Abstract
In neurons, the proteins derived from mRNAs localized in dendrites have been implicated in synaptic plasticity. The cytoplasmic polyadenylation element (CPE), a cis element in the 3′-UTRs of specific dendritic mRNAs, promotes cytoplasmic polyadenylation-induced translation in response to synaptic stimulation. Here, we demonstrate that the CPE and its binding protein CPEB facilitate mRNA transport to dendrites. In rat hippocampal neurons infected with recombinant viruses, the CPE is sufficient to direct a reporter RNA into dendrites. CPEB-GFP protein forms RNA-containing particles that are transported into dendrites in a microtubule-dependent fashion at an average velocity of 4–8 μm/min. Such particles also contain maskin, a CPEB-associated factor that mediates cap-dependent translational repression of CPE-containing mRNA, and the molecular motors dynein and kinesin. Overexpression of CPEB in neurons promotes the transport of CPE-containing endogenous MAP2 mRNA to dendrites, whereas overexpression of a mutant CPEB that is defective for interaction with molecular motors inhibits this transport. In neurons derived from CPEB knockout mice, the dendritic transport of a CPE-containing reporter RNA is reduced. These results suggest a mechanism whereby CPE-containing mRNAs can be transported to dendrites in a translationally dormant form, but activated at synapses in response to NMDA receptor stimulation.
Keywords: synapse, CPEB, maskin, mRNA transport
The morphological and biochemical changes associated with experience-dependent plasticity of neuronal synapses may underlie long-term memory storage (Miller and Mayford 1999; Scannevin and Huganir 2000; Hering and Sheng 2001). For example, a neuron distinguishes naive from stimulated synapses by establishing a tag at a synapse once it is stimulated; its response to this stimulation (i.e., plasticity) is based on the recognition of this tag (Frey and Morris 1997; Martin et al. 1997). The nature of the tag(s) is (are) unknown, but for at least two forms of plasticity, long-lasting long-term potentiation (L-LTP) and long-term depression (LTD), protein synthesis in dendrites (Kang and Schuman 1996; Huber et al. 2000), possibly in the vicinity of the synapse (Bagni et al. 2000; Scheetz et al. 2000; Aakalu et al. 2001), is required.
Cytoplasmic polyadenylation is one mechanism that might govern mRNA translation in dendrites (Steward and Schuman 2001). In Xenopus oocytes, where a number of mechanistic details are known, several dormant mRNAs have small poly(A) tails; in response to developmental cues, the poly(A) tails are elongated and translation ensues. Polyadenylation is controlled by two cis-acting 3′-UTR elements, the CPE (cytoplasmic polyadenylation element; UUUUAU or similar) and AAUAAA. Polyadenylation is initiated when aurora (Eg2) phosphorylates CPEB, the CPE-binding factor (Mendez et al. 2000a). This phosphorylation induces CPEB to interact and possibly stabilize CPSF (cleavage and polyadenylation specificity factor) on the AAUAAA (Mendez et al. 2000b), which is probably necessary for the recruitment of poly(A) polymerase. Polyadenylation stimulates translation through maskin, a protein that interacts with both CPEB and the cap-binding factor eIF4E (Stebbins-Boaz et al. 1999). A maskin–eIF4E interaction inhibits translation by precluding an eIF4E–eIF4G interaction; the eIF4E–eIF4G complex is required to position the 40s ribosomal subunit on the mRNA. Polyadenylation leads to the dissociation of maskin from eIF4E and the association of eIF4G with eIF4E, thereby stimulating translation (Cao and Richter 2002).
Neurons appear to utilize a similar process to regulate translation in dendrites. CPEB and the other polyadenylation/translation factors noted above are expressed in the mammalian brain, particularly the hippocampus and probably the visual cortex as well (Wu et al. 1998; Huang et al. 2002). Synaptic stimulation results in polyadenylation and translation of the CPE-containing αCaMKII mRNA, but not of the CPE-lacking neurofilament mRNA (Wu et al. 1998). Polyadenylation occurs at synapses since glutamate or N-methyl-D aspartate (NMDA) treatment of synaptosomes isolated from rat hippocampal neurons also stimulates αCaMKII mRNA polyadenylation (Huang et al. 2002). Moreover, the translation of a reporter RNA appended with the αCaMKII 3′-UTR is stimulated when hippocampal neurons are treated with glutamate (Wells et al. 2001).
Synaptic activity not only stimulates mRNA translation in dendrites, it also induces the transport of mRNAs such as activity-regulated cytoskeletal protein (Arc; Steward et al. 1998), αCaMKII (Thomas et al. 1994), BC1 (Muslimov et al. 1998), brain-derived neurotrophic factor (BDNF), and trkB (Tongiorgi et al. 1997) to that region. As demonstrated by the experiments of Miller et al. (2002), this transport is important for synaptic plasticity, because transgenic mice harboring an αCaMKII mRNA that is restricted to the soma have impaired L-LTP and memory consolidation.
For mRNAs found in dendrites both prior to and after synaptic stimulation, premature translation would appear to have an adverse effect on synaptic plasticity. That is, if mRNAs undergoing transport are simultaneously translated, one might expect that newly made proteins would not specifically tag stimulated synapses, because they would be widely distributed in the dendrite. Krichevsky and Kosik (2001) suggested that RNA-containing particles en route to their destinations in dendrites are translationally silent because they do not contain key factors such as eIF4G, an initiation factor, and tRNA. It is plausible that tight regulation of mRNA transport, silencing, and activation might be coordinated by a small group of factors that are involved in each of these processes.
We have investigated whether the CPE and its binding protein CPEB, which regulate mRNA translation at synapses, might also facilitate mRNA transport in dendrites. In cultured hippocampal neurons infected with recombinant viruses, a reporter RNA with a CPE is transported more efficiently than one that lacks the CPE. Indeed, when placed within a polylinker sequence that is devoid of regulatory information, the CPE is sufficient for dendritic RNA transport. CPEB forms ribonucleoprotein (RNP) particles in cultured hippocampal neurons and neuroblastoma cells, is transported both anterograde and retrograde at an average velocity of 4–8 μm/min, and resides in a complex with both kinesin and dynein. CPEB also colocalizes with maskin, suggesting a mechanism whereby mRNA can be transported in a dormant form. Moreover, overexpression of CPEB enhances RNA transport, whereas overexpression of a CPEB mutant protein that is unable to associate with kinesin and dynein inhibits transport. Finally, in neurons derived from CPEB knockout mice, the transport of CPE-containing RNA is diminished. Taken together, these data show that CPEB facilitates mRNA transport as well as translation in neurons.
