Abstract
Genomic imprinting in mammals marks the two parental alleles resulting in differential gene expression. Imprinted loci are characterized by distinct epigenetic modifications such as differential DNA methylation and asynchronous replication timing. To determine the role of DNA methylation in replication timing of imprinted loci, we analyzed replication timing in Dnmt1- and Dnmt3L-deficient embryonic stem (ES) cells, which lack differential DNA methylation and imprinted gene expression. Asynchronous replication is maintained in these ES cells, indicating that asynchronous replication is parent-specific without the requirement for differential DNA methylation. Imprinting centers are required for regional control of imprinted gene expression. Analysis of replication fork movement and three-dimensional RNA and DNA fluoroscent in situ hybridization (FISH) analysis of the Igf2-H19 locus in various cell types indicate that the Igf2-H19 imprinting center differentially regulates replication timing of nearby replicons and subnuclear localization. Based on these observations, we suggest a model in which cis elements containing nonmethylation imprints are responsible for the movement of parental imprinted loci to distinct nuclear compartments with different replication characteristics resulting in asynchronous replication timing.
Keywords: Asynchronous replication, genomic imprinting, nuclear localization, DNA methylation, Igf2-H19, replication fork
Genomic imprinting is an epigenetic mechanism that marks the two parental alleles during gametogenesis, resulting in differential gene expression in the developing embryo and adult. The epigenetic modifications responsible for establishing and maintaining imprinted gene expression are thought to involve DNA methylation, chromatin modifications, and asynchronous replication timing. For several imprinted loci specific cis control elements, imprinting centers (ICs), have been identified which are required for regional control of imprinting, imprinted gene expression, and asynchronous replication timing. All identified ICs harbor regions that contain differentially methylated DNA sequences. These methylation imprints are established in the germline and are resistant to global demethylation during the early cleavages of the preimplantation embryo. To date, DNA methylation is the only epigenetic mark shown to be required for the maintenance of imprinted gene expression. Embryos deficient for the DNA maintenance methyl transferase gene, Dnmt1, lack imprints and die early during development (Li et al. 1992). Moreover, embryos derived from females deficient for Dnmt3L, a newly identified de novo DNA methyl transferase gene that lacks the catalytic domain shared by the other DNA methyl transferases, were shown to lack maternal imprints and fail to develop past 9.5 days postcoitum (dpc; Bourc'his et al. 2001; Hata et al. 2002). A similar phenotype was observed in embryos generated by fertilization of oocytes deficient for the de novo DNA methyl transferase Dnmt3a, suggesting that these proteins act in a complex to initiate methylation imprints in the germline (Hata et al. 2002).
Asynchronous replication, like differential DNA methylation, is an epigenetic mark specific for all imprinted regions (Kitsberg et al. 1993). DNA fluorescent in situ hybridization (FISH) and S-phase fractionation analysis revealed that for a given imprinted region it is either the paternal or the maternal allele which is replicated first in S phase. Asynchronous replication timing at these loci is already present in the embryo immediately after fertilization, and is reset during late gametogenesis (Simon et al. 1999). Differences in replication timing between the parental alleles could be a consequence of local or broad differences in chromatin structure and/or nuclear localization. Alternatively, replication timing may play an important role in the establishment or maintenance of the differential expression pattern of imprinted genes. In support of this idea, recent experiments have shown that DNMT1 can specifically target a chromatin remodeling complex containing HDAC2 and DMAP1, to replication foci in late S phase (Rountree et al. 2000). This interaction could help maintain silent chromatin, which is predominantly being replicated in late S phase. Transient interactions between the replication machinery and chromatin remodeling complexes at specific time points during S phase may therefore account for the maintenance of chromatin features following DNA replication.
Besides imprinted genes, several other monoallelically expressed genes display asynchronous replication timing, such as the immunoglobulin genes (Mostoslavsky et al. 2001), olfactory receptor gene loci (Chess et al. 1994), and X-linked genes in female somatic cells (Takagi 1974). In contrast to asynchronous replication timing of imprinted regions, the choice of which allele is replicated first in S phase appears to be random and clonal and is established around the time of preimplantation (Mostoslavsky et al. 2001). Several recent findings indicate that replication timing may play a role in initiation and/or maintenance of epigenetic changes. In mammals, dosage compensation of X-linked genes is achieved by inactivation of one of the X chromosomes in female somatic cells. At the onset of X inactivation, the noncoding Xist RNA coats the future inactive X. The first detectable epigenetic change after Xist accumulation is a shift to late replication of the X chromosome (Keohane et al. 1996). Conditional deletion of Xist after completion of X inactivation does not result in reactivation of genes located on the inactive X chromosome (Csankovszki et al. 1999). In addition, the chromosome remains replicating late in S phase, indicating that replication timing may play a role in maintaining the inactive state after the initial inactivation has been established. Replication timing of the immunoglobulin κ-light chain locus was recently found to correlate with rearrangement, with the functional and recombined allele being replicated prior to the unrecombined allele (Mostoslavsky et al. 2001). Interestingly, asynchronous replication timing of the immunoglobulin κ-light chain locus is already present before the recombination event takes place and therefore may play an important role in the allelic choice for recombination.
In this paper we have attempted to define the role of asynchronous replication timing in genomic imprinting. We analyzed replication timing in several DNA methylation-deficient embryonic stem (ES) cell lines that lack imprinted gene expression. ICs are required for asynchronous replication timing, genomic imprinting, and imprinted gene expression. We determined the role of ICs in asynchronous replication timing by analyzing replication fork movement and nuclear localization of the Igf2-H19 locus in wild-type and parthenogenetic cell lines, and cell lines with a mutated IC. Our observations suggest a model in which nonmethylation imprints present in ICs are required for the movement of the two parental imprinted alleles to different subnuclear compartments with different replication characteristics, resulting in asynchronous replication.
Results
Asynchronous replication timing of imprinted genes in ES cells
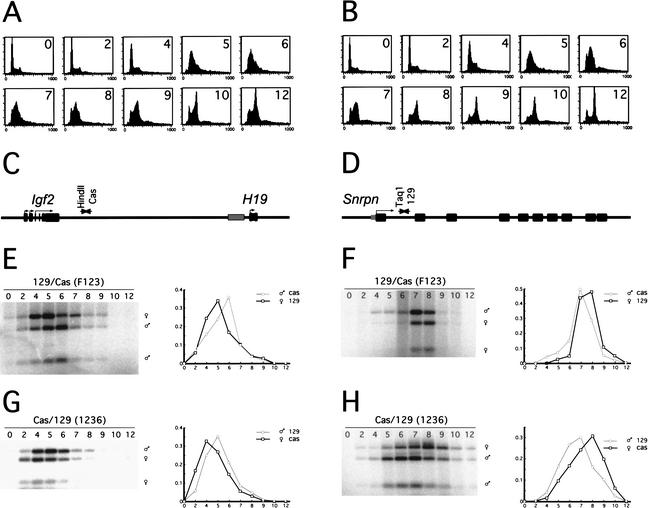
PCR amplification of restriction fragment length polymorphisms (RFLP) on S-phase fractionated DNA was shown to be a sensitive method to monitor replication timing of imprinted genes (Simon et al. 1999). This assay measures the relative amount of newly synthesized DNA of paternal and maternal origin at different points in S phase. We used this method to determine whether imprinted genes replicate asynchronously in ES cells. To synchronize cells, we reversibly arrested ES cells in late G1 phase of the cell cycle with the plant amino acid mimosine (Krude 1999). After washing the cells with Hepes buffer and resuspending them in normal medium, the ES cells started entering S phase as a synchronized population, most of them reaching G2 10 h after release. Four different polymorphic ES cell lines were tested. All cell lines behaved similarly after release of the mimosine block (two cell lines are shown in Fig. 1A,B). BrdU-labeled DNA of individual S-phase fractions was isolated and RFLPs in the paternally imprinted Igf2-H19 locus (Fig. 1C) and the maternally imprinted Snrpn locus (Fig. 1D) were PCR-amplified. Amplification of a HindII polymorphism on the castaneus allele of the Igf2-H19 locus shows that the maternal Igf2-H19 allele is replicated prior to the paternal allele in a 129Sv/Jae×Mus castaneus F1 ES cell line (Fig. 1E, line F123) and in an M. castaneus×129Sv/Jae F1 ES cell line (Fig. 1G, line 1236). In all cell lines tested, the difference in replication timing between the maternal and paternal allele ranges from 1 to 1.5 h. Analysis of a TaqI polymorphism in the Snrpn locus shows the reverse, with the paternal allele being replicated prior to the maternal allele (Fig. 1F,H). We conclude that imprinted gene loci replicate asynchronously in ES cells. These results also demonstrate that there is no parental bias towards the paternal or maternal allele replicating first in S phase.
