSV-1 infection does result in activation of PKR (13), but its effect is probably more than countered by the ≈3000-fold increase in eIF-2α-P phosphatase. This marked increase in eIF-2α-P phosphatase activity is due to the expression of the γ134.5 gene product, because infection with the γ134.5− mutant is associated with a decrease in eIF-2α-P phosphatase activity, that, upon activation of PKR, results in phosphorylation of eIF-2α and inhibition of protein synthesis (13).
(iii) In the yeast two-hybrid system the carboxyl terminus of MyD116 interacted with the human PP1α. The interaction of PP1α and MyD116 or with γ134.5 protein was also demonstrable in pulldown experiments in vitro. The hypothesis that the eIF-2α phosphatase activity measured in cytoplasmic fractions of mock-infected or infected cells is derived from PP1 is strengthened by the observation that it was sensitive to inhibitor 2. Nevertheless, two lines of evidence suggest that this activity may represent a modified form of PP1α redirected to eIF-2α. Specifically, the eIF-2α-P phosphatase activity was less sensitive to inhibitor 2 than that of mock-infected cells, and furthermore, the phosphorylase phosphatase activity, a known function of PP1, was more potent in uninfected cells than in infected cells.
Our studies also shed light on the putative function of GADD34. As noted above, the carboxyl terminus of the murine GADD34 (MyD116) is homologous to and can substitute for the corresponding domain of γ134.5 (12). In other studies, human GADD34 functionally replaced the HSV-1(F) γ134.5 genes (B.H. and B.R., unpublished studies). We have also shown that PP1α interacted with both γ134.5 and MyD116 chimeric proteins in vitro. These results argue that at least one of the functions of the GADD34 gene is similar to that of the γ134.5 gene, and that the γ134.5 gene may be useful in replacing its mammalian cell homolog to sustain protein synthesis in the face of stress that would naturally abolish protein synthesis by phosphorylation of eIF-2α.
Lastly, most viruses studied to date block phosphorylation of eIF-2α by encoding proteins which bind double stranded RNA [e.g., E2L of vaccinia (22–24), σ3 protein of Reo and rotaviruses (25–28), and NS1 protein of influenza (29)], or proteins or RNA that inactivate PKR by binding to it [e.g., vaccinia K3L, Epstein–Barr virus EBERS 1 and 2 (30), adenovirus VA1 RNA (30, 31), influenza virus-induced cellular p58 protein (32)], or degrade PKR [poliovirus (33)]. HIV-1 TAR sequence appears to bind to a cellular TAR binding protein and PKR (34–36). The exceptions may be hepatitis delta virus that does not activate PKR even though in vitro it consists largely of double-stranded RNA (37), and simian viurs 40 whose large T antigen acts at a step postactivation of PKR (38). This report suggests that HSV-1 and HSV-2 use a very different mechanism to preclude the shutoff of protein synthesis by targeting phosphorylated eIF-2α rather than either PKR or double-stranded RNA. The precise mechanism by which PP1 is directed to perform this task by γ134.5 protein remains to be resolved.
Herpesviruses form a large and diverse family. The available data indicate that the γ134.5 gene is conserved in but a few members of the family, suggesting that other herpesviruses employ a different mechanism to block either activation of PKR or its effect on eIF-2α.
Acknowledgments
We thank Suzanne Hessefort and Annette Olin for technical assistance. These studies were aided by grants from the National Cancer Institute (CA47451) and the National Institute for Allergy and Infectious Diseases (AI124009) to B.R., and the National Heart, Lung and Blood Institute (HL30121) to M.G. from the U.S. Public Health Service.
Footnotes
Abbreviations: HSV-1, herpes simplex virus 1; eIF-2α, the α subunit of the eukaryotic translation initiation factor 2; PKR, double-stranded RNA-dependent protein kinase; PP1α, protein phosphatase 1α; GST, glutathione S-transferase; GADD34, growth arrest and DNA damage protein 34; pfu, plaque-forming units.