Results
The CPE facilitates mRNA transport to dendrites
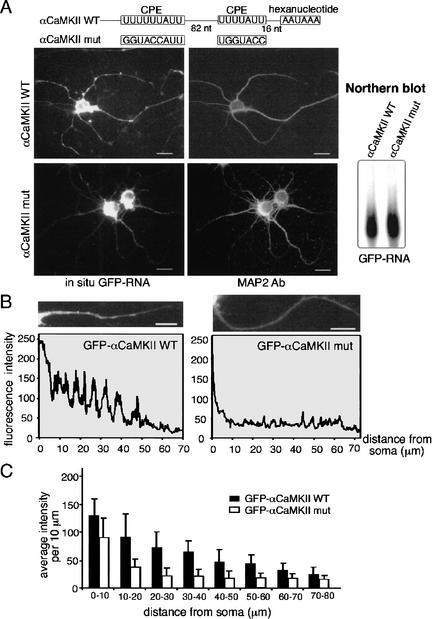
To assess whether the CPE is involved in dendritic mRNA transport, neurons were infected with Sindbis virus carrying GFP fused to the most distal 170 nucleotides of the αCaMKII 3′-UTR; this region contains two CPEs that are necessary for CPEB binding, polyadenylation, and translation (Wu et al. 1998; Wells et al. 2001; Huang et al. 2002). As a control, an αCaMKII 3′-UTR with mutated CPEs was also appended to GFP (αCaMKIImut). The subcellular localization of the fusion transcripts was examined by in situ hybridization to the GFP-coding sequence. Figure 1A shows that the transcript with the wild-type CPEs was present in the soma and in particles in the dendrites, which was confirmed by an immunostaining for the MAP2 dendritic marker. In contrast, a GFP reporter appended with a CPE-mutated αCaMKII 3′-UTR was only weakly detectable in dendrites. The steady-state levels of both RNAs in cells were similar (Fig. 1A, Northern blot). In addition, there was negligible signal in uninfected cells, or when the sense strand was used to probe GFP-expressing neurons (data not shown). Under both low and higher magnification, it is evident that the hybridization signal formed patches (Fig. 1A,B), which could represent RNA-protein particles (Knowles et al. 1996; Ainger et al. 1997). To quantify the RNA in dendrites, images from infected cells were taken under the same exposure setting that maximized the dynamic range of pixel intensity. Although the fluorescence in the soma was saturated, that in the dendrites was not, and consequently the fluorescence signal in this region should reflect the relative amount of GFP RNA that is present. Figure 1B shows one set of dendrites in which pixel intensity, in 10 μm bundles, was plotted against distance from the soma. The data show that the GFP appended with a CPE-containing αCaMKII 3′-UTR was transported more efficiently than its CPE-lacking counterpart. An analysis of several such experiments reveals that the CPEs from αCaMKII 3′-UTR mediate significantly (p < 0.05, Student's t test) enhanced transport of RNA as far as 60 μm from the cell soma (Fig. 1C).
Figure 1.
Facilitation of mRNA transport by the CPE. (A) Hippocampal neurons were infected with Sindbis virus carrying GFP fused to a 170-bp 3′-UTR of wild-type αCaMKII that contains two CPEs, or a mutated αCaMKII 3′-UTR in which the CPEs have been mutated. Seven hours after infection, the neurons were fixed for in situ hybridization with GFP sequences, followed by the MAP2 immunostaining. The Northern blot shows comparable expression of the two constructs in infected neurons. (B) Quantification of dendritic GFP mRNA as assessed by in situ hybridization. The average GFP RNA signal (wild-type or mutant αCaMKII 3′-UTR) in each segment of one major dendrite from each neuron was measured (NIH-Image) and plotted against the distance of the signal from the soma. (C) The mean intensity of each 10-μm dendritic segment was calculated from neurons expressing GFP-αCaMKIIwt (31 dendrites) or GFP-αCaMKIImut (27 dendrites) and presented in histogram form; error bars show S.E.M. The difference in the amount of dendritic GFP hybridization signal between the two RNAs is significant (p < 0.05, Student's t test). Bars, 10 μm.
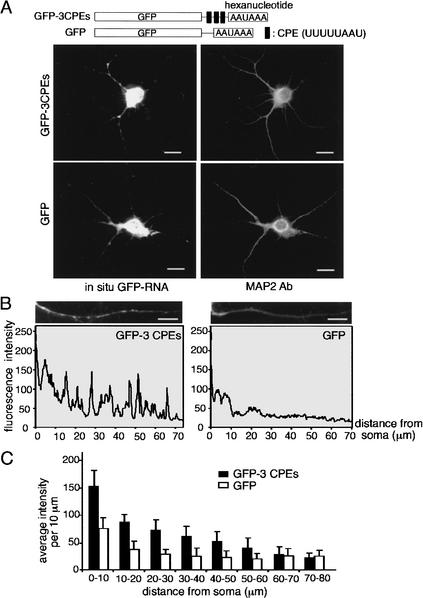
To assess whether the CPE is sufficient for mRNA transport to dendrites, we generated additional GFP-reporters where the 3′-UTR was composed of a polylinker sequence only (GFP), or a polylinker containing a triplicate CPE (GFP-3CPEs). Neurons infected with viruses carrying these constructs were analyzed in a similar way as described above. Figure 2A,B demonstrates that the CPE alone promoted efficient mRNA transport into dendrites. Indeed, except for the 60–80 μm segment, the CPE induced a significant increase (p < 0.05, Student's t test) in the amount of dendritic GFP mRNA (Fig. 2C).
Figure 2.
The CPE is sufficient for dendritic RNA transport. (A) Hippocampal neurons were infected with recombinant Sindbis virus as noted above, but in this instance, the GFP 3′-UTRs consisted of a polylinker sequence (GFP), or a polylinker containing three CPEs. The neurons were subjected to in situ hybridization for GFP RNA, and also costained with MAP2 antibody. (B) The quantification of the in situ hybridization signal for GFP was performed as in Figure 1B. (C) The mean intensity of each 10-μm dendritic segment was calculated from neurons expressing GFP-3CPE (11 dendrites) and GFP (10 dendrites) and presented in histogram form; error bars show S.E.M. The difference in hybridization signal between the two RNAs is significant (Student's t test). Bars, 10 μm.
Colocalization of CPEB with RNA and maskin
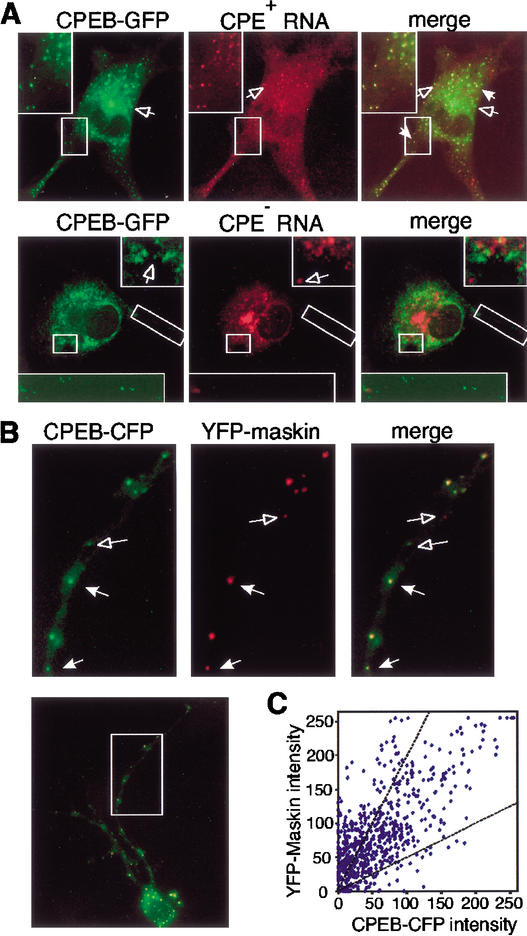
To investigate whether CPEB is involved in targeting CPE-containing mRNAs to dendrites, we used neurite-forming B104 neuroblastoma cells. They were injected with a plasmid encoding a CPEB-GFP fusion protein together with a fluorophore-labeled CPE-containing RNA (nontranslatable truncated GFP RNA fused to the 3CPE-containing polylinker noted in Fig. 2). The CPEB-GFP was assembled into particles that were detected in both the cell body and neurites and were frequently colocalized with the RNA (Fig. 3A). Little colocalization was evident when CPE-lacking RNA was employed (Fig. 3A). These results suggest that the CPEB-GFP particles can form complexes with RNA in vivo.
Figure 3.
