Abstract
Yeast protein kinase GCN2 stimulates the translation of transcriptional activator GCN4 by phosphorylating eIF2α in response to amino acid starvation. Kinase activation requires binding of uncharged tRNA to a histidyl tRNA synthetase-related domain in GCN2. Phosphorylation of serine 577 (Ser 577) in GCN2 by another kinase in vivo inhibits GCN2 function in rich medium by reducing tRNA binding activity. We show that rapamycin stimulates eIF2α phosphorylation by GCN2, with attendant induction of GCN4 translation, while reducing Ser 577 phosphorylation in nonstarved cells. The alanine 577 (Ala 577) mutation in GCN2 (S577A) dampened the effects of rapamycin on eIF2α phosphorylation and GCN4 translation, suggesting that GCN2 activation by rapamycin involves Ser 577 dephosphorylation. Rapamycin regulates the phosphorylation of Ser 577 and eIF2α by inhibiting the TOR pathway. Rapamycin-induced dephosphorylation of Ser 577, eIF2α phosphorylation, and induction of GCN4 all involve TAP42, a regulator of type 2A-related protein phosphatases. Our results add a new dimension to the regulation of protein synthesis by TOR proteins and demonstrate cross-talk between two major pathways for nutrient control of gene expression in yeast.
Keywords: TOR, TAP42, translational control, eIF2α kinase, GCN2
General amino acid control (GAAC) is a major pathway for regulating amino acid biosynthesis in the yeast Saccharomyces cerevisiae. The transcriptional activator GCN4 is induced at the translational level by limitation for any amino acid and activates the transcription of >500 genes, including the majority involved in amino acid biosynthesis (Natarajan et al. 2001). The protein kinase (PK) GCN2 is responsible for the increased translation of GCN4 mRNA in starved cells and functions by phosphorylating the α subunit of translation initiation factor 2 (eIF2α; Hinnebusch 1996; Hinnebusch and Natarajan 2002). The eIF2 is responsible for delivery of charged methionyl initiator tRNA to the initiation codon in the form of a ternary complex (TC) with GTP. Phosphorylation of eIF2α converts eIF2 from substrate to inhibitor of its guanine nucleotide exchange factor, eIF2B. The inhibition of GDP–GTP exchange on eIF2 reduces the GTP-bound form of eIF2 and impedes TC formation. Although the decrease in TC levels reduces general protein synthesis, it specifically stimulates translation of GCN4 mRNA. A specialized reinitiation mechanism involving four short open reading frames (uORFs) in the GCN4 mRNA leader serves to repress GCN4 translation under nonstarvation conditions and derepress it in response to eIF2α phosphorylation in starved cells (Hinnebusch 2000).
Uncharged tRNAs that accumulate during amino acid starvation activate GCN2 by binding to a histidyl-tRNA synthetase (HisRS)-related domain located C-terminal to the PK domain (Wek et al. 1995; Hinnebusch 1996; Zhu et al. 1996). Physical interactions of the PK domain with the HisRS and extreme C-terminal domain of GCN2 are thought to prevent binding of uncharged tRNA and kinase activation by basal concentrations of uncharged tRNA in nonstarved cells (Dong et al. 2000). Autophosphorylation of threonines 882 and 887 in the activation loop of the PK domain is essential for GCN2 function in vivo (Romano et al. 1998). A two-step activation mechanism has been proposed in which tRNA binding eliminates an autoinhibitory structure in the PK domain and elicits autophosphorylation of T882 and T887, with ensuing activation of the eIF2α kinase function of GCN2 (Qiu et al. 2002).
Serine 577 (Ser 577) in GCN2 was identified by mass spectrometry as a site of phosphorylation by another kinase in vivo, and genetic evidence suggests that phosphorylation of this residue down-regulates GCN2 activity. Ser 577 is phosphorylated under nonstarvation conditions, and its replacement with nonphosphorylatable alanine (S577A mutation) results in partial activation of GCN2 in the absence of amino acid limitation. The S577A mutation also increases tRNA binding by GCN2 in vitro, suggesting that Ser 577 phosphorylation decreases the affinity of GCN2 for uncharged tRNA. As Ser 577 was only transiently and partially dephosphorylated during starvation for histidine, we speculated that its dephosphorylation would occur under starvation or stress conditions distinct from amino acid limitation in which GCN2 must be activated without an increase in levels of uncharged tRNA (Garcia-Barrio et al. 2002).
GCN2 activity is also induced in response to starvation for purines or glucose (Rolfes and Hinnebusch 1993; Yang et al. 2000), growth on nonfermentable carbon sources (Yang et al. 2000), and environmental stresses including high salinity (Goossens et al. 2001) and the alkylating agent methyl methanesulfonate (MMS; Natarajan et al. 2001). Consistently, all of these conditions elicit increased synthesis of GCN4 and derepressed transcription of genes subject to GAAC. GCN4 (or its target genes) also are induced by hydroxyurea (HU), an inhibitor of DNA replication and repair, by tunicamycin, an inducer of the unfolded protein response, and by rapamycin, an inhibitor of the target of rapamycin (TOR) proteins (Hughes et al. 2000; Valenzuela et al. 2001). However, it was unknown whether the responses to these last three drugs are dependent on activation of GCN2 and increased eIF2α phosphorylation.
To understand the physiological role of Ser 577 phosphorylation in controlling GCN2 activity, we investigated whether it becomes dephosphorylated in response to purine starvation or treatment of cells with various drugs known to induce the GAAC response. Only treatment with rapamycin led to stimulation of eIF2α phosphorylation coincident with dephosphorylation of Ser 577 in vivo. Our genetic analysis suggests that activation of GCN2 and induction of GCN4 translation by rapamycin involves dephosphorylation of Ser 577 and requires the tRNA binding activity of GCN2 and TOR proteins.
The TOR signaling pathway in yeast negatively regulates genes involved in utilization of poor nitrogen sources, many of which are targets of the transcriptional activator GLN3 (Cardenas et al. 1999; Schmelzle and Hall 2000). The TOR proteins also stimulate protein synthesis by controlling the abundance of ribosomes and translation initiation factor eIF4F (Berset et al. 1998; Powers and Walter 1999). Hence, inactivation of TOR by rapamycin elicits a variety of responses characteristic of nutrient-deprived cells. The TOR proteins control several downstream effectors in the pathway, including GLN3 and NPR1, a Ser/Thr PK that regulates nutrient transporters (Schmidt et al. 1998), by preventing their dephosphorylation by the type 2A-related protein phosphatase SIT4. It appears that TOR inhibits SIT4 by promoting its interaction with the negative regulatory protein TAP42 (Di Como and Arndt 1996; Schmidt et al. 1998; Beck and Hall 1999). Interestingly, we found that TAP42 is involved in dephosphorylation of Ser 577 in GCN2 and consequent induction of eIF2α-P and GCN4 translation by rapamycin. By inhibiting GCN2 via TAP42-dependent phosphorylation of Ser 577, the TOR pathway maintains repression of GCN4 and its target genes, some of which are subject to dual regulation by GCN4 and GLN3 (Valenzuela et al. 2001). Together, our findings unveil an intricate interplay between the two major pathways that couple nutrient metabolism and rates of protein synthesis to the abundance and quality of nutrients in the environment.