Figure 1.
Imprinted loci replicate asynchronously in wild-type ES cells. FACS analysis of mimosine-treated 129Sv/Jae×M. castaneus (A, line F123) and M. castaneus×129Sv/Jae (B, line 1236) F1 ES cells after block and release into S phase (numbers indicate hours after release: x-axis, DNA fluorescence; y-axis, number of cells,). (C,D) Schematic diagrams of the murine Igf2-H19 and Snrpn loci, and the location of PCR primers and RFLPs. Gray boxes indicate DMRs. (E–H) PCR analysis of BrdU-positive DNA fractions isolated at different time points (in hours) after release into S phase. The plots represent the relative amount of paternal and maternal replicated DNA over time. (E,G) The maternal Igf2-H19 locus replicates prior to the paternal locus. (F,H) The Snrpn locus shows the opposite pattern with the paternal allele being replicated first.
Asynchronous replication in a DNA methylation-deficient background
DNA methylation plays an essential role in the imprinting process, as mouse embryos deficient for the maintenance DNA methyl transferase Dnmt1, and offspring from females deficient for the de novo DNA methyl transferases Dnmt3a and/or Dnmt3L lack imprinted gene expression (Li et al. 1992; Bourc'his et al. 2001; Hata et al. 2002). We used ES cells deficient for these three DNA methyl transferases to determine the role of DNA methylation in setting up and/or maintaining the asynchronous replication timing of imprinted genes.
Because all mutant ES cell lines were made in a nonpolymorphic background, we used DNA FISH analysis to determine the replication timing status of different loci in these cells. ES cells were stained for BrdU followed by DNA FISH. Three types of nuclei can be detected in the S-phase/BrdU-positive fraction of cells (Fig. 2): nuclei with two single spots, indicating that both alleles have not been replicated yet (SS); nuclei with one single spot and one double spot, indicating that one allele has replicated and the other has not (SD); and nuclei with two double spots, indicating that both alleles have been replicated (DD). In general, when using FISH probes that detect synchronously replicating loci, 10%–20% of the nuclei show an SD pattern. However, for imprinted loci this percentage is much higher and ranges from 25% to 40% (Kitsberg et al. 1993).
Figure 2.
Analysis of replication timing by DNA FISH. The top three panels show representative ES cell nuclei, which were hybridized with a probe specific for the Igf2R locus. The nucleus in the left panel shows two unreplicated alleles (SS), the middle panel shows a nucleus with one replicated and one unreplicated allele (SD), and the right panel shows a nucleus in which both alleles are replicated (DD). The bottom three panels show the same nuclei stained with anti-BrdU antibody.
DNA FISH experiments using probes specific for several imprinted loci show that all these loci replicate asynchronously in wild-type ES cells (Table 1). These results are consistent with our mimosine replication timing analysis. Probes specific for nonimprinted loci such as the α-globin locus and the L23mrp locus, which is located downstream of the H19 gene (Greally et al. 1998), detect significantly lower numbers of SD nuclei, indicating synchronous replication timing. Next we analyzed homozygous Dnmt1−/− ES cells that lack differential DNA methylation and imprinted gene expression (Li et al. 1992). Interestingly, imprinted loci remained replicating asynchronously in S phase in these cells (Table 1). Similarly, compound Dnmt3a−/−3b−/− ES cells also exhibited asynchronous replication timing of imprinted loci. These experiments suggest that DNA methylation or these methyl transferases are not required for the maintenance of asynchronous replication. However, because the Dnmt1−/− ES cells as well as the Dnmt3a−/− 3b−/− ES cells were derived from wild-type ES cells through consecutive gene targeting, it is possible that the timing of replication of imprinted loci is properly set in the germline or immediately after fertilization by methyl transferases and this pattern is maintained by epigenetic means other than DNA methylation. To test whether methylation imprints established in the germ cells are required for asynchronous replication, we analyzed the replication timing of imprinted loci in Dnmt3L(mat−/−)/+ ES cells that were isolated from a cross of a Dnmt3L−/− female and a wild-type male. These ES cells lack maternal imprinting because the maturing Dnmt3L−/− oocytes fail to establish methyl imprints. Consistent with the results obtained with the Dnmt1 and Dnmt3a/3b mutant ES cell lines, the paternally imprinted Igf2-H19 locus replicated asynchronously in Dnmt3L(mat−/−)/+ ES cells (Table 1). Interestingly, the maternally imprinted loci also remained replicating asynchronously (Table 1), indicating that differential DNA methylation is not required to set up asynchronous replication timing. To test whether asynchronous replication timing of imprinted genes was specific for genomes of biparental origin, we analyzed replication timing in uniparental parthenogenetic ES cells, which carry two maternal chromosome sets. In agreement with a previous report (Simon et al. 1999), parthenogenetic ES cells showed synchronous replication timing for all tested imprinted genes (Table 1). These results suggest that replication timing is parent-specific without the requirement for differential DNA methylation.
Table 1.
Replication timing analysis in wild-type and mutant ES cell lines
| Imprinted loci
|
Wildtype
|
Dnmt1−/−
|
3a−/−, 3b−/−
|
3Lmat−/−/+
|
Parth. ♀♀
|
|---|---|---|---|---|---|
| H19pat | 37% | 39% | 38% | 36% | 20% |
| Igf2pat | 34% | 28% | 40% | 39% | 18% |
| Snrpnmat | 40% | 38% | 39% | 40% | 17% |
| Igf2rmat | 29% | 34% | 41% | 39% | 13% |
| Kvlqt1mat | 32% | 41% | 36% | 33% | 12% |
| P57kip2mat | 34% | 31% | N.D. | 37% | 15% |
| Not imprinted | |||||
| L23mrp | 12% | 13% | 13% | 16% | 18% |
| a-globin | 18% | 19% | N.D. | 16% | 15% |
Relative amount of BrdU positive nuclei, with one replicated and one unreplicated allele (SD) in wildtype, Dnmt1−/−, Dnmt3a−/−b−/−, Dnmt3L(mat−/−)/+ and parthenogenetic ES cells, for several imprinted and nonimprinted loci (pat: Paternally imprinted methylation mark, mat: Maternally imprinted methylation mark).
Replication fork direction at the Igf2-H19 locus
Mimosine S-phase fractionation analysis revealed that the maternal Igf2-H19 allele replicates before the paternal allele in S phase. Interestingly, maternal inheritance of a mutant H19 allele that lacks the H19 gene sequence (ΔH19) and the upstream differential methylated domain (DMR), results in synchronous replication timing of this locus in splenocytes (Greally et al. 1998). Presumably, the mutated maternal allele replicates later in S phase, suggesting that the deleted area may be involved in setting up early replication timing of the maternal allele, either by affecting the timing of firing of nearby origins or by harboring an origin of replication. To distinguish between these possibilities, we analyzed the direction of the replication fork movement at the Igf2-H19 locus. Wild-type ES cells were treated overnight with BrdU in the presence or absence (control) of emetine. Emetine treatment blocks lagging strand synthesis and therefore results in BrdU-labeled leading strands (Burhans et al. 1991). BrdU-labeled leading strands and BrdU-labeled control DNA were isolated by immunoprecipitation and spotted in duplicate onto slot blots. The direction of the replication fork movement was then determined by using strand-specific riboprobes. Detection of more signal with the antisense probe than with the sense probe compared to the control samples, indicates that the replication fork is moving in the sense direction towards the L23mrp gene (Fig. 3A). If more signal is detected with the sense probe than the antisense probe compared to the control samples, this indicates that the replication fork is moving in the antisense direction towards the insulin gene (Fig. 3B). We found that in wild-type ES cells (Fig. 3C) and in mouse embryonic fibroblasts (MEFs; data not shown), the locus contains at least two replicons, one covering the Igf2 gene, the other overlapping with the H19 gene. Interestingly, the second replicon appears to start downstream of the H19 gene in the nonimprinted L23mrp locus, indicating that the DMR and the downstream H19 gene do not themselves contain a bidirectional origin of replication. DNA FISH analysis with a cosmid probe covering L23mrp showed that this region replicates synchronously in S phase (Table 1, Fig. 4A; Greally et al. 1998), suggesting that the replicon that contains the H19 gene is also replicating synchronously. Although such a finding would contradict the DNA FISH result with the H19 cosmid probe (Table 1, Fig. 4A), which showed that H19 is replicated asynchronously in S phase, it could be explained by the fact that the H19 FISH probe covers or is very close to the transition from an asynchronous replicon to a synchronous replicon.