CPEB-GFP forms particles and colocalizes with RNA and maskin. (A) Colocalization of CPEB-GFP and CPE-containing RNA in B104 cells. CPEB-GFP plasmid DNA and Alexa-546-labeled 3CPE-containing or -lacking polylinker RNA were coinjected into B104 cells and imaged by dual-channel confocal microscopy. Particles containing either CPEB-GFP (green) or Alexa-546-labeled RNA (red) are denoted by open arrows; particles containing both CPEB-GFP and RNA (yellow) are denoted by solid arrows. (B) Hippocampal neurons were cotransfected with CPEB-CFP (green) and YFP-maskin (red) and imaged by confocal microscopy. The arrows in the magnified regions denote particles that contain both CPEB-CFP and YFP-maskin proteins. (C) Colocalization of CPEB-CFP and YFP-maskin in 620 RNA particles from dendrites of eight double-transfected cells was quantified by single-particle ratiometric analysis. The two broken lines define the region in which CPEB and maskin in particles are present within 1:2 and 2:1 intensity ratios.
In Xenopus oocytes, CPEB-mediated translation requires the activity of maskin, a CPEB-associated protein (Stebbins-Boaz et al. 1999; Cao and Richter 2002). To determine whether such an interaction could occur in neurons, primary hippocampal neurons were cotransfected with plasmids encoding CPEB-CFP (cyan fluorescent protein) and YFP (yellow fluorescent protein)-maskin, and the distribution of the expressed proteins was examined by confocal microscopy (Fig. 3B). The open arrows in the figure indicate particles containing either CPEB (rendered in green) or maskin (rendered in red); the solid arrows indicate colocalization of both proteins (yellow). Six hundred-twenty particles from the dendrites of eight transfected cells were subjected to single-particle ratiometric analysis (Kwon et al. 1999) by plotting the intensities of YFP-maskin and CPEB-CFP in each particle (Fig. 3C). The majority of the dendritic particles (∼94%) contained both CPEB and maskin signals, ∼61% of which fell within diagonal lines denoting ratios of these two proteins of 1:2 or 2:1. These results suggest that CPEB and maskin might be transported to dendrites together, possibly with CPE-containing RNA. Because maskin functions as a translational repressor through its interaction with eIF4E, colocalization of maskin and CPEB would suggest a plausible mechanism for maintaining the translational silence of CPE-containing RNA during transport. We examined whether eIF4E was also colocalized in CPEB/maskin particles; it was distributed uniformly throughout the cells (data not shown).
CPEB particles: Movement, microtubules, and motors
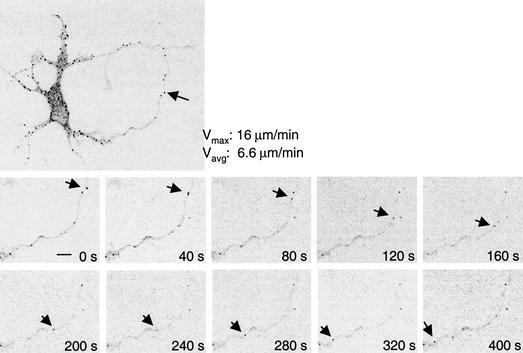
The detection of CPEB particles in dendrites implies that they may have been actively transported to that region. To investigate this possibility, time-lapse confocal images of CPEB-GFP particles in hippocampal neurons were recorded every 20 sec for 20 frames. This procedure revealed that although the majority of the particles oscillated bidirectionally within confined spaces, some traveled in either anterograde or retrograde directions over longer distances at an average velocity of 4–8 μm/min (Fig. 4). In some instances, depolarization of neurons with glutamate or KCl induced the splitting or merging of particles, although the velocity was unaffected (data not shown).
Figure 4.
Transport of CPEB-GFP particles. Confocal time-lapse images of CPEB-GFP particles in transfected hippocampal neurons depicting a retrograde movement. Vmax and Vavg are the maximal and average velocities, respectively, of indicated particle movement. Bar, 5 μm.
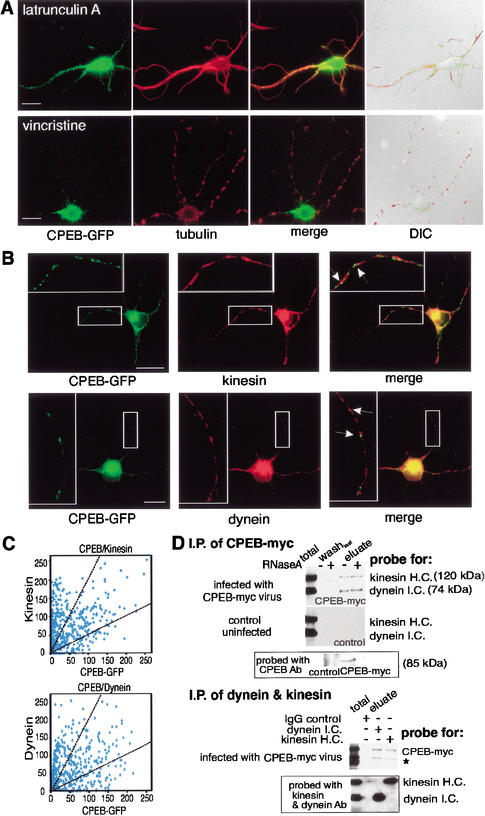
The directionality and velocity of CPEB-GFP particle movement are consistent with translocation along microtubules (Bassell et al. 1994). To examine the role of cytoskeletal elements in particle movement, neurons expressing CPEB-GFP were treated with latrunculin A, which disrupts actin filaments, or with vincristine, which depolymerizes microtubules (Allison et al. 2000), and then labeled with rhodamine phalloidin to detect actin polymers or immunostained with tubulin antibody to detect microtubules. Latrunculin A treatment, while disrupting actin polymers (data not shown), had no effect on microtubule staining or CPEB-GFP fluorescence in dendrites (Fig. 5A, merge). On the other hand, vincristine destroyed much of the microtubule network, such that only isolated patches of tubulin could be detected; it also inhibited CPEB-GFP transport into dendrites (Fig. 5A). The dendrites remained intact with both treatments (Fig. 5A, DIC). Thus, CPEB-GFP particle transport is microtubule-dependent.
Figure 5.
CPEB-GFP particles contain dynein and kinesin and are transported in a microtubule-dependent manner. (A) CPEB-GFP transfected hippocampal neurons were treated with latrunculin A, which disrupts microfilaments, or vincristine, which disrupts microtubules, and examined for GFP fluorescence (green) as well as tubulin immunofluorescence (red). Only vincristine disrupted most microtubules by leaving a patchwork of these structures, whereas neither treatment affected dendrite integrity (merged DIC image). Vincristine, but not latrunculin A, also inhibited GFP-CPEB transport. (B) CPEB-GFP-expressing neurons were immunostained with kinesin or dynein antibody and imaged by confocal microscopy. The magnified images (insets) show particles containing CPEB-GFP colocalized with motor proteins (solid arrows). (C) The green and red fluorescence intensities representing CPEB-GFP and either kinesin or dynein were analyzed from 407 and 403 dendritic CPEB-GFP-containing particles, respectively. The intensity of kinesin or dynein in each particle was plotted against the intensity of CPEB-GFP in the same particle. The two broken lines define the region in which CPEB and the molecular motors in particles were detected in 1:2 and 2:1 intensity ratios. Bars, 10 μm. (D) Coimmunoprecipitation of kinesin and dynein with CPEB-myc13. Cytoplasmic extracts prepared from hippocampal neurons, either uninfected or infected with Sindbis virus-expressing myc-tagged CPEB, were immunoprecipitated with myc antibody in the absence or presence of RNase A. The antibody beads were washed, eluted with 2 M KCl, and probed on a Western blot for kinesin and dynein, as well as for CPEB. For reciprocal immunoprecipitation experiments, the extract from CPEB-myc13-infected neurons was incubated with monoclonal antibodies against cytoplasmic dynein intermediate chain or kinesin heavy chain; mouse IgG served as a negative control. The immunoblots were probed with antibodies against CPEB-myc13, dynein, and kinesin. The asterisk denotes the breakdown product of CPEB-myc13.