Results
Rapamycin stimulates eIF2α phosphorylation on Ser 51 coincident with dephosphorylation of Ser 577 in GCN2
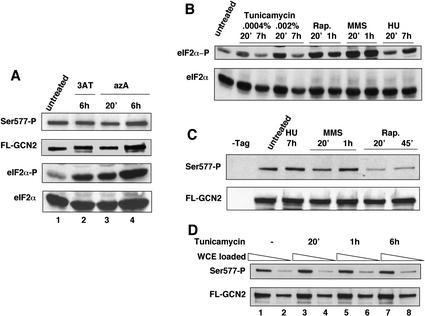
Ser 577 in GCN2 is phosphorylated under nonstarvation conditions and is only partially and transiently dephosphorylated in response to histidine starvation (Garcia-Barrio et al. 2002). Here we asked whether starvation for purines or glucose, conditions that induce GCN4 translation (Rolfes and Hinnebusch 1993; Yang et al. 2000), would elicit dephosphorylation of Ser 577. Adenine starvation was imposed by addition of 8-azaadenine (azA) to the medium. Whole cell extracts (WCEs) were prepared from gcn2Δ cells expressing a functional, Flag epitope-tagged form of GCN2 (FL-GCN2) produced at native levels (Garcia-Barrio et al. 2002) and subjected to Western analysis with antibodies specific for eIF2α phosphorylated on Ser-51 (eIF2α-P); subsequently, the blots were stripped and reprobed with polyclonal antibodies against total eIF2α. As expected (Rolfes and Hinnebusch 1993), addition of azA led to increased phosphorylation of eIF2α on Ser 51 after 20 min of treatment, as judged by a large increase in the amount of phosphorylated eIF2 (eIF2α-P) relative to total eIF2α (Fig. 1A, bottom two panels, cf. lanes 3,4 and 1). The increased eIF2α-P content produced by azA was comparable to that seen in response to histidine starvation imposed by 3-aminotriazole (3AT; Fig. 1A, lanes 2–4). To detect phosphorylation of Ser 577, the FL-GCN2 was immunoprecipitated with anti-Flag resin from the same extracts and subjected to Western analysis with antibodies specific for phosphorylated Ser 577 (Ser 577-P; Garcia-Barrio et al. 2002). The blots were stripped and reprobed with polyclonal antibodies against GCN2. As shown in Figure 1A (top panels), azA treatment for 20 min or 6 h did not diminish the Ser 577-P content of immunoprecipitated FL-GCN2, as the ratio of Ser 577-P to FL-GCN2 Western signals was essentially unchanged. A similar result was obtained after 6 h of histidine starvation (Fig. 1A), in agreement with previous findings (Garcia-Barrio et al. 2002). Thus, it appears that dephosphorylation of Ser 577 is not critically involved in activation of GCN2 by purine or histidine starvation. Starvation for glucose also did not elicit dephosphorylation of Ser 577 (data not shown).
Figure 1.
Effect of purine starvation and drug treatments on eIF2α phosphorylation on Ser 51 and GCN2 phosphorylation on Ser 577. (A) Transformants of gcn2Δ strain H2511 bearing centromeric plasmid pDH101, encoding Flag epitope-tagged GCN2 (FL-GCN2), were grown to early logarithmic phase in SD medium, and the cultures were split and cultured either untreated or treated with 50 μg/mL azaadenine (azA) for 20 min or 6 h. WCEs were prepared and phosphorylation of Ser 51 in eIF2α was detected by Western analysis with antibodies specific for eIF2α phosphorylated on Ser 51 (eIF2α-P) and compared with the total amount of the protein detected with antibodies against eIF2α. To detect phosphorylated Ser 577 in GCN2, we immunopurified FL-GCN2 from WCEs with anti-Flag M2 agarose affinity gel, resolved it by SDS-PAGE, and subjected it to Western analysis using antibodies specific for GCN2 phosphorylated on Ser 577 (Ser 577-P) or antibodies against total GCN2 (FL-GCN2). (B–D) Transformants described in A were grown in SC medium lacking uracil and treated with 0.0004% or 0.002% tunicamycin, 200 ng/mL rapamycin (Rap.), 0.07% MMS, or 50 mM HU for the indicated times, or were cultured without treatment to early logarithmic phase. Phosphorylation of Ser 51 in eIF2α (B) and Ser 577 in GCN2 (C,D) was measured as described in A. In C the first lane contained WCE from an isogenic transformant harboring empty plasmid pRS316. The odd-numbered lanes in D contained twice as much WCE as for the subsequent even-numbered lanes.
We next investigated the effects on eIF2α and Ser 577 phosphorylation of treating cells with tunicamycin, rapamycin, MMS, and HU, all shown previously to induce expression of GCN4 or its target genes (Hughes et al. 2000; Natarajan et al. 2001; Valenzuela et al. 2001). Except for long-term treatment with tunicamycin (7 h) and short-term exposure to HU (20 min), all of the other treatments increased the levels of eIF2α-P in WCEs (Fig. 1B). Importantly, only the treatment with rapamycin led to dephosphorylation of Ser 577 (Fig. 1C,D), consistent with the possibility that rapamycin activates GCN2 by promoting dephosphorylation of Ser 577.
Increased phosphorylation of eIF2α induced by rapamycin depends strongly on Ser 577 and requires the tRNA binding activity of GCN2
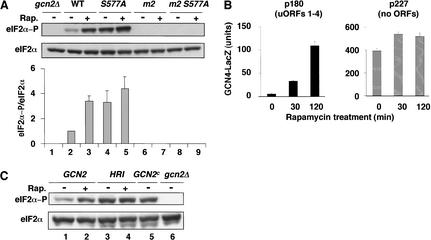
Activation of GCN2 by various starvation or stress conditions requires the tRNA binding activity of its HisRS-like domain (for review, see Hinnebusch and Natarajan 2002). To determine if this was true for rapamycin, we asked whether substitutions in the m2 motif of the HisRS-like domain (Y1119L and R1120L) would prevent induction of eIF2α phosphorylation by rapamycin. These mutations abolish tRNA binding by GCN2 in vitro and its kinase activity in vivo (Wek et al. 1995; Dong et al. 2000). The results in Figure 2A (lanes 6,7) show that no eIF2α-P was detectable in the presence or absence of rapamycin in WCEs prepared from the gcn2-Y1119L,R1120L strain (m2 mutant). Thus, it appears that tRNA binding by GCN2 is absolutely required for its activation by rapamycin, as observed previously for amino acid limitation (Wek et al. 1995).
Figure 2.
(A) Effect of rapamycin on phosphorylation of eIF2α is strongly dependent on dephosphorylation of Ser 577 and the tRNA binding activity of GCN2. Transformants of gcn2Δ strain H2511 bearing plasmid pDH101 containing GCN2-FL (WT), pCB149 containing GCN2-S577A-FL (S577A), pCB150 containing GCN2-m2-FL (m2), pCB151 containing GCN2-S577A-m2-FL (m2 S577A), or empty vector (gcn2Δ) were grown to early logarithmic phase in SC medium lacking uracil. The cultures were split in half and cultured either untreated (Rap.−) or treated with 200 ng/mL rapamycin for 20 min (Rap.+). WCEs were prepared, resolved by SDS-PAGE, and subjected to Western analysis using antibodies against total eIF2α or phosphospecific antibodies against eIF2α phosphorylated on Ser 51 (eIF2α-P). Western signals were quantified by video densitometry using NIH Image software (version 1.61), and the average ratios of eIF2α-P/eIF2α signals measured in three independent experiments are shown graphically below the relevant lanes with standard errors shown as error bars. The ratio in wild type was assigned a value of unity. (B) Induction of GCN4 expression by rapamycin is dependent on the uORFs. Transformants of wild-type strain BY4741 harboring CEN plasmids containing the GCN4–lacZ reporter with all four uORFs (p180) or lacking the upstream AUG codons of the uORFs (p227) were grown to early logarithmic phase in SC medium lacking uracil. The cultures were split in half and either cultured untreated (0 min) or treated with 200 ng/mL rapamycin for 30 or 120 min. Units of β-galactosidase activity (defined as nmole of o-nitrophenol β-D-galactopyranoside cleaved per minute per milligram of protein) were measured in WCEs from three or more independent transformants and the mean values and standard errors are depicted graphically. (C) Evidence that rapamycin activates GCN2 rather than inhibiting an eIF2α phosphatase. Transformants of GCN2 strain F113 bearing empty vector pRS316 (lanes 1,2), or transformants of gcn2Δ strain H2511 bearing p1246 containing HRI under the GAL promoter (lanes 3,4), or p1056 containing GCN2c-E537K,E1522K (lane 5), or empty vector pRS316 (lane 6) were grown to early logarithmic phase in SC medium lacking uracil and containing raffinose as the carbon source. The cultures were split in half, and cultured either untreated or treated with 200 ng/mL rapamycin for 20 min. WCEs were prepared and analyzed as described for A.
In agreement with previous findings, the S577A substitution in GCN2 led to increased eIF2α phosphorylation in the absence of rapamycin (Fig. 2A, cf. lane 4 and 2). Importantly, treatment of GCN2-S577A cells with rapamycin increased the eIF2α-P content only slightly compared with that observed in GCN2 cells (Fig. 2A cf. lanes 4,5 and 2,3). Quantification of multiple experiments of the type shown in Figure 2A revealed an increase in eIF2α-P by a factor of 1.1 ± 0.15 (S.E.) in GCN2-S577A cells compared with the 3.4 ± 0.4 (S.E.) increase observed in GCN2 cells. This difference is consistent with the idea that rapamycin activates GCN2 by a mechanism involving dephosphorylation of Ser 577. We also verified that the increased eIF2α-P content in GCN2-S577A versus GCN2 cells seen in the absence of rapamycin was abolished by the m2 mutations (Fig. 2A, cf. lane 8 and 4). Thus, the constitutive activity of GCN2-S577A is dependent on tRNA binding to the HisRS domain.