Figure 3.
Replication fork analysis of the Igf2-H19 locus in wild-type ES cells. Cells were treated with (+) or without (−) emetine in the presence of BrdU. Emetine blocks lagging strand synthesis and therefore results in BrdU-labeled leading strands. DNA of emetine-treated cells and control DNA was immobilized on slot blots and hybridized to strand-specific riboprobes. (A) Detection of more signal with the antisense (AS) than sense (S) probe compared to the control DNA sample indicates that the replication fork is moving in the sense direction. (B) Detection of more signal with the sense than antisense probe indicates that the replication fork is moving in the antisense direction. (C) Sense probes were transcribed from the top strand in the Insulin to L23mrp direction; and antisense probes were transcribed from the bottom strand in the L23mrp to Insulin direction (probes are indicated by bars). The bias ratio was calculated as (antisense: emetine/control)/(sense: emetine/control); a ratio >1 implies that the replication fork is moving in the sense direction. Ratios <1 imply that the replication fork is moving in the antisense direction.
Figure 4.
Replication timing analysis of a sequence 5′ of the DMR of the Igf2-H19 locus. (A) Location of the primers that amplify a length polymorphism between M. castaneus and 129/Sv alleles is marked with arrows. Location and size of the DNA FISH probes used for the replication timing analysis are indicated with gray bars, and the direction of the replication fork in wild-type ES cells is indicated with arrows. (B,C) PCR analysis of BrdU-positive DNA fractions isolated at different time points (in hours) after release into S phase shows no significant difference of replication timing between the maternal and paternal sequences in 129Sv/Jae×M. castaneus (B, line F123) and M. castaneus×129Sv/Jae (C, line 1021) F1 ES cell lines. The plots represent the relative amount of paternal and maternal replicated DNA over time.
In order to test this hypothesis, we performed mimosine S-phase fractionation analysis, using PCR primers located upstream of the DMR, a sequence that is located in the same replicon as the nonimprinted L23mrp gene (Fig. 4A). We tested two ES cell lines and found that this area is replicated synchronously in S phase (Fig. 4B,C), indicating that the DMR is embedded in a synchronously replicating replicon but affects asynchronous replication timing in the replicon containing Igf2.
We next analyzed the direction of the replication fork movement in the Igf2-H19 locus in cell lines in which this locus replicates synchronously as measured by DNA FISH. We therefore generated wild-type and mutant ΔH19/+ and +/ΔH19 MEFs and isolated leading strands. As was reported for splenocytes (Greally et al. 1998), in ΔH19/+ mutant MEFs the Igf2-H19 locus replicated synchronously in S phase, whereas the Igf2-H19 locus in +/ΔH19 and wild-type MEFs replicated asynchronously in S phase (data not shown). Using probes specific for the two different replicons (Fig. 5A), we found that the bias ratio is comparable between the different cell lines, indicating that the movement of the replication fork in the Igf2-H19 locus does not differ between the asynchronously replicating wild-type and +/ΔH19 MEFs and the synchronously replicating ΔH19/+ MEFs (Fig. 5B). Similar to the ΔH19/+ MEFs, parthenogenetic ES cells exhibited synchronous replication of the Igf2-H19 locus (Table 1). Examination of these parthenogenetic ES cells showed that the bias ratio in these cells is similar to those of wild-type and Dnmt1−/−-deficient ES cells, indicating that the replication fork direction in the Igf2-H19 locus is identical in all three cell lines (Fig. 5C). These results suggest that replicons in the Igf2-H19 locus are fixed and do not change when replication timing is shifted on one of the alleles. The data also show that replication forks move in the same direction on both alleles.
Figure 5.
The direction of the replication fork in the Igf2-H19 locus in asynchronously and synchronously replicating fetal liver and ES cells. (A) Location of the riboprobes and the direction of the replication fork in wild-type (WT) ES cells. Sense probes were transcribed from the top strand in the Igf2 to H19 direction. Antisense probes were transcribed from the bottom strand in the H19 to Igf2 direction. (B) Replication fork analysis of wild-type (WT), mutant ΔH19/+, and +/ΔH19 mouse embryonic fibroblasts. (C) Replication fork analysis of wild-type (WT), Dnmt1−/−, and parthenogenetic ES cells.
Nuclear localization of the Igf2-H19 locus
We have demonstrated that the Igf2-H19 DMR element affects timing of origin firing in a different replicon over a relatively large distance. A similar phenomenon has been described in the human β-globin locus. This locus contains one bidirectional origin located between the two adult δ and β genes, which fires early in red cells and late in all other cell types. Early firing in S phase requires a 40-kb element including the locus control region (LCR; Forrester et al. 1990; Simon 2001) and affects origin firing over a distance of more than 50 kb. The same element is required for localization of the β-globin locus away from centromeres in red cells (Schubeler et al. 2000), indicating that there is a close correlation between nuclear localization and replication timing. Similar observations have been made in yeast where late replicating origins were found to be enriched in the nuclear periphery, whereas early-replicating origins were found to be distributed randomly (Heun et al. 2001). However, this enrichment of late-replicating origins in the nuclear periphery is lost as cells progress into S phase. We wanted to know whether the same correlation between replication timing and nuclear localization exists for the imprinted Igf2-H19 locus.
We isolated wild-type fetal liver cells, which express both Igf2 and H19 (Biniszkiewicz et al. 2002), and performed RNA FISH to detect primary transcripts. This approach allowed for the discrimination of the parental alleles, since the paternal allele expresses Igf2 and the maternal allele expresses H19. Using two different fluorescent dyes to detect Igf2 and H19 transcripts, we found that only 8% of the cells expressed both alleles in cis, showing that only 4% of the maternal alleles expressed Igf2 and 4% of the paternal alleles expressed H19 (Biniszkiewicz et al. 2002). These low percentages indicate that there is a minor error rate in determining the parental allele using a single probe for RNA FISH. In order to measure the subnuclear localization of transcripts, we performed RNA FISH with probes specific for Igf2 or H19 in conjunction with immunostaining for nuclear pore membrane proteins (Fig. 6A,B). Confocal images were taken and the relative distance of the transcription spot to the nuclear membrane was measured in 3D. To exclude the possibility that the subnuclear organization is lost after entering S phase, we only measured nuclei that showed clearly defined single spots which are most likely in G1 phase of the cell cycle. Each nucleus was divided into five compartments of equal volume, and transcription foci were assigned to one of these compartments based on the relative distance to the nuclear membrane (Fig. 6H). With this approach we found that the distribution of the Igf2-expressing alleles was significantly higher in the periphery compared to the H19-expressing alleles that were more located toward the center of the nucleus (Fig. 6I; Table 2). Analysis of the nuclear localization of the β-globin locus, using a probe detecting the highly expressed βmajor gene, shows that the distribution of the globin locus closely overlaps with the distribution of the maternal H19-expressing Igf2-H19 locus (Fig. 6I, N = 42). S-phase fractionation analysis indicates that the maternal allele is replicated prior to the paternal allele in ES cells. These results therefore suggest that the late-replicating paternal allele is localized more toward the periphery of the nucleus than the early-replicating maternal allele. In contrast to the late-replicating β-globin gene in nonerythroid cells, Igf2 or H19 transcription foci do not colocalize with centromeres (data not shown). In addition, we did not find colocalization of Igf2 or H19 transcription foci and heterochromatic regions by costaining with M31/HP1 protein-positive areas in the nucleus (data not shown). Next, we analyzed the nuclear localization of the Igf2-H19 locus in ΔH19/+ fetal liver cells (Fig. 6C). These cells show synchronous replication of the Igf2-H19 locus and express Igf2 biallelically (Leighton et al. 1995; Greally et al. 1998). Based on the mimosine S-phase fractionation analysis, we assume that in this mutant the maternal allele is replicating later than in wild-type cells. Indeed, when we analyzed the localization of the two Igf2 transcription spots in nuclei of ΔH19/+ cells, we found that the distribution of both alleles is similar to the distribution of the single Igf2-expressing allele in wild-type nuclei (Fig. 6I; Table 2).