The bidirectional microtubule-dependent movement of CPEB-GFP particles suggests that plus-end and minus-end motor proteins, such as kinesin and dynein, might both be involved in this translocation. Using an antibody specific against conventional kinesin heavy chain and cytoplasmic dynein intermediate chain, we employed confocal imaging to analyze ∼400 CPEB-GFP particles in the dendrites of five neurons and found that ∼43% also contained these two motor proteins in ratios of 1:2 or 2:1 (Fig. 5B,C). To examine the interaction of CPEB with kinesin and dynein, neurons were infected with Sindbis virus expressing CPEB-myc13 (i.e., 13 myc epitope repeats fused to the C terminus of CPEB) and subjected to myc antibody immunoprecipitation in the presence or absence of RNase A. Figure 5D shows that both kinesin and dynein were coimmunoprecipitated with CPEB-myc13 irrespective of the RNase A, whereas in uninfected cells, this procedure did not result in the detection of these same proteins. The figure also shows that heterologous CPEB was indeed immunoprecipitated with the myc antibody. To confirm the interaction of CPEB and motor proteins in the presence of RNase A, the CPEB-myc13-expressing neuronal extract was reciprocally immunoprecipitated with antibodies against dynein or kinesin. The CPEB-myc13 protein was coimmunoprecipitated with the motors (Fig. 5D) but not with mouse IgG control antibody. These data indicate that kinesin and dynein are associated with CPEB-containing complexes.
A region of CPEB required for particle transport: Possible use as a dominant negative mutation
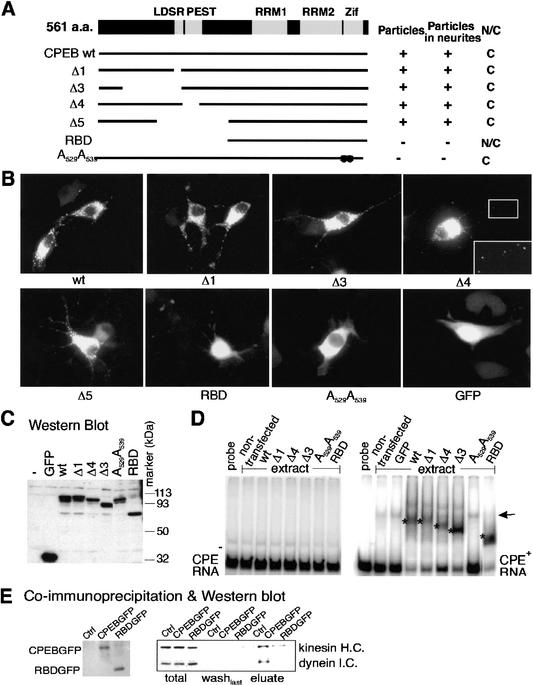
With the goal of identifying a mutant CPEB that could serve as a dominant negative factor for RNA transport, we delineated a region of this protein required for particle transport. DNA encoding CPEB truncation mutants fused to GFP was transfected into B104 cells, and the resulting distribution and structure of the proteins were examined by fluorescence microscopy. Figure 6A depicts key features of CPEB including the LDSR aurora phosphorylation site (Mendez et al. 2000a), the PEST protein destruction box (Mendez et al. 2002), two RNA recognition motifs (RRMs), and two zinc fingers (Zif; Hake et al. 1998); the RRMs and zinc fingers are required for optimal RNA binding. Most of the mutated CPEB proteins that retained the ability to bind RNA (Δ1–Δ5) formed particles that were transported to neurites; the mutant protein that does not bind RNA (A529A539; Hake et al. 1998) was retained in the cytoplasm and distributed in a diffuse manner. The CPEB mutant protein that contained an intact RNA binding region (RBD) was present in the cytoplasm as well as in the nucleus, but failed to form particles (Fig. 6A,B). A Western blot (Fig. 6C) shows that the CPEB-GFP fusion proteins remained intact following transfection. RNA gel shifts confirmed that CPEB mutant proteins Δ1, Δ3, Δ4, and RBD specifically bind CPE-containing RNA (Fig. 6D). This assay also indicates that the GFP tag at the C terminus of CPEB did not affect its RNA binding specificity. Thus, the RNA binding domain of CPEB is necessary but not sufficient for particle formation.
Figure 6.
A CPEB mutant is defective for kinesin and dynein interaction. (A) Salient features of CPEB showing the LDSR aurora phosphorylation site, a PEST instability region, two RNA recognition motifs (RRMs) and two zinc fingers (Zif). The structures of various CPEB mutant proteins are also shown, as well as whether they form granules, are transported to neurites, or are nuclear or cytoplasmic. (B) Localization of wild-type and mutant CPEB-GFP fusion proteins in transfected B104 cells. (C) Western blot of wild-type and mutant CPEB-GFP proteins in transfected B104 cells. (D) Gel shift using extracts from B104 cells transfected with wild-type or mutant CPEB-GFP fusion proteins with CPE-lacking (left) or CPE-containing (right) RNA. The asterisks denote specific shifts attributed to CPEB proteins. The arrow denotes a nonspecific band caused by an unknown protein from B104 cells. (E) Coimmunoprecipitation of wild-type or RBD mutant CPEB-GFP fusion proteins with motor proteins. Cells infected with DNA encoding the heterologous proteins noted above were immunoprecipitated, washed, and eluted with SDS loading buffer. The eluates were probed for CPEB-GFP and RBD-GFP (left) as well as kinesin heavy chain and dynein intermediate chain.
We focused on the RBD mutant of CPEB because although it retained the ability to bind RNA, it was not transported to neurites. To determine why it might be defective for transport, extracts prepared from hippocampal neurons infected with viruses harboring this mutant protein as well as wild-type CPEB (both fused to GFP) were immunoselected and probed on a Western blot for kinesin and dynein. Although both motor proteins were immunoselected with wild-type CPEB, as shown in Figure 5, the RBD mutant did not bind any kinesin or dynein above background (Fig. 6E). Thus, the RBD protein could serve as a useful tool to investigate CPEB-mediated RNA transport to dendrites.
Transport of CPE-containing RNA to dendrites: Control by CPEB
We have employed three methods to determine whether CPEB is involved in RNA transport to dendrites. In the first approach, synaptoneurosomes prepared from neurons infected with viruses expressing GFP, CPEB-GFP, or RBD-GFP were used to detect endogenous RNA by reverse transcription-coupled real-time PCR. Thus, if the CPE and CPEB are involved in RNA transport and if the transport machinery is not saturated, overexpressed CPEB-GFP might enhance the amount of CPE-containing mRNAs in synaptoneurosomes, whereas the RBD-GFP mutant might reduce this amount.