Derepression of GCN4 expression by rapamycin depends strongly on Ser 577 and requires phosphorylation of Ser 51 in eIF2α
Treatment of cells with rapamycin increases the expression of a GCN4–lacZ reporter (Valenzuela et al. 2001), but it was unknown whether this occurs at the translational level and requires GCN2. Accordingly, we compared expression of a GCN4–lacZ construct containing all four uORFs required for translational control (p180) to that given by a construct lacking the ATG codons of the uORFs (p227), in the presence and absence of rapamycin. As expected (Valenzuela et al. 2001), rapamycin treatment for 30 min or 2 h produced 6- and 20-fold inductions, respectively, of the construct containing uORFs (Fig. 2B, p180). In contrast, the construct lacking uORFs showed high expression in the absence of rapamycin that increased only slightly (∼1.3-fold) with drug treatment. The latter finding is at odds with the possibility that transcriptional induction of GCN4–lacZ accounts for the large increase in fusion enzyme expressed from the p180 construct. The dependence on uORFs provides strong evidence that GCN4 expression is induced by rapamycin at the translational level.
We next asked whether derepression of GCN4–lacZ expression by rapamycin is dependent on GCN2 and involves dephosphorylation of Ser 577. In the strain background used for these experiments, rapamycin treatment for 3 h produced a 5.9-fold increase of GCN4–lacZ expression in the wild-type strain, comparable to the 5.7-fold derepression seen in response to histidine starvation by 3AT (Table 1, row 1). The derepression of GCN4–lacZ by rapamycin was eliminated by deletion of GCN2 (Table 1, row 2) or the SUI2-S51A mutation (substituting Ser 51 in eIF2α with nonphosphorylatable alanine; Table 1, row 3), indicating complete dependence on Ser 51 phosphorylation by GCN2 for response to the drug. As shown previously (Garcia-Barrio et al. 2002), the GCN2-S577A mutation partially derepressed GCN4–lacZ in untreated cells (Table 1, cf. rows 1 and 4 under repressing conditions). Importantly, the response to rapamycin and 3AT was dampened in the GCN2-S577A strain (Table 1, row 4), showing derepression ratios of only 2.1 and 1.4, respectively, compared with the corresponding ratios of 5.7 and 5.9 seen in the GCN2 strain (Table 1, row 1). This is consistent with the idea that GCN2 activation and consequent derepression of GCN4 translation by rapamycin involves dephosphorylation of Ser 577. As expected, the SUI2-S51A mutation eliminated the derepressing effect of the GCN2-S577A mutation (Table 1, row 5). The fact that GCN2-S577A also dampened the derepression response to 3AT agrees with the idea that this mutation enhances tRNA binding by GCN2, reducing the dependence of kinase activation on elevated levels of uncharged tRNA in amino acid-starved cells.
Table 1.
Derepression of GCN4-lacZ expression by rapamycin involves dephosphorylation of Ser577 and requires phosphorylation of Ser51 in eIF2α
| Genotype
|
R
|
GCN4-lacZ activity (U)
|
|||
|---|---|---|---|---|---|
| 3AT
|
Rapamycin
|
||||
| DR
|
DR/R
|
DR
|
DR/R
|
||
| GCN2 SUI2 | 13 ± 0.55 | 74 ± 5 | 5.7 | 77 ± 7 | 5.9 |
| gcn2Δ SUI2 | 6 ± 0.8 | 11 ± 1.7 | 1.8 | 9.5 ± 1.4 | 1.6 |
| GCN2 SUI2-S51A | 6 ± 0 | 9 ± 1.4 | 1.5 | 8.5 ± 0.7 | 1.4 |
| GCN2-S577A SUI2 | 66 ± 5.2 | 91 ± 4.9 | 1.4 | 136 ± 8.2 | 2.1 |
| GCN2-S577A SUI2-S51A | 5.7 ± 1.5 | 6.3 ± 1.5 | 1.1 | 7 ± 1.9 | 1.2 |
The gcn2Δ SUI2 strain H1816, with an integrated GCN4-lacZ fusion, was transformed with empty vector pRS316 (gcn2Δ SUI2), pB107 (GCN2 SUI2), or pB111 (GCN2-S577A SUI2). The isogenic gcn2Δ SUI2-S51A strain H1817 was transformed with pB107 (GCN2 SUI2-S51A) or pB111 (GCN2-S577A SUI2-S51A). Cells were grown to exponential phase in SD medium containing minimal supplements (repressing conditions, R), the cultures were split in three, and 3AT (40 mM) or rapamycin (20 ng/mL) was added to one-third of each culture (DR). Cells were harvested after 3 h and WCEs were prepared and assayed for β-galactosidase activity. (DR/R) The ratio of β-galactosidase activities measured under derepressing and repressing conditions.
Rapamycin stimulates GCN2 rather than inhibiting an eIF2α phosphatase
Our results thus far suggested that rapamycin stimulates eIF2α phosphorylation by activation of GCN2, at least partly through dephosphorylation of Ser 577. To address the possibility that rapamycin additionally inhibits the phosphatase that dephosphorylates eIF2α-P (most likely GLC7; Wek et al. 1992), we asked whether rapamycin would increase the level of eIF2α-P in a gcn2Δ strain expressing the mammalian eIF2α kinase HRI. In accordance with previous results (Marton et al. 1993), this strain exhibited a high level of eIF2α-P under normal growth conditions relative to the isogenic GCN2 strain, comparable to that observed in cells containing the constitutively activated GCN2c-E537K,E1522K protein (Fig. 2C, lanes 1,3,5). Whereas rapamycin produced the expected increase in eIF2α-P in GCN2 cells, it had no effect on the eIF2α-P content in the cells containing HRI in place of GCN2 (Fig. 2C, lanes 1–4). These results suggest that rapamycin does not induce eIF2α-P levels by inhibiting an eIF2α-P phosphatase and, hence, must act primarily by stimulating GCN2 function.
Rapamycin stimulates GCN2 by inhibiting the TOR proteins
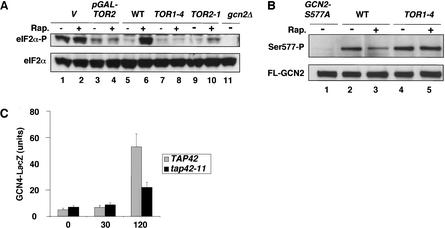
Rapamycin elicits many responses of nutrient-deprived cells because it inhibits the TOR1 and TOR2 proteins. To show that rapamycin stimulates GCN2 through inactivation of TOR proteins, we measured phosphorylation of eIF2α and Ser 577 in strains harboring dominant rapamycin-resistant TOR alleles (TOR1-4 and TOR2-1). Treatment of TOR1-4 and TOR2-1 mutants with rapamycin produced no or little increase in eIF2α phosphorylation, respectively, whereas the isogenic wild-type showed high-level eIF2α-P when treated with the drug (Fig. 3A, lanes 5–10). Similarly, overexpression of wild-type TOR2 from the GAL promoter reduced the induction of eIF2α phosphorylation by rapamycin (Fig. 3A, lanes 3,4), although growth on galactose alone induced eIF2α-P levels and thus diminished the effect of rapamycin in the isogenic wild-type strain (Fig. 3A, lanes 1,2). The results in Figure 3B show that the dominant TOR1-4 allele also abolished the reduction in Ser 577-P levels that occurs in cells with wild-type TOR when treated with rapamycin. We conclude that inhibition of TOR proteins is responsible for dephosphorylation of Ser 577 and increased eIF2α phosphorylation by GCN2 in cells treated with rapamycin.
Figure 3.