References
- 1.Ackermann M, Chou J, Sarmiento M, Lerner R A, Roizman B. J Virol. 1986;58:843–850. doi: 10.1128/jvi.58.3.843-850.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chou J, Roizman B. J Virol. 1986;57:629–637. doi: 10.1128/jvi.57.2.629-637.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou J, Roizman B. J Virol. 1990;64:1014–1020. doi: 10.1128/jvi.64.3.1014-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou J, Kern E R, Whitely R J, Roizman B. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 5.Whitley R J, Kern E R, Chatterjee S, Chou J, Roizman B. J Clin Invest. 1993;91:2837–2843. doi: 10.1172/JCI116527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou J, Roizman B. Proc Natl Acad Sci USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou J, Roizman B. Proc Natl Acad Sci USA. 1994;91:5247–5251. doi: 10.1073/pnas.91.12.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGeoch D J, Barnett B C. Nature (London) 1991;353:609. doi: 10.1038/353609b0. [DOI] [PubMed] [Google Scholar]
- 9.Lord K A, Hoffman-Liebermann B, Liebermann D A. Nucleic Acids Res. 1990;18:2823. doi: 10.1093/nar/18.9.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan Q M, Lord K A, Alamo I, Jr, Hollander M C, Carrier F, Ron D, Kohn K W, Hoffman B, Liebermann D A, Fornace A J., Jr Mol Cell Biol. 1994;14:2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fornace A J, Jr, Nebert D W, Hollander M C, Luethy J D, Papathanasiou M, Fargnoli J, Holbrook N J. Mol Cell Biol. 1989;9:4196–4203. doi: 10.1128/mcb.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He B, Chou J, Liebermann D A, Hoffman B, Roizman B. J Virol. 1996;70:84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou J, Chen J J, Gross M, Roizman B. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Proud C G. Trends Biochem Sci. 1995;20:241–246. doi: 10.1016/s0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- 15.Ejercito P M, Kieff E D, Roizman R. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 16.Durfee T K, Bechere K, Cheng P L, Li S H, Yang Y Z, Kilburn A E, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 17.Gross M, Kaplansky D A. J Biol Chem. 1980;255:6270–6275. [PubMed] [Google Scholar]
- 18.Gross M, Rabinovitz M. Biochem Biophys Res Commun. 1973;50:832–838. doi: 10.1016/0006-291x(73)91320-x. [DOI] [PubMed] [Google Scholar]
- 19.Gross M, Kaplansky D A. Biochim Biophys Acta. 1983;740:255–263. doi: 10.1016/0167-4781(83)90134-3. [DOI] [PubMed] [Google Scholar]
- 20.Song Q, Khanna K K, Lu H, Lavin M F. Gene. 1993;129:291–295. doi: 10.1016/0378-1119(93)90282-8. [DOI] [PubMed] [Google Scholar]
- 21.Cohen P. Methods Enzymol. 1991;201:389–398. doi: 10.1016/0076-6879(91)01035-z. [DOI] [PubMed] [Google Scholar]
- 22.Davies M V, Chang H-W, Jacobs B L, Kauffman R J. J Virol. 1993;67:1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll K, Elroy-Stein O, Moss B, Jagust R. J Biol Chem. 1993;268:12837–12842. [PubMed] [Google Scholar]
- 24.Yuwen H, Cox J H, Yedell J W, Bennick J R, Moss B. Virology. 1993;195:732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]
- 25.Langland J O, Pettiford S, Jian B, Jacobs B L. J Virol. 1994;68:3821–3829. doi: 10.1128/jvi.68.6.3821-3829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beattie E, Denzler K L, Tartaglia J, Perkus M E, Paoletti E, Jacobs B L. J Virol. 1995;69:499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imani F, Jacobs B L. Proc Natl Acad Sci USA. 1988;85:7887–7891. doi: 10.1073/pnas.85.21.7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd R M, Sharkin A J. J Virol. 1992;66:6878–6884. doi: 10.1128/jvi.66.12.6878-6884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, Wambach M, Katze M G, Krug R M. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 30.Sharp T V, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud C G, Hilese K, Clemens M J. Nucleic Acids Res. 1993;21:4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghadge G D, Malhotra P, Furtado M R, Dhar R, Thimmapaya B. J Virol. 1994;68:4137–4151. doi: 10.1128/jvi.68.7.4137-4151.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee T G, Tang N, Thompson S, Miller J, Katze M G. Mol Cell Biol. 1994;14:2331–2342. doi: 10.1128/mcb.14.4.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Black T L, Barber G N, Katze M G J. J Virol. 1993;67:71–800. doi: 10.1128/jvi.67.2.791-800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMillan N A J, Chun R F, Sierovski D P, Galabru J, Toone W K, Samuel C E, Mak T W, Hovansessian A G, Jeang K-T, Williams B R G. Virology. 1995;213:413–424. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 35.Park H, Davies M V, Langland J O, Chang H-W, Nam Y S, Tartaglia J, Paoletti E, Jacob B L, Kaufman B L. Proc Natl Acad Sci USA. 1994;91:4713–1717. doi: 10.1073/pnas.91.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consentino G P, Venkatesan S, Serluca F C, Green N R, Mathews M B, Sonenberg N. Proc Natl Acad Sci USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNair A N B, Cheng D, Monjardino J, Thomas H C, Kerr I M. J Gen Virol. 1994;75:1371–1378. doi: 10.1099/0022-1317-75-6-1371. [DOI] [PubMed] [Google Scholar]
- 38.Swaminahan S, Rajan P, Savinova O, Jagus R, Thimmapaya B T. Virology. 1996;219:321–323. doi: 10.1006/viro.1996.0255. [DOI] [PubMed] [Google Scholar]