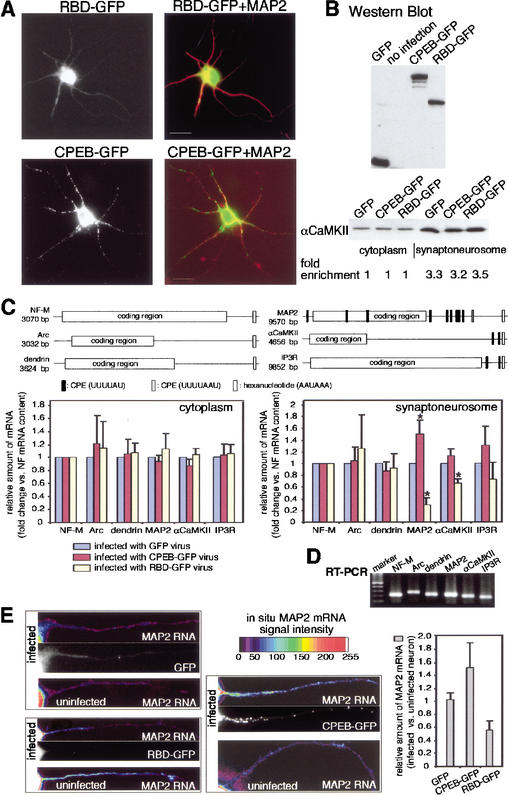
To ensure the accuracy of these experiments, neurons obtained from the same litter of embryos were used for each complete set of experiments, and were infected with nearly the same number of viral particles (4 × 106 neurons and 5 × 106 viral particles/100-mm dish) expressing GFP, CPEB-GFP, or RBD-GFP. The expression patterns of these proteins again demonstrate that CPEB formed particles in dendrites (MAP2 immunostaining positively identified dendrites), whereas RBD remained in the soma (Fig. 7A). The levels of these proteins were also comparable as determined by GFP immunoblotting (Fig. 7B). Synaptoneurosomes isolated from the infected neurons showed a similar degree of enrichment (3.2- to 3.5-fold) of αCaMKII compared to the starting cytoplasmic extract (Fig. 7B). To avoid possible contamination from DNA, RNA extracted from the synaptoneurosome fraction was pretreated with DNase I before reverse transcription. The cDNAs from each set of experiments were PCR-amplified in the same 96-well plate using primer sets for six dendritic RNAs, three of which contain CPEs [MAP2, αCaMKII, inositol 1,4,5-triphosphate receptor (IP3R)], and three of which lack CPEs (NF-M, Arc, dendrin; Fig. 7C). cDNAs derived from RNAs extracted from the cytoplasm and synaptoneurosome fraction were subjected to quantitative PCR using SYBR Green, a DNA amplification indicator that fluoresces only when it is incorporated into double-stranded DNA. The final PCR products were analyzed on agarose gels to assess the specificity of each amplification; in each case, only a single band was detected (Fig. 7D). Finally, each PCR amplification product was gel-isolated and sequenced to confirm the identity of the DNA. The relative amounts of the target mRNAs were calculated using the threshold cycle number (Ct) derived from real-time PCR. From three independent experiments, there was a significant reduction (Student's t test, p < 0.05) in the amount of the CPE-containing RNAs MAP2 and αCaMKII in synaptoneurosomes isolated from RBD-GFP expressing neurons (Fig. 7C). In addition, a significant increase (p < 0.05) in the amount of synaptoneurosome MAP2 mRNA was also observed in CPEB-GFP-expressing neurons (Fig. 7C, right). Although the amount of IP3R mRNA in synaptoneurosomes did not change in a significant manner, there was still a noticeable reduction in cells expressing RBD-GFP, and a noticeable increase in cells expressing CPEB-GFP. In contrast, the levels of CPE-lacking RNAs in synaptoneurosomes barely changed irrespective of the virus that was used for infection (Fig. 7C). Because the levels of the target RNAs in the cytoplasm did not change regardless of the virus used for infection (Fig. 7C, left), the different amounts of the target mRNAs in synaptoneurosomes could not be due to differential stability. Thus, results from this study indicate that the CPE and CPEB facilitate the transport of specific mRNAs to dendrites.
Figure 7.
CPEB facilitates RNA transport to dendrites. (A) Infection of hippocampal neurons with Sindbis virus harboring RBD-GFP (RBD refers to the RNA binding region of CPEB) and CPEB-GFP, which were also immunostained with antibody directed against MAP2. Note that whereas CPEB-GFP was detectable in the soma and dendritic arbors, RBD-GFP was confined to the soma. Bars, 10 μm. (B) The extracts from neurons infected with recombinant Sindbis virus were immunoblotted for GFP and CPEB-GFP fusion proteins. The bottom portion of the panel shows the degree of synaptoneurosome enrichment following infection with recombinant virus. Using a densitometric scan of a Western blot probed for αCaMKII, the degree of enrichment was comparable for each preparation. (C) Key features of neurofilament-M (NF-M), Arc, and dendrin mRNAs, which lack obvious CPEs, and MAP2, αCaMKII, and IP3 receptor mRNAs, which contain multiple putative CPEs. Synaptoneurosomes were prepared from GFP-, CPEB-GFP-, and RBD-GFP-containing Sindbis virus-infected cells, and the amount of the RNAs noted above was quantified by real-time PCR. To take into account small variations of the amount of cDNA in each preparation, the amount of RNA in synaptoneurosomes following GFP-expressing virus infection was used as the normalization standard. Error bars indicate the S.E.M., and the asterisks denote significance (p < 0.05, Student's t test). (D) The gel at the bottom shows the final PCR amplification products. (E) In situ hybridization of MAP2 RNA. Hippocampal neurons were infected with virus containing GFP, CPEB-GFP, or RBD-GFP and processed for in situ hybridization for MAP2 RNA. The intensity of the hybridization signal is color-coded. On the same coverslip, the MAP2 RNA hybridization signals were compared between GFP-infected and uninfected neurons, CPEB-GFP-infected and uninfected neurons, and RBD-GFP-infected and uninfected neurons, quantified, and plotted as a ratio of the two (right). The differences between the MAP2 RNA signals in dendrites are significantly different between CPEB-GFP-infected and uninfected neurons, and between RBD-GFP-infected and uninfected neurons (p < 0.05, Student's t test). The GFP fluorescence in dendrites from infected neurons is also shown.
To confirm these results, we performed in situ hybridization with digoxygenin-labeled antisense oligonucleotides to MAP2 mRNA on cultured hippocampal neurons infected with viruses harboring GFP, CPEB-GFP, or RBD-GFP. We analyzed the amount of hybridization signal (sense oligonucleotides yielded no signal; data not shown), specifically in dendrites, of neurons on the same coverslip that were infected or uninfected. For example, there was no change in the amount of MAP2 mRNA in dendrites from neurons on the same coverslip that were either GFP-infected or uninfected (Fig. 7E). On the other hand, there was more MAP2 RNA in dendrites from neurons on the same coverslip that were infected with CPEB-GFP compared to those that were uninfected. Finally, there was less MAP2 RNA in dendrites from neurons infected with CPEB-RBD compared to those that were uninfected (Fig. 7E). The enhancement (with CPEB-GFP) or reduction (with RBD-GFP) of MAP2 RNA in dendrites compared to uninfected cells was significant (p < 0.05, Student's t test).
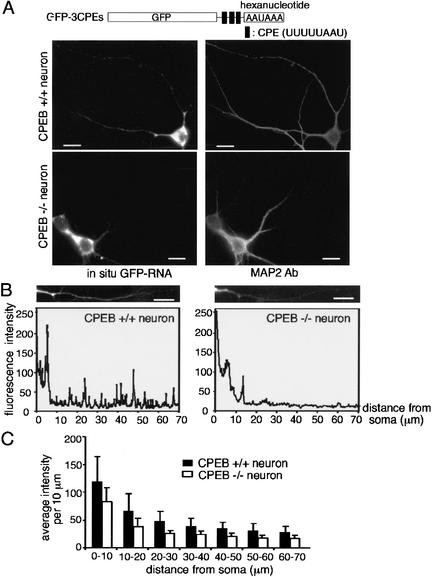
Finally, we examined RNA transport in hippocampal neurons from CPEB knockout mice (Tay and Richter 2001). Cells from these animals as well as from wild-type mice were cultured for 1 wk and then infected with virus expressing GFP-3CPEs RNA, whose transport we have shown is CPE-dependent (Fig. 2). Compared to wild-type neurons, the relative amount of reporter RNA in dendrites, which was monitored by in situ hybridization to GFP, was reduced in the CPEB knockout (KO) neurons by a significant amount (p < 0.05, Student's t test; Fig. 8). These studies demonstrate that the CPE, and CPEB, promote the transport of RNA to dendrites.
Figure 8.