Figure 6.
The nuclear localization of the Igf2-H19 locus in different cell types. Representative confocal images of wild-type (WT) fetal liver cells after RNA FISH with Igf2- (A) and H19-specific (B) probes (FITC). (C) RNA FISH on ΔH19/+ fetal liver cell using an Igf2-specific probe (FITC). (D,E) DNA FISH on wild-type (WT; D) and ΔH19/+ (E) fetal liver cells using a cosmid probe (FITC), which detects the Igf2-H19 locus. Nuclear pore protein is stained in red. (F,G) Same analysis on wild-type (WT; F) and parthenogenetic (G) ES cells. (H) Nuclei were divided into five compartments of equal volume, and loci were grouped according to the relative distance of the spot to the nuclear membrane. (I) Localization of Igf2 RNA foci (light blue), H19 RNA foci (dark blue), and βmajor-globin RNA foci (gray) in wild-type (WT) fetal liver cells, and Igf2 RNA foci (orange) in ΔH19/+ fetal liver cells. (J,K) Localization of the Igf2-H19 locus in different cell types as determined by DNA FISH. The distance of both alleles to the nuclear membrane of a nucleus was measured in 3D, and then the difference between the two parental alleles per nucleus was calculated and plotted by subtracting the two measurements. (J) Box plots representing the distribution of the difference in distance relative to the nuclear membrane between the two parental Igf2-H19 alleles in wild-type (blue) and ΔH19/+ fetal liver cells (orange). (K) Box plots representing the distribution of the difference in distance relative to the nuclear membrane between the two parental Igf2-H19 alleles in wild-type (WT; blue) and parthenogenetic ES cells (orange).
Table 2.
Measurements of the nuclear localization of the Igf2-H19 locus in different cell types
The average distance of foci to the nuclear membrane in different cell types is given in micrometers with standard deviation. The difference per nucleus was calculated by subtracting the distance of the two parental alleles to the nuclear membrane, and then averaged for the whole population of measurements per cell type and presented in micrometers with standard deviation. P values were calculated by t-test analysis.
To further confirm these results, we examined both wild-type and ΔH19/+ mutant fetal liver cells by DNA FISH analysis (Fig. 6D,E). Because the parental alleles cannot be discriminated by DNA FISH, we measured the distance of the two spots to the nuclear membrane, and then subtracted these distances for each nucleus, yielding the difference in subnuclear localization of the two parental loci relative to the nuclear membrane. In wild-type fetal liver cells, the difference in the nuclear localization between the two parental alleles, relative to the nuclear membrane, is much larger than in the ΔH19/+ mutant fetal liver cells (Fig. 6J; Table 2), indicating that in the mutant cells both alleles are localized in the same area in the nucleus. As for the RNA FISH analysis, we found that in the mutant cells both alleles are located closer to the periphery than in the wild-type fetal liver cells (Table 2). Although the difference in distance to the nuclear membrane between the two cell types seems relatively small, this can be explained by the fact that the distance measurement in the wild-type fetal liver cells is an average of both parental alleles. These results together with the RNA FISH data, demonstrate that in cells that express Igf2 and H19 there is a clear correlation between subnuclear localization and replication timing.
In order to test whether this correlation persists in cells that do not express Igf2 or H19, we analyzed the localization of the Igf2-H19 locus by 3D imaging in wild-type and parthenogenetic ES cells (Fig. 6F,G). Similar to fetal liver cells, in wild-type ES cells we found that the difference in the nuclear localization between the two parental alleles, relative to the nuclear membrane, is much larger than in parthenogenetic ES cells (Fig. 6K; Table 2). In parthenogenetic ES cells, in which the Igf2-H19 locus replicates synchronously in S phase, both parental alleles are located more toward the nuclear membrane, akin to ΔH19/+ mutant fetal liver cells (Table 2). However, parthenogenetic ES cells contain two maternally imprinted Igf2-H19 loci, which presumably both replicate early in S phase based on our mimosine S-phase fractionation analysis. By comparing the relative amount of DD nuclei in wild-type and parthenogenetic ES cells, we found that the amount of DD nuclei is much higher in parthenogenetic ES cells (parthenogenetic: SS, 26%; SD, 20%; DD, 54%; wild-type: SS, 26%; SD, 37%; DD, 37%), which is another indication that both alleles replicate early in S phase. We conclude that in contrast to fetal liver cells, the early-replicating allele of ES cells is localized more to the periphery of the nucleus than the late-replicating allele. These results suggest that the different parental chromosomes are restricted to different nuclear compartments, and that localization of these compartments is cell-type-specific.
Discussion
To date, DNA methylation is the only epigenetic mark known to be required for imprinted gene expression (Li et al. 1992; Bourc'his et al. 2001; Hata et al. 2002). Here we found that asynchronous replication timing is dependent on a biparental origin without the requirement for parent-specific DNA methylation. The fact that differential DNA methylation and replication timing both are parent-specific but separable processes is consistent with the finding that erasure of DNA methyl imprints and asynchronous replication timing take place at different stages during gametogenesis (Simon et al. 1999). Our results also show that asynchronous replication timing of imprinted genes is not a consequence of differential DNA methylation and gene expression, but may be a process important in setting up imprinting in the germline. In addition, the presence of asynchronous replication timing in a DNA methyl transferase-deficient background may explain why some genes retain their imprinted expression status in Dnmt1−/− embryos (Caspary et al. 1998). Interestingly, asynchronous replication timing alone is not sufficient to initiate methyl imprints in ES cells, as was shown by rescue experiments of Dnmt1−/− ES cells. Dnmt1−/− ES cells lack DNA methylation, and the introduction of a bacterial atrificial chromosome (BAC) containing the wild-type Dnmt1 gene into these Dnmt1−/− ES cells results in the restoration of overall genomic methylation. However, imprinted genes do not regain differential DNA methylation or gene expression (Biniszkiewicz et al. 2002), despite the presence of asynchronous replication timing of imprinted loci. Interestingly, the only two proteins shown to be required for establishing imprints in the germline, Dnmt3L and Dnmt3a, are highly expressed in ES cells, but apparently not sufficient to restore differential methylation patterns. Similarly, restoration of Dnmt3L protein levels in the early embryo after fertilization of a Dnmt3L−/− oocyte with wild-type sperm does not initiate methyl imprints on the maternal genome. This suggests that other cofactors, which may only be present in the germline, are required for restoring methyl imprints and imprinted gene expression.
S-phase fractionation versus DNA FISH
Mimosine S-phase fractionation analysis and DNA FISH analysis on wild-type ES cells both show that imprinted genes replicate asynchronously in S phase. The mimosine S-phase fractionation analysis indicates that the maximum difference in replication timing is 1.5 h, with S phase lasting about 6 h. Comparable and even smaller differences in replication timing between the two parental IGF2, Igf2r and SNRPN alleles, have been reported by others, using fluorescent-activated cell sorter (FACS)-sorted S-phase fractions (Kawame et al. 1995; Simon et al. 1999). Based on these findings, one would expect to see 25% of the BrdU-positive cells to have an SD pattern after DNA FISH. We find much higher numbers that range from 30% to 40%. Many other groups have reported similar percentages of SD nuclei for different imprinted loci in various cell types (Kitsberg et al. 1993; Knoll et al. 1994; Greally et al. 1998; Simon et al. 1999). This may indicate that the FISH assay detects differences in chromatid segregation caused by epigenetic differences other than replication timing. Differences in chromatin structure, for instance, could influence the rate of chromatid segregation, and differences in compaction of nearby chromatin may influence the DNA FISH result. Alternatively, the discrepancy between these results could be explained by the fact that the mimosine S-phase fractionation analysis detects replication timing as it progresses at a specific point in a locus, whereas the DNA FISH assay only detects a replication event when the entire replicon has been replicated. The two segregated alleles as detected by DNA FISH therefore reflect the replication state of the sequence, within one or possibly more replicons, with the largest difference in replication timing. Another explanation for the different results may be the difference in resolving power between DNA FISH and S-phase fractionation analysis. DNA FISH analysis shows the sharp endpoint of a replication event on a cellular level, whereas the S-phase fractionation analysis is based on pooled cells of different S-phase fractions that inevitably will have some cross-contamination. None of these explanations are mutually exclusive and may all have contributed to the different outcomes between the two methodologies used to monitor replication timing.