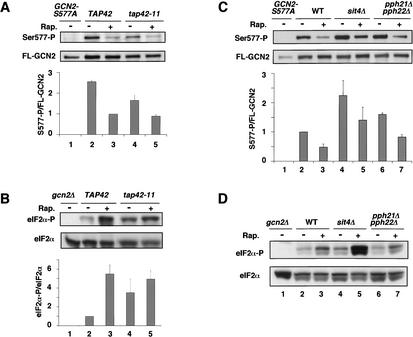
The rapamycin-induced phosphorylation of Ser 51 in eIF2α and dephosphorylation of Ser 577 in GCN2 are both mediated by TOR proteins. (A) Overexpression of TOR2 from the GAL promoter or dominant TOR mutations that confer rapamycin resistance overcome the stimulation of eIF2α phosphorylation by rapamycin. Transformants of wild-type strain F113 with empty vector (lanes 1,2) or pJK5 expressing TOR2 under the GAL promoter (lanes 3,4) growing exponentially on SC medium lacking uracil were induced with galactose for 5 h and then treated with 200 ng/mL rapamycin for 20 min or were left untreated. (Lanes 5–11) Isogenic WT, TOR1-4, and TOR2-1 strains (MLY41, MLY90-1, and MLY152, respectively) were grown to early logarithmic phase on YPD medium and treated with rapamycin at 200 ng/mL for 20 min or left untreated. The gcn2Δ strain is H2511. Levels of eIF2α phosphorylation on Ser 51 in WCEs were measured as described in Figure 1A. (B) The TOR1-4 mutation abolishes dephosphorylation of Ser 577 in response to rapamycin. Transformants of strains MLY41 (WT) and MLY90-1 (TOR1-4) expressing FL-GCN2 from plasmid pDH101 (lanes 2–5) or MLY41 expressing GCN2-S577A-FL from pCB149 (GCN2-S577A) were grown in SC medium lacking uracil and treated with 200 ng/mL rapamycin for 20 min or were left untreated. Levels of FL-GCN2 phosphorylated on Ser 577 were measured as described in Figure 1A. Phosphorylation of Ser 577 in GCN2 was detected with antibodies that specifically recognize phosphorylated Ser 577, as it was described in the Figure 1 legend. (C) The tap42-11 mutation reduces induction of GCN4 expression by rapamycin. Transformants of TAP42 strain CY1077 or tap42-11 strain CY1078 harboring p180 containing the GCN4–lacZ reporter with all four uORFs were grown for the indicated times in the presence of rapamycin and levels of β-galactosidase activity in the WCEs were measured, all as described in Figure 2B.
Involvement of TAP42 in regulation of Ser 577 phosphorylation, eIF2α phosphorylation, and GCN4 translation by TOR
The ability of TOR proteins to regulate GLN3 and NPR1 is dependent on inactivation of the type 2A-related protein phosphatase SIT4, and there is evidence that TOR inhibits SIT4 by promoting its interaction with TAP42 (Di Como and Arndt 1996; Schmidt et al. 1998; Beck and Hall 1999; Jacinto et al. 2001). Fractions of the type 2A phosphatases (PP2As) PPH21 and PPH22 also are associated with TAP42 (Di Como and Arndt 1996) and may be regulated by this interaction. Hence, we wished to determine whether TAP42 is required for the effects of rapamycin on Ser 577 and eIF2α phosphorylation. Because TAP42 is essential, we analyzed the temperature-sensitive tap42-11 allele that confers semidominant resistance to rapamycin at 25°C and recessive lethality at 37°C. Presumably, the tap42-11 product at the permissive temperature can bind to SIT4 and inhibit its activity even in the presence of rapamycin (Di Como and Arndt 1996).
If the TOR proteins inhibit GCN2 by TAP42-dependent antagonism of SIT4 or PP2A, then tap42-11 should diminish the induction of GCN4 translation in response to rapamycin. In accordance with this prediction, induction of the GCN4–lacZ fusion containing uORFs by rapamycin was significantly reduced in tap42-11 versus TAP42 cells (Fig. 3C). The residual induction of GCN4–lacZ in tap42-11 cells may reflect leakiness of the mutation or a TAP42-independent mechanism for controlling GCN4 expression.
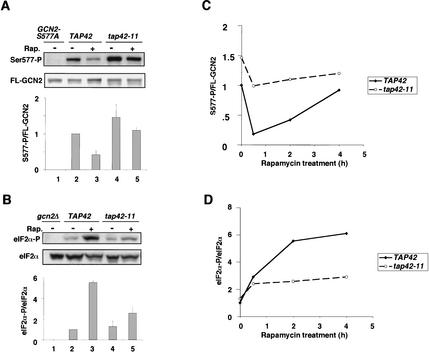
We next asked whether tap42-11 would reduce the dephosphorylation of Ser 577 elicited by rapamycin. Indeed, we found that tap42-11 cells at 25°C had a higher level of Ser 577-P in the presence or absence of rapamycin compared with TAP42 cells, and the difference in Ser 577-P levels was more pronounced in the presence of drug (Fig. 4A, lanes 2–5). The fact that tap42-11 dampens the effect of rapamycin in reducing Ser 577-P levels is consistent with the idea that TAP42 promotes TOR-dependent phosphorylation of this residue. Having found that Ser 577-P levels are elevated in tap42-11 cells, we expected to find lower levels of eIF2α phosphorylation under these conditions as the result of decreased GCN2 activity. In agreement with this prediction, rapamycin produced a smaller increase in eIF2α-P in the tap42-11 versus TAP42 cells after treatment for 2 h at 25°C (Fig. 4B, cf. lanes 4,5 and 2,3). Thus, TAP42 is required for a strong decrease in Ser 577-P and a large increase in eIF2α-P after 2 h of rapamycin treatment.
Figure 4.
The tap42-11 mutation diminishes the effects of rapamycin in promoting dephosphorylation of Ser 577 in GCN2 and increased phosphorylation of Ser 51 in eIF2α at 25°C. (A) Transformants of strains CY1077 (TAP42) and CY1078 (tap42-11) containing GCN2-FL on pDH101 (lanes 2–5), or CY1077 transformed with pCB149 containing GCN2-S577A-FL (lane 1), were grown on SC medium lacking uracil to early logarithmic phase at 25°C and either treated with 200 ng/mL rapamycin for 2 h or left untreated. WCE extracts were prepared and analyzed for phosphorylation of Ser 577 in FL-GCN2 as described in Figure 1. (B) Strains CY1077 (TAP42) and CY1078 (tap42-11) were cultured in SC medium and analyzed for eIF2α phosphorylation as described in Figure 1. The gcn2Δ strain ESY6053 was analyzed in parallel (lane 1). (C,D) Multiple experiments analogous to those described in A and B were carried out identically except that the rapamycin treatment was for 30 min or 4 h. The eIF2α-P/eIF2α and S577-P/FL-GCN2 ratios were calculated as described in Figure 2A and plotted against the hours of rapamycin treatment.
We conducted additional experiments similar to that just described except that cells were treated with rapamycin for 30 min or 4 h instead of 2 h. The data from multiple experiments for all three regimes were quantified and plotted in Figure 4C and D. The level of Ser 577-P decreased by only ∼30% in the tap42-11 cells after 30 min and remained relatively constant for up to 4 h of drug treatment. In contrast, the TAP42 cells showed a dramatic decrease (∼80%) by 30 min, and displayed a gradual recovery in the level of Ser 577-P after 2 or 4 h of drug treatment (Fig. 4C). The results in Figure 4D show that the levels of eIF2α-P were substantially lower in tap42-11 versus TAP42 cells after 2 or 4 h of drug treatment, consistent with the higher Ser 577-P content in the mutant cells at these time points (Fig. 4C). However, there was little difference in eIF2α-P content between the tap42-11 and TAP42 strains after only 30 min of drug treatment (Fig. 4D), even though the difference in Ser 577-P levels between the two strains was greatest at this time point (Fig. 4C). This discrepancy may indicate the existence of an alternative pathway that can activate GCN2 in rapamycin-treated tap42-11 cells independently of Ser 577 dephosphorylation. This putative pathway cannot provide strong, steady-state activation of GCN2 in the absence of TAP42, because the eIF2α-P level in tap42-11 cells was <50% of that seen in TAP42 cells after 2 or 4 h of rapamycin treatment (Fig. 4D).
The tap42-11 cells grow slowly at 30°C and fail to divide at 37°C (Di Como and Arndt 1996). Interestingly, the mutant cells at 30°C had lower levels of Ser 577-P in the absence of rapamycin compared with TAP42 cells, and they displayed the same low level of Ser 577-P in the presence of drug that occurs in wild-type cells (Fig. 5A). Consistently, the eIF2α-P content was higher in the absence of rapamycin in tap42-11 versus TAP42 cells at 30°C (Fig. 5B). Similar results were observed at 37°C (data not shown). To explain the results in Figure 5, A and B, we propose that the tap42-11 product dissociates from SIT4 or PP2A at both 30°C and 37°C, even in the absence of rapamycin. This results in constitutively elevated phosphatase activity and dephosphorylation of Ser 577, with attendant activation of GCN2 and increased phosphorylation of eIF2α. The idea that tap42-11 dissociates from SIT4 at 30°C or 37°C is consistent with the growth defect at 30°C, and the strong reduction in translation initiation and G1 arrest observed at 37°C in tap42-11 cells, as both phenotypes are observed in mutants with low levels of functional TOR proteins (Barbet et al. 1996).
Figure 5.