Deficient transport of CPE-containing RNA in neurons from CPEB knockout mice. (A) Wild-type (CPEB+/+) and knockout (CPEB−/−) hippocampal neurons were infected with Sindbis virus expressing GFP-3CPEs. The neurons were subjected to in situ hybridization for GFP RNA, and also costained with MAP2 antibody. (B) The quantification of the in situ hybridization signal for GFP was performed as in Figure 1B. (C) The mean intensity of each 10-μm dendritic segment was calculated from 20 CPEB+/+ and 20 CPEB−/− neurons expressing GFP-3CPE and presented in histogram form; error bars show the S.E.M. The difference in hybridization signal between the two RNAs is significant (p < 0.05, Student's t test). Bars, 10 μm.
Discussion
The results presented in this study demonstrate that two factors that control polyadenylation-induced translation (Wu et al. 1998; Mendez and Richter 2001; Huang et al. 2002) can assemble in neurons into RNA particles that are transported to dendrites. These particles, which include CPEB and perhaps maskin as well, move bidirectionally along microtubules that are probably driven by kinesin and dynein, two motor proteins that coimmunoprecipitate with CPEB. Analysis of reporter RNAs appended with various heterologous 3′-UTRs shows that the CPE is sufficient for transport to dendrites. The transport process is enhanced by overexpression of wild-type CPEB but repressed by overexpression of a dominant negative mutant CPEB that is unable to bind the motor proteins. These results suggest a model for CPE-containing RNA transport to dendrites. In the soma, we envisage that RNA is assembled into particles containing CPEB, maskin, possibly eIF4E, kinesin, and dynein. The particles are transported in a translationally repressed state along microtubules. When NMDA receptors are stimulated, calcium entry presumably causes the activation of aurora, a kinase that in turn phosphorylates and activates only the CPEB that is local, in the vicinity of the stimulated synapse (Huang et al. 2002). This phosphorylation event then induces cytoplasmic polyadenylation and translation of local CPE-containing mRNAs; the proteins derived from these newly translated mRNAs might then influence synaptic plasticity. This general model could account for the repression of specific mRNAs in dendrites and their rapid activation at stimulated synapses.
Overexpression of the CPEB RNA binding domain (RBD-GFP) reduced the distribution of endogenous CPE-containing RNAs (MAP2, αCaMKII), but not CPE-lacking RNAs (Arc, dendrin, NF-M) to the synaptodendritic compartment of dendrites. We infer that the RBD protein displaced endogenous CPEB(s) from the CPE, and because the RBD protein could not respond to the appropriate transport signals, it inhibited the RNAs from traveling to dendrites. Thus, we propose that the CPEs of MAP2 and αCaMKII RNAs are important for transport to dendrites. However, other dendritic targeting elements for both of these RNAs were identified previously, and appear to be at odds with the results presented here. For example, although all investigators agree that the 3′-UTR of αCaMKII mRNA mediates dendritic localization (Mayford et al. 1996; Rook et al. 2000; Aakalu et al. 2001), Mori et al. (2000) suggested that the most proximal 94 nucleotides of the 3′-UTR are essential, whereas Blichenberg et al. (2001) stated that an ∼1300-nucleotide sequence within the 3′-UTR is required. Neither of these sequences contains an obvious CPE. One key difference between our results and those noted above is that we examined endogenous as well as exogenous mRNAs, whereas the other investigators focused exclusively on reporter mRNAs overexpressed in neurons. Moreover, in our analysis of reporter RNA transport, we measured the amount of RNA along the length of the dendrite, which yields many data points from which statistical significance can be derived. In contrast, Mori et al. (2000) and Blichenberg et al. (2001) scored RNAs as being positive for transport if they were detected only 10–20 μm away from the soma. Finally, in our studies we demonstrated that the CPE, when placed within a polylinker sequence that presumably lacks trafficking information, was sufficient to stimulate RNA transport. On the other hand, both Mori et al. (2000) and Blichenberg et al. (2001) used truncated 3′-UTRs to measure transport; such RNAs may contain or lack inhibitory sequences that could affect transport efficiencies (Mori et al. 2000). It is noteworthy that in transgenic mice, an αCaMKII RNA that contains the 94-base element identified by Mori et al. (2002) but lacking other sequences, including the CPE, remains in the cell soma (Miller et al. 2002). In any case, the results presented here demonstrate that the CPEs in the 3′-UTR of αCaMKII RNA are sufficient to mediate dendritic transport.
Complex RNP transport particles
Neuronal RNP transport particles are macromolecular structures composed of a large number of diverse proteins and probably several RNA molecules (Mouland et al. 2001). Whether the RNPs contain different mRNAs is a matter of debate, although at least some contain tRNAs and ribosomes (Krichevsky and Kosik 2001). Two proteins thought to be involved in dendritic RNA transport in neurons are staufen (Kiebler et al. 1999; Monshausen et al. 2001; Tang et al. 2001) and MARTAs (Rehbein et al. 2002). Staufen is a double-stranded RNA binding protein first identified in Drosophila as mediating bicoid RNA localization in embryos. In neurons, Staufen is thought to be a component of most RNP transport particles (Krichevsky and Kosik 2001), and its overexpression drives bulk neuronal RNA (i.e., cytoplasmic ethidium bromide-stained material) into dendrites (Tang et al. 2001). These data suggest that staufen does not play a specificity role in discriminating among mRNAs for transport to dendrites. MARTA 1 and MARTA 2 are members of the KSRP/FUSE binding protein (FBP) family of DNA/RNA binding proteins that are involved in transcription and nuclear pre-mRNA processing (Duncan et al. 1994; Min et al. 1997). The MARTA proteins bind to a sequence of MAP2 RNA that is thought to enhance dendritic transport (Rehbein et al. 2002). Although a truncated form of KSRP is thought to regulate β-actin mRNA transport to growth cones of developing neurons (Gu et al. 2002), there is little direct evidence that the MARTA proteins are necessary for MAP2 RNA transport. Thus, although RNP particles containing these proteins all exhibit a similar morphology and velocity of transport (∼4–10 μm/min; Knowles et al. 1996), CPEB so far appears to be unique in that it modulates specific endogenous RNA transport to neuronal dendrites.
Glutamate stimulation of CPEB activity
Glutamate stimulation of synapses induces CPEB-dependent polyadenylation (Huang et al. 2002). Although glutamate- or KCl-induced depolarization induced the splitting and merging of CPEB-GFP particles, it did not enhance the rate of transport, as has been observed for other particles (Knowles et al. 1996). Although the function of this splitting/merging is unknown, it could be a reflection of the change in RNP particle morphology as shown by Krichevsky and Kosik (2001). Those investigators identified a large and dense RNP particle that sediments faster than polysomes in sucrose gradients, which is replete with ribosomes and mRNA but devoid of other components of the translational apparatus such as tRNA and the initiation factor eIF4G, and thus presumably is not actively translated. The morphology of this RNP particle seems to change when neurons are treated with KCl; it becomes less dense and distributes more uniformly in dendrites. Commensurate with this change in dispersion pattern is a shift into polysomes of several dendritic mRNAs (Krichevsky and Kosik 2001). These data are consistent with our observations that glutamate stimulates polyadenylation at synapses (Huang et al. 2002) and CPE-dependent mRNA translation (Wells et al. 2001).
Expression of multiple CPEBs in neurons
Neurons derived from a CPEB knockout mouse show reduced, but not ablated, CPE-containing RNA transport, suggesting that other proteins can also drive this process. In this regard, we note that the mouse genome contains four CPEB genes; the protein analyzed here is CPEB-1, the founding member of this multigene family. Two other CPEB-like proteins, CPEB-2 and CPEB-3, are similar to CPEB-1 in the RNA binding region (Mendez and Richter 2001); they bind CPE-containing RNA in vitro and are present in the mammalian brain (Y.-S. Huang and J. Richter, unpubl.). We suspect that CPEB-2 and/or CPEB-3 might functionally overlap with CPEB-1 and facilitate dendritic RNA transport.