S-phase fractionation analysis showed that the maternal Igf2-H19 locus is replicated before the paternal Igf2-H19 locus in four different murine ES cell lines. In contrast, employing a double-label DNA FISH assay detecting the human IGF2-H19 locus in combination with a nearby deletion in a β-thalassemic cell line, Kitsberg et al. (1993) reported that the paternal human IGF2-H19 locus is replicated prior to the maternal locus. Although puzzling, this discordance could be explained by the different techniques applied by us and Kitsberg et al. (1993) to determine allele-specific replication timing. As mentioned above, S-phase fractionation analysis determines allele-specific replication timing at a single local and temporal point at the locus, whereas DNA FISH analysis only detects the endpoint of a replication event. Therefore, allele-specific replication timing as determined by DNA FISH does not necessarily reflect the local replication characteristics of the regions that we analyzed using S-phase fractionation analysis. However, comparison of replication timing of the Igf2-H19 locus in parthenogenetic and wild-type ES cells indicated that, for the regions analyzed by us, both S-phase fractionation and DNA FISH gave consistent results. Alternatively, the different results may be due to species-specific or cell-type-specific effects at the Igf2-H19 locus, or allele-specific replication patterns may have changed upon immortalization of the β-thalassemia cell line used by Kitsberg et al. (1993) and may thus be different from replication timing results obtained by us using ES cell lines.
A clear example that replication timing and chromosome segregation do not always correlate is shown by analysis of a region 5′ of the H19 DMR. DNA FISH analysis of this region revealed a high number of SD nuclei comparable to the SD value of a probe detecting the Igf2 gene. However, mimosine S-phase fractionation analysis showed that in contrast to the Igf2 gene, the 5′ DMR region replicates synchronously in S phase. This is rather surprising but could be explained by the fact that part of the H19 FISH probe overlaps with or is located very close to the start of the asynchronously replicating replicon, and is not far enough away to resolve segregation by DNA FISH.
The Igf2-H19 DMR affects origin firing over large distances
Earlier studies showed that the DMR of the Igf2-H19 locus is required for imprinted gene expression and asynchronous replication timing of the locus. Similarly, random introduction in the genome of a 1.2-kb transgene containing the SNRPN short region of overlap is sufficient to initiate parent-specific replication timing of this transgene (Shemer et al. 2000). Assessment of the replication fork direction in the Igf2-H19 locus revealed that the DMR is not itself a bidirectional origin of replication, but rather affects replication timing of a nearby replicon. Analysis of the replication fork direction in the Igf2r locus also showed that region 2 has no origin function (J. Gribnau and R. Jaenisch, unpubl.), indicating that this is not a general property of a DMR. Interestingly, we found that the direction of the replication fork in a replicon is unchanged whether it replicates synchronously or asynchronously. This suggests that clear boundaries may exist between replicons.
Changes in local and broad chromatin structure appear to affect replication timing. In yeast, silent telomeric origins can be activated by mutations in SIR3, which is a basic component of telomeric heterochromatin (Stevenson and Gottschling 1999). Recently, it has been reported that deletion of the histone deacetylase RPD3 or specific recruitment of the histone acetyltransferase Gcn5p in yeast affects replication timing, indicating that histone acetylation is a direct determinant of timing of origin firing (Vogelauer et al. 2002). In Xenopus sperm nuclei, somatic histone H1, which is required for higher-order chromatin structure, appears to reduce the number of replication forks, indicating that H1 plays a role in regulating origin usage (Lu et al. 1998). The human β-globin locus provides another example of chromatin structure affecting replication timing. This locus contains one bidirectional origin that fires early in S phase in erythroid cells and late in S phase in all other cell types. Replication timing correlates well with DNase I sensitivity of the locus, and both epigenetic characteristics also correlate with histone H3/H4 acetylation and nuclear localization (Cimbora et al. 2000; Schubeler et al. 2000). A large deletion of the β-globin LCR results in a late-replicating, hypoactetylated locus which localizes closely to centromeric heterochromatin, indicating that this element affects replication timing over a distance of at least 50 kb. In addition, transgenic studies show that the LCR is sufficient for directing globin-specific replication timing of transgenes and integration sites (Simon et al. 2001).
How do elements affect the timing of firing of an origin over such large distances? Like the LCR of the globin locus, the DMR 5′ of the H19 gene may be responsible for broad chromatin changes on one of the parental alleles. Different chromatin states may themselves be sufficient for differential firing of origins; alternatively, origin firing may be similar on both parental alleles but replication fork progression could be affected by chromatin differences and therefore result in asynchronous replication timing. Although parental-specific local differences in DNase I hypersensitivity (Hark and Tilghman 1998; Khosla et al. 1999), histone acetylation, and histone methylation (Grandjean et al. 2001; Fournier et al. 2002) have been reported, we have not yet been able to detect differences in general DNase I sensitivity between the two parental Igf2-H19 alleles (J. Gribnau and R. Jaenisch, unpubl.). Nevertheless, local differences in chromatin structure between the parental alleles could be sufficient to initiate origin firing at different points in S phase, or affect replication fork progression through the locus. Alternatively, origin firing may be regulated by direct interaction in trans between the prereplication complex and the DMR, as has been proposed for LCR-globin gene activation (Choi and Engel 1988). However, it is difficult to envision how the DMR of the Igf2-H19 locus, and possibly other loci, regulates multiple origins by a direct interaction mechanism over large distances in a coordinate fashion.
Nuclear localization correlates with replication timing
Several recent studies report that subnuclear localization correlates with the time of origin firing in S phase. BrdU labeling of DNA in cells at different time points in S phase shows that there are distinct domains of replication timing in the nucleus. Early-replicating DNA is predominantly found in the nuclear interior, whereas late-replicating heterochromatin is mostly located in the nuclear periphery (Ferreira et al. 1997). Supporting this view is the finding that late-replicating origins appear to be enriched near the nuclear envelope in yeast, whereas the distribution of early firing origins seems to be random (Heun et al. 2001). The human β-globin locus replicates late in S phase and associates with pericentric heterochromatin in cells that do not express β globin. In contrast, in erythrocytes replication timing of the locus is shifted to an earlier point in S phase and the locus localizes to areas that are excluded from pericentric heterochromatin (Epner et al. 1988; Brown et al. 2001).
We found that unlike the globin locus, neither of the two Igf2-H19 alleles associates with centromeric heterochromatin. However, RNA and DNA fluorescent in situ hybridization on fetal liver cells that do express Igf2 and H19 showed that the two parental alleles are localized in different subnuclear compartments (Table 3). Because it is unlikely that replication timing of imprinted loci switches alleles between different cell types, the data suggest that the early-replicating allele is localized more toward the interior of the nucleus relative to the late-replicating locus. Surprisingly, we observed the opposite in nonexpressing ES cells, suggesting that it is not the proximity to the nuclear membrane that determines replication timing of the Igf2-H19 locus (Table 3). The correlation between replication timing and localization for each cell type, however, does indicate that the paternal and maternal alleles are restricted to different fixed compartments within the nucleus. The subnuclear localization of these compartments, like chromosomal territories, appears to differ among cell types and thus explains the heterogeneity of our results.
Table 3.