(A,B) The tap42-11 mutation alters the effects of rapamycin on phosphorylation of Ser 577 in GCN2 and Ser 51 in eIF2α at 30°C. Experiments analogous to those described in Figure 4A and B were carried out identically except that the strains were grown to early logarithmic phase at 25°C in SC medium lacking uracil or SC complete and transferred to the same medium prewarmed to 30°C for 30 min, after which they were treated with 200 ng/mL rapamycin for 20 min. (C,D) Effects of inactivating TAP42-regulated phosphatases on dephosphorylation of S577 and eIF2α phosphorylation. Transformants of strains BY4741 (WT), #3744 (sit4Δ), and CY1090 (pph21Δ pph22Δ) containing GCN2-FL on pDH101, were grown on SC medium lacking uracil to early logarithmic phase at 30°C and treated with 200 ng/mL rapamycin for 20 min, or were left untreated. Phosphorylation of Ser 577 in FL-GCN2 (C) or Ser 51 in eIF2α (D) was detected and quantified as described in Figure 1. Strain BY4741 transformed with pCB149 containing GCN2-S577A-FL (C) or gcn2Δ strain ESY6053 transformed with empty vector (D) was analyzed in parallel (lane 1).
If TAP42 is involved in TOR-mediated dephosphorylation of Ser 577, we might expect that inactivation of SIT4 or the PP2A isoforms PPH21 and PPH22 would increase the level of Ser 577-P. This result was observed in sit4Δ cells, and to a lesser extent in the pph21Δpph22Δ mutant, although rapamycin still produced reductions in Ser 577-P levels in both mutants (Fig. 5C). To explain these last findings, we propose that SIT4 and PP2A have overlapping functions in Ser 577 dephosphorylation. Inactivation of either one leads to elevated Ser 577-P, but the remaining enzyme is capable of rapamycin-sensitive dephosphorylation of Ser 577. Although the pph21Δpph22Δ double mutant showed somewhat higher levels of Ser 577-P, it did not exhibit a significant reduction in eIF2α-P levels produced by rapamycin versus that seen in the isogenic wild type (Fig. 5D, cf. lanes 6,7 and 2,3). Moreover, the level of eIF2α-P in the sit4Δ strain was even greater than that seen in the wild-type strain (Fig. 5D, cf. lanes 4,5 and 2,3). To explain these discrepancies, we suggest that complete inactivation of SIT4 or PP2A has multiple, offsetting effects on GCN2 activity (see Discussion).
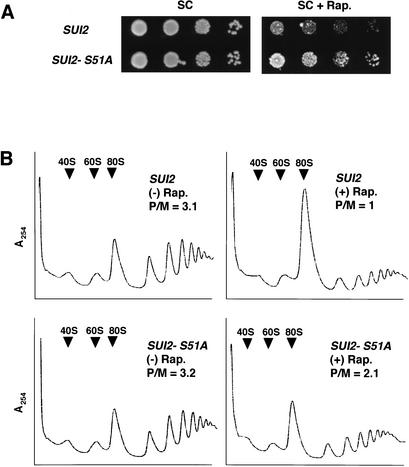
Finally, we wished to determine the extent to which induction of eIF2α phosphorylation is responsible for a general inhibition of protein synthesis and cell growth by rapamycin. As shown in Figure 6A, cells containing the nonphosphorylatable form of eIF2α containing alanine at position 51 (SUI2-S51A) were less sensitive to rapamycin than were isogenic wild-type cells. In contrast, isogenic gcn4Δ and GCN4 cells displayed no differences in rapamycin sensitivity (data not shown; Valenzuela et al. 2001). Thus, it appears that down-regulation of general translation initiation, and not induction of GCN4 and its target genes, is responsible for the growth inhibition associated with increased eIF2α phosphorylation in cells treated with rapamycin. It was shown previously (Barbet et al. 1996) that rapamycin elicits a pronounced inhibition of translation initiation in yeast. To evaluate the contribution of eIF2α phosphorylation to this inhibition, we compared polysome profiles of wild-type and SUI2-S51A cells treated with rapamycin. As expected, rapamycin reduced the polysome content of wild-type cells, as manifested by a threefold decrease in the polysome:monosome (P/M) ratio. In contrast, rapamycin produced only an ∼1.5-fold reduction in the P/M ratio in SUI2-S51A cells (Fig. 6B). Similar results were obtained when comparing GCN2 versus gcn2Δ strains (data not shown). Thus, it appears that phosphorylation of eIF2α by GCN2 is responsible for about 50% of the inhibition of translation initiation by rapamycin.
Figure 6.
(A) Phosphorylation of eIF2α on Ser 51 contributes to the inhibition of cell growth by rapamycin. Strains CY1100 (SUI2) and CY1101 (SUI2-S51A) were grown to saturation in liquid SC medium and serial dilutions were spotted to SC or SC plates containing 100 ng/mL rapamycin and incubated at 30°C for 2 and 4 d, respectively. (B) Phosphorylation of eIF2α on Ser 51 contributes to the inhibition of translation initiation by rapamycin. Strains CY1100 (SUI2) and CY1101 (SUI2-S51A) were grown in SC medium to early logarithmic phase and either treated with 200 ng/mL rapamycin for 2 h [(+) Rap.] or left untreated [(−) Rap.]. Fifty micrograms per milliliter cycloheximide was added 5 min before harvesting. WCEs were prepared and resolved on 5%–45% sucrose gradients. (P/M) Polysome-to-monosome ratio of the A254 in all polysome fractions to that in the 80S peak.
Discussion
Evidence that rapamycin-induced eIF2α phosphorylation and induction of GCN4 translation by GCN2 involves dephosphorylation of Ser 577
GCN2 is activated in response to amino acid starvation and the resulting phosphorylation of eIF2α leads to a reduction in general protein synthesis and specific stimulation of GCN4 translation. It was known that phosphorylation of eIF2α by GCN2 is activated by purine or glucose starvation in addition to amino acid limitation (Rolfes and Hinnebusch 1993; Yang et al. 2000). We found that treatment with tunicamycin, HU, MMS, and rapamycin also induced eIF2α phosphorylation. Among the latter conditions, the involvement of GCN2 in the GAAC response was established only for MMS treatment (Natarajan et al. 2001). We showed that GCN4–lacZ expression is induced by rapamycin at the translational level dependent on both the uORFs and eIF2α phosphorylation by GCN2, and that stimulation of eIF2α phosphorylation by rapamycin requires the tRNA binding activity of GCN2. Hence, the contribution of GCN4 to transcriptional activation of a subset of genes induced by rapamycin (Valenzuela et al. 2001) involves GCN2-dependent derepression of GCN4 translation. The fact that rapamycin did not increase eIF2α phosphorylation in cells expressing the heterologous eIF2α kinase HRI in place of GCN2 suggests that rapamycin elevates eIF2α-P levels primarily through GCN2 activation rather than inhibition of an eIF2α-P phosphatase.
Among the numerous stress or starvation conditions that elicit increased eIF2α phosphorylation, only treatment with rapamycin was associated with dephosphorylation of Ser 577 (Fig. 1). Both the increased eIF2α phosphorylation and derepression of GCN4–lacZ expression produced by rapamycin were diminished by the S577A mutation in GCN2, consistent with the idea that rapamycin activates GCN2 by a mechanism involving Ser 577 dephosphorylation. The substitution of Ser 577 with alanine elevates GCN2 kinase function in nonstarved cells and increases the tRNA binding activity of the purified protein (Garcia-Barrio et al. 2002). Here we showed that activation of GCN2 by rapamycin is dependent on its tRNA binding activity. Accordingly, we propose that GCN2 is activated by rapamycin in amino acid-replete cells at least partly because dephosphorylation of Ser 577 allows GCN2 to bind uncharged tRNAs at basal concentrations. For other stress and starvation conditions that increase eIF2α-P levels in the presence of abundant amino acids, it remains to be seen whether other mechanisms exist to increase the affinity of GCN2 for uncharged tRNAs without Ser 577 dephosphorylation.
Sequence alignments of yeast GCN2 with its Drosophila melanogaster, mouse, and human homologs show significant variability in the region containing Ser 577 (Olsen et al. 1998; Berlanga et al. 1999). Thus, the mechanism we described here may be restricted to S. cerevisiae. It is possible, however, that a similar mechanism for activating GCN2 exists in higher eukaryotes and involves a different phosphorylation site(s).