Is the CPE a default transport signal?
The results presented in this study indicate that at least two endogenous mRNAs, αCaMKII and MAP2, are targets of the CPEB transport machinery. Although the CPE might be expected to be a feature that distinguishes these dendritic mRNAs from those in the cell body, it appears that the regulatory mechanism for mRNA transport might be more complicated and require multiple RNA binding proteins. That is, the CPE is composed only of U and A residues, but because eukaryotic 3′-UTRs are very UA-rich, it seems likely that a number of other mRNAs, perhaps several that are confined to the soma, would also harbor this sequence. If so, then how could the CPE drive the dendritic transport only of certain mRNAs, but not others? We think there are two possibilities. First, the CPE might have to be present in a certain “open” context that allows easy access to CPEB; RNAs with strong secondary structure in the 3′-UTR might not associate with CPEB and the rest of the transport machinery. Second, consider that when the CPE is placed within a polylinker sequence, which presumably lacks any regulatory information, the RNA is transported to dendrites, demonstrating the sufficiency of this element for transport. This sufficiency might indicate that the CPE is a default sequence for dendritic transport, and that CPE-containing RNAs that remain in the soma might have an additional sequence (and associated binding protein) that is essential for cell body retention. Further investigations will address these possibilities.
Materials and methods
Plasmid construction and riboprobe preparation
To create fusion proteins with GFP, the stop codon of full-length CPEB was mutated to a cysteine by PCR amplification; the PCR product was digested with HindIII and BglII and cloned into HindIII/BamHI-digested pEGFP-N1 (Clontech). The BglII and HindIII fragments excised from wild-type and several deletion mutant CPEB proteins in pBluescript (Mendez et al. 2002) were cloned into the above vector digested with BglII and HindIII. A plasmid containing the CPEB point mutations A529A539 (Hake et al. 1998) was constructed in a similar way. Maskin was fused to the C terminus of GFP. Full-length maskin was PCR amplified, digested with XhoI and BamHI, and cloned into XhoI/BamHI-digested pEYFPC3, a derivative of pEYFPC1 (Clontech) to produce pEYFP-maskin.
To construct recombinant Sindbis virus, CPEB-GFP and RBD-GFP were digested with BglII and NotI, blunt-ended, and ligated to StuI-digested pSinRep5 (Invitrogen). To generate GFP-reporter viruses for in situ hybridization, the NheI-XbaI fragment of pEkGFP-αCaMKII 3′UTR wild type or mutant (Wells et al. 2002) was cloned into XbaI-cleaved pSinRep5 plasmid. The 110-bp PstI-KpnI DNA containing 3CPE and hexanucleotide was excised from pBSSK-3CPE (de Moor and Richter 1999) and cloned into pEkGFP to yield the pEkGFP-3CPE. This plasmid was cleaved with BamHI, blunt-ended, GFP-3CPE DNA excised following NheI digestion, and ligated into the XbaI-StuI-digested pSinRep5.
RNAs used for the gel shift assay have been described (de Moor and Richter 1999). For microinjection, the nontranslated (NT) truncated GFP sequence was fused to the linker sequence containing no CPE or three CPEs to increase the size of injected RNA to prevent diffusion Depending on the experiment, linearized plasmids were transcribed in the presence of 32P-UTP, Alexa-546-labeled UTP (Molecular Probes), or digoxigenin (dig)-UTP (Roche). Further details of the cloning procedures are available upon request.
In situ hybridization and immunostaining
In situ hybridization of reporter RNA in primary hippocampal neurons was performed as described (Knowles et al. 1996). To detect the digoxigenin-labeled probes, as well as immunolabeling of dendrites, the cells were incubated with the MAP2 antibody (AP20, Chemicon), followed by the secondary Alexa-488-labeled goat α-mouse IgG (Molecular Probes) and rhodamine-conjugated anti-digoxigenin Fab fragment (Roche). For some experiments, transfected B104 cells were fixed with 4% formaldehyde and mounted onto microscope slides. For immunostaining, CPEB-GFP-expressing cells were fixed with methanol, permeabilized with 0.1% Triton X-100 in PBS, blocked with 10% horse serum and 1% goat serum in PBS for 1 h, and treated with primary antibodies directed against kinesin heavy chain (MAB1614), or cytoplasmic dynein intermediate chain (MAB1618, both from Chemicon), or tubulin (Sigma), followed by Alexa-546-labeled goat α-mouse secondary antibody (Molecular Probes).
In situ hybridization of endogenous MAP2 mRNA with digoxygenin (dig)-labeled antisense oligonucleotide was performed according to the manufacturer's protocol (Roche). To detect the dig-labeled probes, the cells were incubated with fluorescein-conjugated anti-dig antibody and subsequently amplified with Alexa-594-conjugated antifluorescein antibody (Molecular Probes). The GFP expression was detected by anti-GFP antibody (Novus), followed by Alexa-488-labeled goat α-rabbit secondary antibody.
Cell culture, transfection, and Sindbis virus infection
Cultures of rat hippocampal neurons (Banker and Goslin 1988) were grown on coverslips at a cell density of 10,000/cm2 for 1 wk, infected with Sindbis virus for 1 h, and then placed in dishes containing a glial cell layer and incubated for an additional 4 h before immunostaining or in situ hybridization. Because of the infertility of CPEB knockout mice, the hippocampi from individual embryos obtained from CPEB heterozygous matings were trypsinized and plated separately on coverslips. During the 1-wk culture period, the tails collected from those embryos were used for genotyping (Tay and Richter 2001); the neurons from wild-type and CPEB null animals were then analyzed.
B104 neuroblastoma cells were grown in DMEM supplemented with 2.5% FBS and plated onto poly-L-ornithine (50 μg/mL)-coated coverslips a day before the transfection (via Effectene, QIAGEN) or injection.
Immunoprecipitation
Hippocampal neurons cultured for 1 wk in Neurobasal medium with B27 supplement (Invitrogen) were infected with Sindbis virus expressing CPEB-myc13. Three hours after infection, the neurons were harvested and homogenized in buffer (10 mM Hepes at pH 7.4, 50 mM KCl, 1 mM MgCl2, 0.1 mM ZnCl2, 0.5 mM DTT, 1 mM PMSF, 0.1% NP40) and centrifuged for 5 min at 1000g. The supernatant was incubated with antibodies for myc, kinesin heavy chain, or dynein intermediate chain in the presence or absence of 50 μg/mL RNaseA. After several washes, the coimmunoprecipitated proteins were eluted with 2 M KCl, TCA-precipitated, and boiled in SDS sample buffer.
Gel retardation and Western blotting
Each 60-mm dish of B104 cells transfected with various CPEB-GFP constructs was harvested in 120 μL of the buffer containing 10 mM Hepes at pH 7.4, 50 mM KCl, 1 mM MgCl2, 0.1 mM ZnCl2, 0.5 mM DTT, 1 mM PMSF, and 10% glycerol, and frozen and thawed 3× to break the cytoplasmic membranes. Ten microliters of this extract was used for immunoblotting with the GFP antibody and for RNA gel shift analysis (de Moor and Richter 1999).
Microinjection and image acquisition and analysis
CPEB-GFP DNA (0.3 μg/μL) and Alexa-546-labeled RNA (0.3 μg/μL) were coinjected into the cytoplasm of B104 cells; 4–5 h later, dual channel confocal images in both the GFP and Alexa546 channels were collected. For the time-lapse study, CPEB-GFP-expressing neurons were recorded every 15 or 20 sec for 30 or 20 frames with a Zeiss LSM 510 confocal microscope fitted with an oil-immersion 63×/NA1.4 objective. Fixed cell images were taken with the confocal microscope or in some cases, with a Nikon Eclipse E600 microscope (60×/NA 1.4 oil-immersion objective) equipped with a Spot digital camera. For colocalization studies, exposures were determined using single wavelength laser excitation to ensure that only the appropriate fluorophore could be detected in each channel. For RNA quantification, images were acquired with parameters that maximized the dynamic range of pixel intensity for the dendritic signal. For all experiments, identical acquisition parameters and settings were used for both control and experimental dendrites.