Summary of expression, replication timing, and nuclear localization status of the Igf2-H19 locus in different cell types
The expression status is indicated with a male or female symbol if the gene is expressed; symbols between brackets indicate the potential of these genes to be expressed in differentiated ES cells. The localization of the paternal Igf2-H19 loci are shown as horizontal box plots with the scale bar representing the distance in micrometers to the nuclear membrane. The localization of the paternal allele is colored in black, and the localization of the maternal allele is in white. The localization of alleles for which no distinction could be made is colored in gray. Wild-type fetal liver cells express H19 from the maternal allele and Igf2 from the paternal allele. The locus is replicated asynchronously in these cells and RNA FISH analysis shows that the two parental loci are located in distinct places in the nucleus. In mutant ΔH19/+ fetal liver cells the Igf2-H19 locus is replicated synchronously and Igf2 is expressed from both parental alleles. RNA and DNA FISH analysis comparing wild-type and mutant fetal liver cells shows that the mutated locus is shifted toward the periphery of the nucleus, indicating that the paternal allele is located more in the periphery of the nucleus when compared to the maternal allele. Wild-type and parthenogenetic ES cells do not express H19 and Igf2. In wild-type ES cells the maternal allele replicates first in S phase and DNA FISH analysis shows that the parental loci are located in a relatively broad area of the nucleus. However, in parthenogenetic ES cells in which the locus is replicated synchronously localization of the two maternally derived loci is more confined and shifted toward the periphery, indicating that the maternal locus is located more in the periphery compared to the paternal locus.
It was recently reported that the spatial organization of chromosomal territories is conserved between mouse and human, suggesting a functional significance (Mahy et al. 2002). S-phase-labeling experiments with thymidine analogs show that domains with specific replication timing characteristics within a chromosomal territory remain stable at all stages during the cell cycle, even through multiple cell divisions. This indicates that replication timing of subchromosomal domains is highly organized, and that assembly of specific chromatin remodeling complexes at different time points in S phase may provide a mechanism to maintain this organization (Rountree et al. 2000). The two parental Igf2-H19 loci may be confined to separate domains with different replication timing characteristics within the reciprocal chromosomal territories and therefore replicate asynchronously in S phase. The ICs appear to be responsible for these differences in nuclear localization, because deletion of the Igf2-H19 IC of the early-replicating maternal allele results in a change in localization to the nuclear periphery, and synchronous replication timing. Depending on the imprinting state, these regions could specifically recruit factors involved in setting up the local architecture within a chromosomal territory. Differential DNA methylation of sequences in the IC would have been the most likely mechanism to differentially attract factors involved in setting up the nuclear architecture of the two parental alleles. However, we found that differential DNA methylation does not play a key role in setting up asynchronous replication, suggesting that other epigenetic modifications between the two parental alleles are sufficient to do so. The chromosomal compartmentalization model would explain why imprinted loci replicate asynchronously, but the question remains which precise role replication timing plays in imprinted gene expression.
Materials and methods
Mimosine S-phase fractionation
ES cells were cultivated in Dulbecco's modified eagle medium (DMEM) medium containing 1000 U of leukemia inhibitory factor (LIF). To block ES cells in late G1 phase of the cell cycle, 5 × 106 cells per time point were incubated for 16 h in 0.5 mM mimosine (Krude 1999). The cells were released into S phase by two washes with Hepes buffer and subsequently resuspended in normal DMEM medium + LIF. One hour prior to harvesting of each time point, medium was supplemented with 50 μM 5-bromo-2‘-deoxyuridine (BrdU) and 10 μM 5-fluoro-2‘-deoxyuridine (FdU). Cells were harvested by trypsinization, and digested in lysis buffer (100 mM Tris-HCl at pH 8.5, 5 mM EDTA, 0.2% SDS, 200 mM NaCl, 100–300 μg/mL Proteinase K) for several hours at 55°C, and phenol:chloroform extracted. DNA was precipitated with 0.6 volumes of isopropanol, resuspended in 20 mM Tris at pH 7.5, and sonicated to an average size of 1 kb. Light DNA and heavy BrdU-labeled DNA fractions were separated by cesium chloride gradient centrifugation. DNA solution (25 μg) was supplemented with 4 g cesium chloride, 125 μL 2M Tris at pH 7.5, 10 μL 0.5m EDTA at pH 8.0, 500 μL ethidium bromide (10 mg/mL), 3.265 μL of water and 32P-labeled genomic tracer DNA. Samples were spun for 24 h at 54,000 rpm in a VTi65.2 vertical rotor at room temperature. Next, seven drop samples (200 μL) were collected from the bottom of the gradient. Ten microliters of each fraction was temperature-denatured and spotted on a Hybond+ membrane, and the relative amount of tracer “light” DNA was quantitated on a phosphorimager. The same filter was then hybridized with 500 ng of 32-P-labeled genomic DNA in Church buffer (0.5 M NaPO4 at pH 7.5, 7% SDS, 2 mM EDTA) at 65°C, and washed; radioactivity was quantitated on a PhosphorImager. The samples containing uncontaminated “heavy” BrdU-labeled DNA were determined by subtracting the relative amount of tracer DNA from the total amount of DNA. Noncontaminated BrdU-positive fractions were pooled and DNA was precipitated after addition of 10μg yeast tRNA. Pellets were resuspended in 25μL of 20 mM Tris at pH 7.5.
BrdU-labeled DNA of each fraction was subjected to PCR analysis using the following primers, 3′Igf2-forward, GGGTAC CCATTTCTGCTCCT; 3′Igf2-reverse, AGTCGGGACAACCA ACTCAC; 5′DMR-forward, GGTGGTGCACTTATCATCCA; 5′DMR-reverse, TCCCAATGGACGTACACAGA; Snrpn-forward, TCCTCCCTCTTTTCATGAATGT; Snrpn-reverse, GG GAACACCTCACAAAATGG. The 3′Igf2 PCR product contains a HindII polymorphism present on the M. castaneus allele. HindII digestion results in 450-bp and 200-bp fragments. The 5′DMR primers amplify a 50-bp length polymorphism (129/Sv: 650-bp, M. castaneus: 700 bp). The Snrpn PCR product contains a TaqI polymorphism that is present on the 129/Sv allele (Blaydes et al. 1999). TaqI digestion results in 450- and 200-bp fragments. All PCR reactions were carried out for 28–30 cycles. To prevent heteroduplex formation of the product for analysis, we added one more amplification round with 5 μL of the final PCR product using new primers and buffer containing 32P- dCTP. Radioactive PCR products were run on agarose gels, blotted onto Hybond+, and quantitated on a phosphorimager.
Generation of ES cells
Parthenogenetic ES cells were generated by activating unfertilized metaphase II-arrested oocytes with 10 mM Sr2+ in Ca2+-free MCZB medium in the presence of 5 μg/mL cytochalasin B to prevent polar body extrusion. Oocytes were deactivated after 6 h and cultured to the blastocyst stage in KSOM medium. Blastocysts were briefly treated with Acid Tyrode’s solution to remove the zona pellucida and transferred onto γ-irradiated MEF feeder cells in DMEM (15% FBS, 1000 U/mL LIF, 50 μM Mek1 kinase inhibitor, PD98059, Cell Signaling Technology). Inner cell masses were dissociated after 3–4 d with trypsin-EDTA and replated onto feeder cells to establish ES cell lines.
Dnmt1−/− and compound Dnmt3a−/−, Dnmt3b−/− ES cells were generated through consecutive gene targeting (Lei et al. 1996; Okano et al. 1999). Dnmt3L ES cells were isolated from delayed blastocysts of a cross of a Dnmt3L−/− female and a wild-type male. Blastocysts were collected 4 d after ovariectomy, which was performed at 2.5 dpc and transferred onto γ-irradiated MEF feeder cells in DMEM (15% FBS, 1000 U/mL LIF). Inner cell masses were dissociated after 3–4 d with trypsin-EDTA and replated onto feeder cells to establish ES cell lines.
DNA fluorescence in situ hybridization
DNA FISH was performed as described in Selig et al. (1992) with minor modifications. Briefly, medium of exponentially growing cells was supplemented with 10 μM BrdU and incubated for 45 min. Cells were trypsinized, washed with Hepes buffer, and resuspended in 0.75 M KCl. After trypsinization, ES cells were incubated for 10 min on ice, all other cell types for 10 min at 37°C. Cells were fixed for 10 min in ice-cold methanol:acetic acid solution (3:1 ratio), washed three times with methanol:acetic acid and stored at 4°C or spotted onto poly-L-lysine coated slides.