Evidence that rapamycin-induced dephosphorylation of Ser 577 is mediated by the TOR proteins and TAP42
Using dominant TOR alleles that confer resistance to rapamycin, we showed that the increased eIF2α phosphorylation induced by this drug requires inhibition of the TOR proteins. The TOR pathway controls cell growth and metabolism in response to nutrients, in large part through TAP42-mediated regulation of the type PP2A-related phosphatase SIT4, and possibly also PP2A. TAP42 is thought to be an inhibitor of SIT4 because rapamycin leads to dissociation of the TAP42/SIT4 complex and results in SIT4-dependent dephosphorylation of GLN3 and NPR1. Moreover, the latter responses are blocked by the tap42-11 allele that confers rapamycin resistance (Di Como and Arndt 1996; Schmidt et al. 1998; Beck and Hall 1999; Jacinto et al. 2001). Based on this model, an attractive hypothesis to explain our findings would be that SIT4 (or PP2A), freed from association with TAP42, is responsible for the dephosphorylation of GCN2 residue Ser 577 in rapamycin-treated cells.
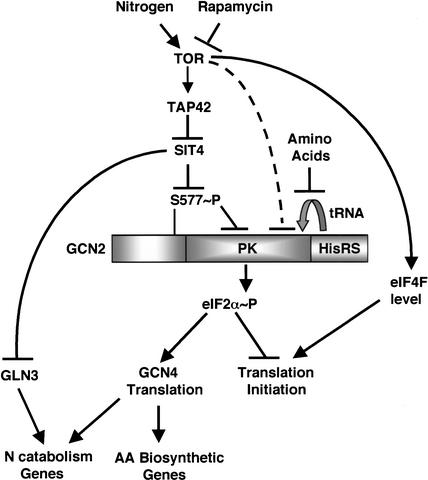
It was proposed that tap42-11 remains associated with SIT4 and PP2A at 25°C in the presence of rapamycin, maintaining the inhibition of these phosphatases and thereby conferring resistance to rapamycin (Di Como and Arndt 1996). We observed a higher level of S577 phosphorylation in tap42-11 versus TAP42 cells treated with rapamycin at 25°C. This finding is consistent with the idea that dephosphorylation of S577 produced by rapamycin requires dissociation of TAP42 from SIT4 or PP2A. In contrast, Ser 577-P levels were reduced in the tap42-11 strain at semipermissive (30°C) and nonpermissive (37°C) temperatures in the absence of rapamycin. To account for these latter findings, we suggested that tap42-11 dissociates partially or completely from SIT4 at 30°C or 37°C, leading to increased phosphatase activity, even in the absence of rapamycin. Deletion of SIT4 or PPH21/PPH22 led to increased Ser 577-P levels without eliminating the effect of rapamycin in reducing S577 phosphorylation. These last results were explained by proposing that SIT4 and PP2A both contribute to dephosphorylation of S577 in a manner stimulated by rapamycin. Together, our results support the idea that the TOR proteins promote S577 phosphorylation and thereby inhibit GCN2 function by negative regulation of SIT4 and PP2A in a manner controlled at least partly by TAP42 (Fig. 7).
Figure 7.
Model depicting the TAP42/Ser 577-dependent pathway and a partially redundant uncharacterized pathway used by TOR to down-regulate GCN2 activity when nitrogen is plentiful. Phosphorylation of Ser 577 inhibits the GCN2 protein kinase (PK) domain by antagonizing tRNA binding. Treatment with rapamycin inhibits TOR, leading to dephosphorylation of Ser 577 by the TAP42-regulated phosphatases SIT4/PP2A. The ensuing activation of GCN2 and eIF2α phosphorylation down-regulates general translation initiation and induces GCN4 translation. The GCN4 thus produced activates amino acid biosynthetic genes and augments the GLN3-dependent activation of genes required for catabolism of poor nitrogen sources. Because rapamycin can partially induce eIF2α phosphorylation in tap42-11 cells at 25°C even though Ser 577 remains phosphorylated, we propose that a redundant pathway exists for TOR-dependent inactivation of GCN2 that is also inhibited by rapamycin. Starvation for amino acids can lead to activation of GCN2 through binding of uncharged tRNA to the HisRS domain even under conditions of TOR-dependent Ser 577 phosphorylation.
Evidence that TOR and SIT4 regulate eIF2α phosphorylation by multiple mechanisms
The induction of eIF2α-P after 2 h of rapamycin treatment was diminished in tap42-11 cells in parallel with the higher levels of Ser 577-P observed in this mutant (Fig. 4). This inverse correlation strongly supports the idea that activation of GCN2 by rapamycin involves dephosphorylation of Ser 577. However, the eIF2α-P level was higher than expected in tap42-11 cells after only 30 min of treatment, especially considering that Ser 577-P levels differed the most between tap42-11 and wild-type cells at this time point (Fig. 4C,D). This quantitative discrepancy suggests that an alternative mechanism exists for activation of GCN2 independently of Ser 577 dephosphorylation and is responsible for the induction of eIF2α-P by rapamycin in tap42-11 cells. Further evidence for another pathway comes from the fact that the eIF2α-P level in rapamycin-treated wild-type cells did not decline after 4 h of treatment even though phosphorylation of Ser 577 had recovered to nearly the same level seen in untreated cells (Fig. 4C,D). Even at the 2-h time point, the steady-state level of eIF2α-P in the tap42-11 cells was greater than that seen in untreated tap42-11 cells (Fig. 4D), although this could reflect leakiness of this mutation in eliminating the effects of rapamycin on GCN2 activity. Finally, the induction of eIF2α-P (Fig. 2A) and GCN4–lacZ expression (Table 1) by rapamycin was not completely eliminated by the S577A substitution in GCN2. The results in Figure 2C imply that the alternative pathway(s) for inducing eIF2α-P in response to rapamycin involves GCN2. It will be interesting to determine whether these Ser 577-independent responses are mediated by additional phosphorylation sites in GCN2.
Another way to explain the residual induction of eIF2α-P that occurs in tap42-11 or GCN2-S577A cells would be to propose that rapamycin impairs the synthesis of one or more amino acids and thus activates GCN2 by the conventional mechanism involving increased binding of uncharged tRNAs to GCN2. Furthermore, it was shown that TOR inhibition by rapamycin elicits TAP42-dependent degradation of the tryptophan permease, which could lead to a tryptophan limitation and uncharged tRNATrp (Schmidt et al. 1998). At odds with this last possibility, all strains used in this study are tryptophan prototrophs, and it is known that withdrawal of external amino acids from cells capable of synthesizing all 20 amino acids does not elicit steady-state activation of GCN2 (Tzamarias et al. 1989). Hence, a defect in tryptophan uptake is not a viable explanation for the Ser 577-independent induction of eIF2α-P by rapamycin.
The inactivation of SIT4 or PP2A led to higher levels of Ser 577-P (Fig. 5C), as predicted by the model in Figure 7; however, we did not observe a commensurate decrease in the eIF2α-P levels in these mutants. This discrepancy is not severe for the pph21Δpph22Δ mutant because the increase in Ser 577-P levels was modest in this strain. However, it is striking that the level of eIF2α-P induced by rapamycin was higher in sit4Δ cells than in wild type. To account for this final discrepancy with the model, we suggest that physiological inhibition of SIT4 and PP2A by TAP42 is not equivalent to complete inactivation of these phosphatases. This proposal is inspired by previous genetic data indicating that TAP42 can act as a positive regulator of SIT4 and PP2A. Thus, high-copy TAP42 suppresses the growth defects of sit4 and pph21 mutations, and also exacerbates the toxic effect of overexpressing PPH21 (Di Como and Arndt 1996). If TAP42 directs SIT4 or PP2A toward specific substrates (Jiang and Broach 1999), then dissociation of TAP42 from these enzymes would affect a more restricted set of substrates than does inactivation of the phosphatases. Although Ser 577-P is elevated in sit4Δ cells, the absence of SIT4 may disrupt cellular metabolism in a way that elicits one of the starvation or stress signals that activate GCN2 independently of Ser 577. Indeed, sit4Δ cells have a pronounced slow-growth phenotype.