For colocalization studies, images in both channels (either GFP and Alexa-546 or CFP and YFP) were analyzed using NIH-Image software where a specific Macro program (written by Frank Morgan, University of Connecticut Health Center, Farmington, CT) was used to measure the fluorescence intensity of selected particles in both channels. Only well-resolved particles in the processes of cells were selected for analysis. To measure the velocity of CPEB-GFP particle movement, the distance a particle traveled between two adjacent time-lapse images was divided by the time between exposures, the largest of which was considered as the maximum rate. The average velocity was calculated by dividing the total traveling distance by the total recording time.
For the in situ hybridization data analysis, images taken under the same exposure conditions were analyzed using NIH-Image. The GFP RNA signals within dendrites were measured and given an average value (≤255) at each single pixel from the base (adjacent to soma) to the end, as assessed by the immunostaining of MAP2, a dendritic marker. To analyze the RNA in many dendrites, they were grouped into 10-μm (=86 pixels) compartments, and the average pixel intensity was calculated. The background intensity from uninfected neurons was subtracted. Group means were established for each segment, and the standard error of mean (S.E.M.) was calculated.
Synaptoneurosome preparation and analysis
Approximately 4 × 106 virus-infected hippocampal neurons were harvested in 1.5 mL buffer (0.32 M sucrose, 0.1 mM EDTA, 0. 25 mM DTT, 2 mM Hepes at pH 7.4) and then homogenized, and the nuclei and cell debris were pelleted by brief centrifugation. The synaptoneurosome-containing fraction was pelleted and then resuspended, treated with RNase A (100 μg/mL) for 20 min, and then filtered (Millipore, 5-μm pore). Synaptoneurosomes were collected by centrifugation, and the RNA was extracted from both the cytoplasm and synaptoneurosome fraction, which were treated with DNase I. The RNAs were subjected to real-time PCR amplification using QuantiTech SYBR Green PCR mixture (QIAGEN) and ABI PRSIM7700 (Perkin Elmer). The data were analyzed with V1.6.3 sequence software (primer sequences available on request). Statistical analysis was performed using Student's t test.
Acknowledgments
We thank Steve Lambert for many helpful discussions and essential advice, Mark Sharky and Mario Stevenson for the use of their real-time PCR thermocycler, and Inge The for the use of her confocal microscope. We thank Rebecca Hodgman for genotyping the embryos. This work was supported by grants from the NIH (NS39321 to J.D.R. and NS15190 to J.H.C.). Support from the Diabetes Endocrinology Research Center Program Project (DK32520) is also gratefully acknowledged. Y.-S.H. was supported by the Charles A. King Trust fellowship from the Medical Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL joel.richter@umassmed.edu; FAX (508) 856-4289.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1053003.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, Carson JH. Transport and localization elements in myelin basic protein mRNA. J Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DW, Chervin AS, Gelfand VI, Craig AM. Postsynaptic scaffolds of excitatory and inhibitory synapses in hippocampal neurons: Maintenance of core components independent of actin filaments and microtubules. J Neurosci. 2000;20:4545–4554. doi: 10.1523/JNEUROSCI.20-12-04545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Mannucci L, Dotti CG, Amaldi F. Chemical stimulation of synaptosomes modulates α-Ca2+/calmodulin-dependent protein kinase II mRNA association to polysomes. J Neurosci. 2000;20:1–6. doi: 10.1523/JNEUROSCI.20-10-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker G, Goslin K. Developments in neuronal cell culture. Nature. 1988;336:185–186. doi: 10.1038/336185a0. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Singer RH, Kosik KS. Association of poly(A) mRNA with microtubules in cultured neurons. Neuron. 1994;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- Blichenberg A, Rehbein M, Muller R, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in the mRNA encoding the α subunit of Ca2+/calmodulin-dependent protein kinase II. Eur J Neurosci. 2001;13:1881–1888. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- Cao Q, Richter JD. Dissolution of the maskin–eIF4E complex by cytoplasmic polyadenylation and poly(A) binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor CH, Richter JD. Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 1999;18:2294–2303. doi: 10.1093/emboj/18.8.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, Levens D. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes & Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- Gu W, Pan F, Zhang H, Bassell GH, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of β-actin mRNA in fibroblasts and neurons. J Cell Biol. 2002;156:41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake LE, Mendez R, Richter JD. Specificity of RNA binding by CPEB: Requirement for RNA recognition motifs and a novel zinc finger. Mol Cell Biol. 1998;18:685–693. doi: 10.1128/mcb.18.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering H, Sheng M. Dendritic spines: Structure, dynamics and regulation. Nat Rev Neurosci. 2001;2:880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and αCaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Hemraj I, Verkade P, Kohrmann M, Fortes P, Marion RM, Ortin J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: Implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Kwon S, Barbarese E, Carson JH. The cis-acting RNA trafficking signal from myelin basic protein mRNA and its cognate trans-acting ligand hnRNPA2 enhance cap-dependent translation. J Cell Biol. 1999;147:247–256. doi: 10.1083/jcb.147.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yapping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of Aplysia sensory to motor synapses: A function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CaMKIIα is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: A means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000a;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000b;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Mendez R, Barnard D, Richter JD. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002;21:1833–1844. doi: 10.1093/emboj/21.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Mayford M. Cellular and molecular mechanisms of memory: The LTP connection. Curr Opin Genet Dev. 1999;9:333–337. doi: 10.1016/s0959-437x(99)80050-1. [DOI] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Johns Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Min H, Turck CW, Nikolic JM, Black DL. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes & Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- Monshausen M, Putz U, Rehbein M, Schweizer M, DesGroseillers L, Kuhl D, Richter D, Kindler S. Two rat brain staufen isoforms differentially bind RNA. J Neurochem. 2001;76:155–165. doi: 10.1046/j.1471-4159.2001.00061.x. [DOI] [PubMed] [Google Scholar]
- Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of α-CaMKII regulate its dendritic targeting. Nat Neurosci. 2000;3:1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- Mouland AJ, Xu H, Cui H, Krueger W, Munro TP, Prasol M, Mercier J, Rekosh D, Smith R, Barbarese E, et al. RNA trafficking signals in human immunodeficiency virus type 1. Mol Cell Biol. 2001;21:2133–2143. doi: 10.1128/MCB.21.6.2133-2143.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov IA, Banker G, Brosius J, Tiedge H. Activity-dependent regulation of dendritic BC1 RNA in hippocampal neurons in culture. J Cell Biol. 1998;141:1601–1611. doi: 10.1083/jcb.141.7.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehbein M, Wege K, Buck F, Schweizer M, Richter D, Kindler S. Molecular characterization of MARTA1, a protein interacting with the dendritic targeting element of MAP2 mRNAs. J Neurochem. 2002;82:1039–1046. doi: 10.1046/j.1471-4159.2002.01058.x. [DOI] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS. CaMKIIα 3′ untranslated region-directed mRNA translocation in living neurons: Visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannevin RH, Huganir RL. Postsynaptic organization and regulation of excitatory synapses. Nat Rev Neurosci. 2000;1:133–141. doi: 10.1038/35039075. [DOI] [PubMed] [Google Scholar]
- Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell. 2001;1:201–213. doi: 10.1016/s1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- Thomas KL, Laroche S, Errington ML, Bliss TV, Hunt SP. Spatial and temporal changes in signal transduction pathways during LTP. Neuron. 1994;13:737–745. doi: 10.1016/0896-6273(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]