If necessary, slides were treated with RNase (100 μg/mL, 2× SSC) for 30 min at 37°C and washed 3 × 5 min in 2× SSC and dehydrated in 70%, 90%, and 100% ethanol. Target sequences were denatured by applying 100 μL of 70% formamide, 10 mM phosphate buffer in 2× SSC under a coverslip and incubated for 3 min on a hotplate (80°C). After removal of the coverslip, slides were washed in 2× SSC (5 min at 4°C), in 70% ethanol (5 min at −20°C), and through 90% and 100% ethanol for 3 min each. Meanwhile, nick-translated BAC and cosmid probe sequences were dissolved in a hybridization mixture containing 50% formamide, 2× SSC, 50 mM phosphate buffer at pH 7.0, 10 mg/mL salmon sperm DNA, 10% dextrane sulfate, and 100 ng/μL mouse Cot DNA to a final concentration of 2 ng/μL. Probe mix was denatured for 5 min, prehybridized for a minimum of 45 min, and then applied onto the slide. Slides were incubated overnight in a humidified chamber at 37°C.
Igf2 (cDig), H19 (cAH), 3′H19 (17.2), and mouse α-globin cosmid probes have been described before (Kielman et al. 1993; Koide et al. 1994; Greally et al. 1998). Snrpn BAC 397F16 (Gabriel et al. 1998) and Kvlqt1 BAC 101N20 (Onyango et al. 2000) have been described and were acquired from Research Genetics. A cosmid covering the p57(kip2) gene was generated by subcloning BAC 124B2 (Onyango et al. 2000) into a cosmid vector (Stratagene). The presence of the p57(kip2) gene was verified by PCR analysis and Southern blotting. All probes were digoxygenin-labeled by nick translation, purified over G50 columns, precipitated, and resuspended in hybridization mix.
After hybridization slides were washed in 2× SSC (5 min at 37°C), in 50% formamide, 2× SSC (3 × 10 min at 37°C) and in 0.1M Tris, 0.15 M NaCl, 0.05% Tween 20 (2 × 5 min at room temperature), then incubated in 2 mg/mL bovine serum albumin (BSA) in 0.1M Tris, 0.15 M NaCl in a humidified chamber (30 min at room temperature). Detection was with subsequent incubation steps with antidigoxygenin (Boehringer), anti-sheep (FITC, Jackson Labs, only when necessary), anti-BrdU (DAKO), anti-mouse (Rhodamine Red, Jackson Labs), antibodies in 0.1 M Tris, 0.15 M NaCl (30 min at room temperature). Slides were washed twice in between each detection step with 0.1 M Tris, 0.15 M NaCl, 0.05% Tween 20 and mounted with Vectashield (Vector Labs) and stored at 4°C. Fluorescence was detected by epifluorescence/CCD. Between 100 and 150 cells were counted per cell line.
Replication fork direction analysis
Medium of 5 × 107 exponentially growing cells was supplemented with 10 μM BrdU, 10 μM FdU with or without 2 μM emetine, and incubated overnight. Cells were tryspsinized and resuspended in lysis buffer (100 mM Tris-HCl at pH 8.5, 5 mM EDTA, 0.2% SDS, 200 mM NaCl, 100 mg/mL RNase A) and incubated for 4 h at 37°C. Lysis buffer was supplemented with 100–300 μg/mL Proteinase K and incubated overnight at 37°C. DNA was phenol:chloroform-extracted and precipitated with 0.6 volumes of isopropanol. The DNA was resuspended in 20 mM Tris at pH 7.5 and sonicated to an average size of 1 kb. An aliquot of 100 μg was heat-denatured, cooled on ice and mixed with 100 μL of anti-BrdU antibody (DAKO, DK) in 1 mM phosphate buffer at pH 7.2, 0.14 M NaCl and 0.05% Triton X-100 and incubated 1 h at room temperature. Next, 100 μL of anti-mouse IgG (Sigma) was added and the mixture was incubated for another hour at room temperature. The precipitate was spun down for 5 min at 4°C and washed three times with precipitation buffer. DNA was phenol/chloroform extracted and 4 μg of DNA treated with or without emetine was denatured and spotted in duplicates on Hybond+ membrane. Filters were hybridized with the following RNA probes: Igf2, 2.4 kb BamHI fragment (accession no. U71085, 22603–25084), 3′Igf2, 1.6 kb BamHI fragment (accession no. AC013548, 161055–162687), intergenic region, 1.9 kb EcoRI fragment (accession no. AC013548, 203490–205402), 5′DMR, 1.2 kb EcoRI-BamHI fragment (accession no. AC013548, 184006–182864), H19, 0.6 kb PCR product (accession no. AF049091, 7550–8132), 5′L23mrp, 1.4 kb SacI fragment (accession no. AF049091, 37571–38965). All fragments were cloned into Bluescript KS+ and the template was transcribed with T3 and T7 RNA polymerase (Promega). The absence of repetitive sequences was verified by Southern blot hybridization. Filters were hybridized overnight in Church buffer (0.5 M NaPO4 at pH 7.5, 7% SDS, 2 mM EDTA,1% BSA) at 65°C, then washed and radioactivity was quantitated on a phosporimager.
3D RNA fluorescence in situ hybridization
Fetal livers of 13.5 dpc embryos were disrupted into a single cell suspension by pipetting up and down with a small pipette tip. The single cell suspension was spotted on poly-L-lysine-coated slides and fixed with 4% formaldehyde/5% acetic acid in phosphate-buffered saline (PBS) for 18 min at room temperature. Slides were washed with PBS (3 × 5 min) and stored in 70% ethanol at −20°C. Pretreatment of slides included washing for 5 min with 70% ethanol and 0.1 M Tris, 0.15 M NaCl. Next, slides were subjected to pepsin digestion (4 min at 37°C; 0.01% pepsin in 0.01M HCl), rinsed in water and post-fixed in 4% formaldehyde/PBS (5 min). Slides were washed with PBS (10 min), dehydrated in 70%, 90%, and 100% ethanol and air-dried. The hybridization mixture (1 ng/μL DNA-probe, 50% formamide, 2× SSC, 200 ng/μL salmon sperm DNA, 5× Denhardt, 50 mM phosphate buffer, 1mM EDTA) was applied (12 μL per 24 × 24 mm coverslip) and slides were incubated at 37°C in a moisturized chamber for 12 h. Igf2 and H19 probe-DNA's were nick-translated (Roche) with digoxygenin- and biotin-conjugated nucleotides. The Igf2 probe was an 8.6-kb Igf2 fragment (accession no. U71085, 8017–16706), the H19 probe was a 582-bp PCR product (accession no. AF049091, 7550–8132), the βmajor probe was a 630-bp BamHI-PstI fragment-spanning intron 2. After hybridization, slides were washed in 2× SSC (4 × 10 min at 37°C) and in 0.1 M Tris, 0.15 M NaCl, 0.05% Tween 20 (2 × 5 min at room temperature), then incubated in 2 mg/mL BSA in 0.1 M Tris, 0.15 M NaCl in a humidified chamber (30 min at room temperature). Double-label digoxygenin/biotin detection was with subsequent incubation steps with antidigoxygenin (Boehringer), anti-sheep (FITC, Jackson Labs), anti-rabbit (FITC, Jackson Labs), and antibiotin (Boehringer), anti-mouse (Rhodamine Red, Jackson Labs), anti-horse (Rhodamine Red, Jackson Labs) antibodies in 0.1 M Tris, 0.15 M NaCl (30 min at room temperature). Detection of the nuclear localization was with subsequent incubation steps with antidigoxygenin (Boehringer), anti-sheep (FITC, Jackson Labs), anti-rabbit (FITC, Jackson Labs), antinuclear pore complex protein (Babco), anti-mouse (Rhodamine Red, Jackson Labs) antibodies in 0.1 M Tris, 0.15 M NaCl (30 min at room temperature). Slides were washed twice in between each detection step with 0.1 M Tris, 0.15 M NaCl, 0.05% Tween 20 and finally mounted with Vectashield (Vector Labs) and stored at 4°C. Fluorescence was detected with a Zeiss confocal microscope; 3D pictures were processed with commercial Zeiss software.