Cross-talk between two global regulatory mechanisms for nutrient control of gene expression
Our finding that inactivation of TOR by rapamycin leads to activation of GCN2 and eIF2α phosphorylation has important implications for how yeast cells coordinate their responses to nutrient limitation. The TOR pathway is a key regulator of gene expression in response to the quality and availability of the nitrogen supply (Cooper 2002). In cells replete with a good nitrogen source, the TOR pathway stimulates protein synthesis by enhancing ribosome production through increased expression of mRNAs encoding ribosomal proteins (Powers and Walter 1999). The TOR proteins also promote translation initiation by increasing the abundance of eIF4F, an essential factor that stimulates mRNA binding to the 40S ribosomal subunit. The latter occurs by an increase in the stability of eIF4G (Berset et al. 1998), a component of the eIF4F complex that mediates recruitment of the 40S ribosome to mRNAs. The TOR pathway may additionally prevent EAP1 protein from inhibiting eIF4E, the subunit in eIF4F that binds to the 5′ cap structures of mRNA (Cosentino et al. 2000). In mammalian cells, the TOR proteins stimulate eIF4F formation by promoting phosphorylation and inactivation of the eIF4E-binding proteins (4E-BPs) that sequester eIF4E in inactive complexes (Gingras et al. 2001). Our results show that TOR additionally stimulates initiation in yeast at the level of TC formation by reducing the phosphorylation of eIF2α by GCN2. The fact that a SUI2-S51A mutant expressing nonphosphorylatable eIF2α is more resistant than wild type to rapamycin and exhibits ∼50% less inhibition of translation initiation in the presence of the drug (Fig. 6) suggests that eIF2α phosphorylation makes a significant contribution to the inhibition of protein synthesis and cell growth on inactivation of TOR. Presumably, the residual inhibitory effect of rapamycin in SUI2-S51A cells results from the reduction in eIF4F levels (Berset et al. 1998; Cosentino et al. 2000; Fig. 7). It was shown previously that cell cycle arrest produced by rapamycin could be suppressed by a CLN3 allele (encoding a G1 cyclin) bearing the 5′UTR from UBI4, which has a low dependence on eIF4E for translation initiation (Barbet et al. 1996). This result suggests that down-regulating eIF4F by inhibition of TOR with rapamycin decreases the translation of native CLN3 below the threshold necessary for G1-S transition. Perhaps the UBI4 5′UTR also reduces the dependence of CLN3 translation on high concentrations of TC, allowing translation of the UBI4-CLN3 mRNA when eIF2α is phosphorylated in rapamycin-treated cells; alternatively, it may be sufficient to overcome the reduction in eIF4F levels to achieve the critical level of CLN3 synthesis needed to exit G1.
Our results and others (Valenzuela et al. 2001) show that the TOR pathway maintains translational repression of GCN4. GCN4 induces many genes under the control of GLN3 required for utilization of poor nitrogen sources (Natarajan et al. 2001) and also enhances the ability of GLN3 to activate certain target genes (Valenzuela et al. 2001). Thus, GCN4 can be viewed as an auxiliary factor that augments the induction of GLN3 targets on a poor nitrogen source in addition to its central role in regulating amino acid biosynthesis (Fig. 7). Hence, to prevent the expression of GLN3 targets on a good nitrogen source, TOR must repress GCN4 expression in addition to preventing nuclear localization of GLN3 (Beck and Hall 1999).
The TOR-mediated repression of GCN4 translation is overridden when cells are starved for an amino acid in the presence of an abundant nitrogen supply. Because S577 is not dephosphorylated in histidine-starved cells, we presume that TOR remains active under these conditions. This conclusion is in accordance with results indicating that TOR is regulated by glutamine but not several other amino acids, reflecting the importance of glutamine as a general indicator of nitrogen availability (Crespo et al. 2002). Thus, TOR in yeast is not a general sensor of amino acid availability. The activation of GCN2 in cells starved for amino acids other than glutamine is mediated by increased levels of the cognate uncharged tRNAs. This permits binding of tRNA to the HisRS domain in GCN2, with ensuing kinase activation, even though Ser 577 remains phosphorylated because of high TOR activity (Fig. 7).
A regulatory network with similarities to that described in Figure 7 may operate in the response to unfolded proteins. We found that eIF2α phosphorylation was stimulated in yeast treated with tunicamycin (Fig. 1B), an inducer of the unfolded protein response (UPR; Patil and Walter 2001). The consequent reduction in translation initiation expected from eIF2α phosphorylation should alleviate protein load on the endoplasmic reticulum (ER) under conditions of ER stress. The predicted derepression of GCN4 may help to induce certain genes required to handle ER stress in conjunction with HAC1, the transcriptional activator dedicated to the UPR (Patil and Walter 2001). This dual regulatory response seems to operate in ER-stressed mammalian cells, but a different kinase, PERK/PEK, is responsible for the bulk of eIF2α phosphorylation under these conditions (Harding et al. 2002). Because yeast lacks PERK/PEK, the GCN2 in yeast may be regulated by a wider range of stress signals than its mammalian counterpart.
An important question for future work is the identity of the PK(s) responsible for phosphorylation of Ser 577. It is possible that TOR1 and TOR2 phosphorylate Ser 577 in addition to inhibiting TAP42-regulated phosphatases. However, we have obtained evidence that at least one other kinase controls the level of Ser 577-P in vivo, and its activity is not known to be inhibited by rapamycin (V. Cherkasova and A. Hinnebusch, unpubl.). Thus, it is possible that phosphorylation of Ser 577 is constitutive and that inhibition of TOR proteins by rapamycin leads primarily to Ser 577 dephosphorylation. It is also unresolved whether TOR or another kinase is responsible for other phosphorylation events in yeast that are sensitive to rapamycin, including those in GLN3 or NPR1 (Cooper 2002). Similarly, the identity of the kinase(s) responsible for rapamycin-sensitive phosphorylation of mammalian 4E-BPs remains controversial (Gingras et al. 2001). Thus, considerable effort will be required to determine how many kinases contribute to Ser 577 phosphorylation, identify which kinases function directly in this reaction, and determine whether or not their activities are inhibited by rapamycin.
Materials and methods
Strains and plasmids
Strains used in this study are listed in Table 2. The TAP42 and tap42-11 strains in the Research Genetics background (CY1077 and CY1078) were made as follows: TAP42 LEU2 CEN and tap42-11 LEU2 CEN plasmids recovered from CY4927 (TAP42) and CY4928 (tap42-11; Di Como and Arndt 1996) were introduced into Research Genetics strain #20603 (a/α TAP42/tap42Δ∷kanMX4) and the resulting transformants were sporulated, the tetrads dissected, and the spores tested for G418 resistance (G418r) and inability to grow at 37°C (Ts− phenotype). Ts− G418r Leu+ spores and Ts+ G418r Leu+ spores were judged to contain plasmid-borne tap42-11 or TAP42, respectively, in a tap42Δ strain. As expected, all Ts− G418r Leu+ spores were rapamycin-resistant. Furthermore, their Ts− phenotypes were complemented by mating to TAP42 strain CY4927 but not to tap42-11 strain CY4928. The SUI2 and SUI2 S51A strains in the Research Genetics background (CY1100 and CY 1101) were made as follows: the sui2Δ/SUI2 heterozygous diploid strain from Research Genetics (#26804) was transformed with plasmid p1097 (SUI2 LEU2) or p1098 (SUI2 S51A LEU2; Dever et al. 1992). The resulting transformants were sporulated, the tetrads were dissected, and the Leu+ spores were selected from complete tetrads containing only two Leu+ spores. As expected, all such spores (eight tested) that originated from the diploid transformant harboring the SUI2 plasmid were resistant to sulfometuron methyl (SM), an inhibitor of isoleucine-valine biosynthesis, whereas all nine spores from the diploid containing the SUI2-S51A plasmid exhibited SM-sensitivity. Resistance to SM is dependent on GCN2-mediated translational induction of GCN4 (Vazquez de Aldana et al. 1994).
Table 2.
Yeast strains
| Strain
|
Genotype
|
Source or reference
|
|---|---|---|
| F113 | MATa ino1 can1 ura3-52 | G. Fink |
| H2511 | MATa ino1 can-r ura3-52 gcn2Δ | C. Vazquez |
| H1816 | MATa ura3-52 leu2-3 leu2-112gcn2Δsui2Δtrp1Δ63 p1108 (GCN4-lacZ, TRP1) p1097 (SUI2, LEU2) | Dever et al. 1992 |
| H1817 | MATa ura3-52 leu2-3 leu2-112gcn2Δsui2Δ trp1Δ63 p1108 (GCN4-lacZ, TRP1) p1098 (SUI2-S51A, LEU2) | Dever et al. 1992 |
| MLY41 | ∑1278b MATa ura3-52 | Lorenz and Heitman 1997 |
| MLY90-1 | ∑1278b MATα ura3-52 TOR1-4 | Cardenas et al. 1999 |
| MLY152 | ∑1278b MATα ura3-52 TOR2-1 | Cardenas et al. 1999 |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Research Genetics |
| #20603 | BY4741/BY4742 TAP42/tap42Δ::KANMX4 | Research Genetics |
| #26804 | BY4741/BY4742 SUI2/sui2Δ::KANMX4 | Research Genetics |
| #249a | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gcn4::KAN | Research Genetics |
| #3744a | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sit4::KAN | Research Genetics |
| ESY6053a | MATa leu2Δ0 met15Δ0 ura3Δ0 gcn2Δ | E. Sattlegger |
| CY1077a | MATa leu2Δ0 lys2Δ0 ura3Δ0 tap42::KAN {TAP42 LEU2 CEN} | This study |
| CY1078a | MATα leu2Δ0 met15Δ0 ura3Δ0 tap42::KAN {tap42-11 LEU2 CEN} | This study |
| CY1090a | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 lys220 pph21::KAN pph22::KAN | This study |
| CY 1100a | MATa leu2Δ0 lys2Δ0 ura3Δ0 sui2::KAN {SUI2 LEU2 CEN} | This study |
| CY1101a | MATa leu2Δ0 lys2Δ0 ura3Δ0 sui2::KAN {SUI2-S51A LEU2 CEN} | This study |
Isogenic to Research Genetics strain BY4741.
Plasmids pB107 and pB111 and pDH101 were described previously (Garcia-Barrio et al. 2002), as were p180 and p227 (Hinnebusch 1985; Mueller and Hinnebusch 1986). Plasmids containing FL-GCN2-S577A (pCB149), FL-GCN2-m2 (pCB150), and FL-GCN2-S577A-m2 (pCB151) were constructed in two steps. First, pDH101 (FL-GCN2) was digested with SnaBI and XbaI, the ends were filled in and religated to eliminate the MunI site upstream of the SnaBI site in the 5′-noncoding region, leaving intact the MunI site in the coding region of GCN2 (pCB148). pCB149 was made by inserting the MunI–PvuII 3.9-kb fragment from pB107 into pCB148 digested with MunI and PvuII. Plasmids pCB150 and pCB151 were constructed by inserting the KpnI–NheI 1.8-kb fragment from p2201 (Wek et al. 1995; m2 mutant) into pCB148. pJK5, containing the pGAL-TOR2 allele on a CEN URA3 plasmid (Kunz et al. 1993), was kindly provided by Dr. Michael Hall.
Biochemical methods
WCEs were prepared from yeast cell cultures grown to mid log-phase in either YPD or selective SC medium (Sherman et al. 1974) lacking uracil as previously described (Garcia-Barrio et al. 2000), except the breaking buffer was 100 mM sodium phosphate (pH 7.4), 150 mM NaCl, 0.1% Triton X-100, 100 mM NaF, EDTA-free protease inhibitor cocktail (Roche), 1 μg/mL leupeptin, aprotonin, and pepstatin. For analysis of eIF2α phosphorylation, aliquots of WCE were resolved by SDS-PAGE and subjected to immunoblot analysis using antibodies specific for phosphorylated Ser 51 in eIF2α (BIOSOURCE International). The blots were stripped and reprobed with polyclonal antibodies against eIF2α, as described previously (Garcia-Barrio et al. 2002). For analysis of Ser 577 phosphorylation, aliquots of 2–0.25 mg of WCE were incubated with 10–30 μL anti-Flag protein A agarose beads (Sigma) at 4°C overnight. The immunoprecipitates were washed four times with 1 mL of the breaking buffer, resolved by 4%–12% SDS-PAGE and subjected to immunoblot analysis using Ser 577-P antibodies. The blots were stripped and probed with polyclonal GCN2 antibodies (serum HL2523; Romano et al. 1998). The immune complexes were visualized with anti-rabbit antibodies conjugated to horseradish peroxidase (Amersham) and the enhanced chemiluminescence system (Amersham). WCEs were prepared and assayed for β-galactosidase activity as described previously (Moehle and Hinnebusch 1991). Preparation of WCEs and subsequent polysome analysis were performed as described previously (Valášek et al. 2001).
Acknowledgments
We thank Minerva Garcia-Barrio, Hongfang Qui, and Jinsheng Dong for helpful advice on techniques; Tom Dever, Evelyn Sattlegger, and Leos Valasek for critical reading of the manuscript; members of the Hinnebusch and Dever laboratories for advice; and Maria Cardenas, Joe Heitman, Michael Hall, Jinsheng Dong, and Evelyn Sattlegger for gifts of strains and plasmids.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ahinnebusch@nih.gov; FAX (301) 496-6828.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1069003.
References
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- Berset C, Trachsel H, Altmann M. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci. 1998;95:4264–4269. doi: 10.1073/pnas.95.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes & Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: Connecting the dots. FEMS Microbiol Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino GP, Schmelzle T, Haghighat A, Helliwell SB, Hall MN, Sonenberg N. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:4604–4613. doi: 10.1128/mcb.20.13.4604-4613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TD, Hinnebusch AG. Phosphorylation of initiation factor 2a by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes & Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- Garcia-Barrio M, Dong J, Ufano S, Hinnebusch AG. Association of GCN1/GCN20 regulatory complex with the conserved N-terminal domain of eIF2a kinase GCN2 is required for GCN2 activation in vivo. EMBO J. 2000;19:1887–1899. doi: 10.1093/emboj/19.8.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barrio M, Dong J, Cherkasova VA, Zhang X, Zhang F, Ufano S, Lai R, Qin J, Hinnebusch AG. Serine-577 is phosphorylated and inhibits the tRNA binding and eIF2 alpha kinase activities of GCN2. J Biol Chem. 2002;227:30675–30683. doi: 10.1074/jbc.M203187200. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- Goossens A, Dever TE, Pascual-Ahuir A, Serrano R. The protein kinase Gcn2p mediates sodium toxicity in yeast. J Biol Chem. 2001;276:30753–30760. doi: 10.1074/jbc.M102960200. [DOI] [PubMed] [Google Scholar]
- Harding H, Calfon M, Urano F, Novoa I, Ron D. Transcription and translational control in the mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. A hierarchy of trans-acting factors modulate translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— . Translational control of GCN4: Gene-specific regulation by phosphorylation of eIF2. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 199–244. [Google Scholar]
- ————— . Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 185–243. [Google Scholar]
- Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot Cell. 2002;1:22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol Cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Broach JR. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton MJ, Crouch D, Hinnebusch AG. GCN1, a translational activator of GCN4 in S. cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol Cell Biol. 1993;13:3541–3556. doi: 10.1128/mcb.13.6.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle CM, Hinnebusch AG. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller PP, Hinnebusch AG. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45:201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4343–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen DS, Jordan B, Chen D, Wek RC, Cavener DR. Isolation of the gene encoding the Drosophila melanogaster homolog of the Saccharomyces cerevisiae GCN2 eIF-2a kinase. Genetics. 1998;149:1495–1509. doi: 10.1093/genetics/149.3.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: The unfolded protein response in yeast and mammals. Cell Biol. 2001;13:349–356. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Dong J, Hinnebusch AG. Mutations that bypass tRNA binding activate the intrinsically defective kinase domain in GCN2. Genes & Dev. 2002;16:1271–1280. doi: 10.1101/gad.979402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfes RJ, Hinnebusch AG. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: Implications for activation of the protein kinase GCN2. Mol Cell Biol. 1993;13:5099–5111. doi: 10.1128/mcb.13.8.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano PR, Garcia-Barrio MT, Zhang X, Wang Q, Taylor DR, Zhang F, Herring C, Mathews MB, Qin J, Hinnebusch AG. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2a kinases PKR and GCN2. Mol Cell Biol. 1998;18:2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence CW. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1974. pp. 61–64. [Google Scholar]
- Tzamarias D, Roussou I, Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989;57:947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Valášek L, Phan L, Schoenfeld LW, Valásková V, Hinnebusch AG. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001;20:891–904. doi: 10.1093/emboj/20.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L, Aranda C, Gonzalez A. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J Bacteriol. 2001;183:2331–2334. doi: 10.1128/JB.183.7.2331-2334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez de Aldana CR, Wek RC, San Segundo P, Truesdell AG, Hinnebusch AG. Multicopy tRNA genes functionally suppress mutations in yeast eIF-2a kinase GCN2: Evidence for separate pathways coupling GCN4 expression to uncharged tRNA. Mol Cell Biol. 1994;14:7920–7932. doi: 10.1128/mcb.14.12.7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Cannon JF, Dever TE, Hinnebusch AG. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2a kinase GCN2. Mol Cell Biol. 1992;12:5700–5710. doi: 10.1128/mcb.12.12.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2a protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Wek SA, Wek RC. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol Cell Biol. 2000;20:2706–2717. doi: 10.1128/mcb.20.8.2706-2717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Sobolev AY, Wek RC. Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J Biol Chem. 1996;271:24989–24994. doi: 10.1074/jbc.271.40.24989. [DOI] [PubMed] [Google Scholar]