3D DNA fluorescence in situ hybridization
Single cell suspensions of undifferentiated ES cells and fetal liver cells were spotted and fixed onto poly-L-lysine coated slides with 4% paraformaldehyde in PBS for 20 min at room temperature. Slides were pretreated as for 3D RNA FISH, and denatured by applying 100 μL of 70% formamide, 10 mM phosphate buffer in 2× SSC under a coverslip and incubated for 3 min on a hotplate (80°C). After removal of the coverslip, slides were washed in 2× SSC (5 min at 4°C), in 70% ethanol (5 min; at −20°C), and through 90% and 100% ethanol for 3 min each. The Igf2 cosmid probe has been described elsewhere (cDig; Koide et al. 1994), and was digoxygenin-labeled by nick translation. Probe sequences were dissolved in a hybridization mixture containing 50% formamide, 2× SSC, 50 mM phosphate buffer at pH 7.0, 10 mg/mL salmon sperm DNA, 10% dextrane sulfate and 100 ng/μL mouse Cot DNA to a final concentration of 2 ng/μL, denatured for 5 min and prehybridized for a minimum of 45 min, and was then applied onto the slide. Slides were incubated overnight in a humidified chamber. Washes and detection was as for 3D RNA FISH.
Acknowledgments
We thank Ted Rasmussen, Kim Monkhorst, Andy Chess, Albrecht Sippel, Sandra Luikenhuis and Anton Wutz for stimulating discussions, Shirley Tilghman for providing H19 mutant mice, Azim Surani for providing cDig and cAH cosmids, John Greally for providing the c17.2 cosmid and Menno Kielman for providing the α globin cosmid. This work is supported by the NWO and the Human Frontier Science Program (J.G.) and NIH grants CA82389 and GM52106 (E.L.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jaenisch@wi.mit.edu; FAX (617) 258-6505.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1059603.
References
- Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, Mastrangelo MA, Jun Z, Walter J, Jaenisch R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol. 2002;22:2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaydes SM, Elmore M, Yang T, Brannan CI. Analysis of murine Snrpn and human SNRPN gene imprinting in transgenic mice. Mamm Genome. 1999;10:549–555. doi: 10.1007/s003359901042. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Brown KE, Amoils S, Horn JM, Buckle VJ, Higgs DR, Merkenschlager M, Fisher AG. Expression of α- and β-globin genes occurs within different nuclear domains in haemopoietic cells. Nat Cell Biol. 2001;3:602–606. doi: 10.1038/35078577. [DOI] [PubMed] [Google Scholar]
- Burhans WC, Vassilev LT, Wu J, Sogo JM, Nallaseth FS, DePamphilis ML. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J. 1991;10:4351–4360. doi: 10.1002/j.1460-2075.1991.tb05013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Choi OR, Engel JD. Developmental regulation of β-globin gene switching. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- Cimbora DM, Schubeler D, Reik A, Hamilton J, Francastel C, Epner EM, Groudine M. Long-distance control of origin choice and replication timing in the human β-globin locus are independent of the locus control region. Mol Cell Biol. 2000;20:5581–5591. doi: 10.1128/mcb.20.15.5581-5591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Epner E, Forrester WC, Groudine M. Asynchronous DNA replication within the human β-globin gene locus. Proc Natl Acad Sci. 1988;85:8081–8085. doi: 10.1073/pnas.85.21.8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J, Paolella G, Ramos C, Lamond AI. Spatial organization of large-scale chromatin domains in the nucleus: A magnified view of single chromosome territories. J Cell Biol. 1997;139:1597–1610. doi: 10.1083/jcb.139.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester WC, Epner E, Driscoll MC, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes & Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- Fournier C, Goto Y, Ballestar E, Delaval K, Hever AM, Esteller M, Feil R. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 2002;21:6560–6570. doi: 10.1093/emboj/cdf655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel JM, Gray TA, Stubbs L, Saitoh S, Ohta T, Nicholls RD. Structure and function correlations at the imprinted mouse Snrpn locus. Mamm Genome. 1998;9:788–793. doi: 10.1007/s003359900868. [DOI] [PubMed] [Google Scholar]
- Grandjean V, O'Neill L, Sado T, Turner B, Ferguson-Smith A. Relationship between DNA methylation, histone H4 acetylation, and gene expression in the mouse imprinted Igf2-H19 domain. FEBS Lett. 2001;488:165–169. doi: 10.1016/s0014-5793(00)02349-8. [DOI] [PubMed] [Google Scholar]
- Greally JM, Starr DJ, Hwang S, Song L, Jaarola M, Zemel S. The mouse H19 locus mediates a transition between imprinted and nonimprinted DNA replication patterns. Hum Mol Genet. 1998;7:91–95. doi: 10.1093/hmg/7.1.91. [DOI] [PubMed] [Google Scholar]
- Hark AT, Tilghman SM. Chromatin conformation of the H19 epigenetic mark. Hum Mol Genet. 1998;7:1979–1985. doi: 10.1093/hmg/7.12.1979. [DOI] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyl transferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Raghuraman MK, Gasser SM. The positioning and dynamics of origins of replication in the budding yeast nucleus. J Cell Biol. 2001;152:385–400. doi: 10.1083/jcb.152.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawame H, Gartler SM, Hansen RS. Allele-specific replication timing in imprinted domains: Absence of asynchrony at several loci. Hum Mol Genet. 1995;4:2287–2293. doi: 10.1093/hmg/4.12.2287. [DOI] [PubMed] [Google Scholar]
- Keohane AM, O'Neill L, P, Belyaev ND, Lavender JS, Turner BM. X-inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Khosla S, Aitchison A, Gregory R, Allen ND, Feil R. Parental allele-specific chromatin configuration in a boundary-imprinting-control element upstream of the mouse H19 gene. Mol Cell Biol. 1999;19:2556–2566. doi: 10.1128/mcb.19.4.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielman MF, Smits R, Devi TS, Fodde R, Bernini LF. Homology of a 130-kb region enclosing the α-globin gene cluster, the α-locus controlling region, and two nonglobin genes in human and mouse. Mamm Genome. 1993;4:314–323. doi: 10.1007/BF00357090. [DOI] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll DJ, Nicholls RD, Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- Knoll JH, Cheng SD, Lalande M. Allele specificity of DNA replication timing in the Angelman/Prader-Willi syndrome imprinted chromosomal region. Nat Genet. 1994;6:41–46. doi: 10.1038/ng0194-41. [DOI] [PubMed] [Google Scholar]
- Koide T, Ainscough J, Wijgerde M, Surani MA. Comparative analysis of Igf-2/H19 imprinted domain: Identification of a highly conserved intergenic DNase I hypersensitive region. Genomics. 1994;24:1–8. doi: 10.1006/geno.1994.1574. [DOI] [PubMed] [Google Scholar]
- Krude T. Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp Cell Res. 1999;247:148–159. doi: 10.1006/excr.1998.4342. [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lu ZH, Sittman DB, Romanowski P, Leno GH. Histone H1 reduces the frequency of initiation in Xenopus egg extract by limiting the assembly of prereplication complexes on sperm chromatin. Mol Biol Cell. 1998;9:1163–1176. doi: 10.1091/mbc.9.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J Cell Biol. 2002;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff BE, Chess A, Cedar H, et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyl transferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Onyango P, Miller W, Lehoczky J, Leung CT, Birren B, Wheelan S, Dewar K, Feinberg AP. Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15 imprinted domain. Genome Res. 2000;10:1697–1710. doi: 10.1101/gr.161800. [DOI] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new corepressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Schubeler D, Francastel C, Cimbora DM, Reik A, Martin DI, Groudine M. Nuclear localization and histone acetylation: A pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes & Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- Selig S, Okumura K, Ward DC, Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992;11:1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer R, Hershko AY, Perk J, Mostoslavsky R, Tsuberi B, Cedar H, Buiting K, Razin A. The imprinting box of the Prader-Willi/Angelman syndrome domain. Nat Genet. 2000;26:440–443. doi: 10.1038/82571. [DOI] [PubMed] [Google Scholar]
- Simon I, Tenzen T, Reubinoff BE, Hillman D, McCarrey JR, Cedar H. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 1999;401:929–932. doi: 10.1038/44866. [DOI] [PubMed] [Google Scholar]
- Simon I, Tenzen T, Mostoslavsky R, Fibach E, Lande L, Milot E, Gribnau J, Grosveld F, Fraser P, Cedar H. Developmental regulation of DNA replication timing at the human β-globin locus. EMBO J. 2001;20:6150–6157. doi: 10.1093/emboj/20.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE. Telomeric chromatin modulates replication timing near chromosome ends. Genes & Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi N. Differentiation of X chromosomes in early female mouse embryos. Exp Cell Res. 1974;86:127–135. doi: 10.1016/0014-4827(74)90657-0. [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]