Abstract
Runx proteins have been implicated in acute myeloid leukemia, cleidocranial dysplasia, and stomach cancer. These proteins control key developmental processes in which they function as both transcriptional activators and repressors. How these opposing regulatory modes can be accomplished in the in vivo context of a cell has not been clear. In this study we use the developing cone cell in the Drosophila visual system to elucidate the mechanism of positive and negative regulation by the Runx protein Lozenge (Lz). We describe a regulatory circuit in which Lz causes transcriptional activation of the homeodomain protein Cut, which can then stabilize a Lz repressor complex in the same cell. Whether a gene is activated or repressed is determined by whether the Lz activator or the repressor complex binds to its upstream sequence. This study provides a mechanistic basis for the dual function of Runx proteins that is likely to be conserved in mammalian systems.
Keywords: Runx, Cut, Groucho, transcriptional regulation
Runx proteins are a family of conserved transcription factors that regulate hematopoeisis, osteogenesis, epithelial development, and neurogenesis in mammals (Cohen 2001), and embryonic pattern formation, sex determination, visual system development, and hematopoiesis in Drosophila (Canon and Banerjee 2000; Wheeler et al. 2000). These proteins can function as either activators or repressors, but the mechanistic detail of how such a switch may occur in vivo has not been clear. In problems of cell fate determination, the ability of a transcription factor to perform diverse regulatory roles leading to the specification of a multitude of unique cell fates is crucial. Loss of proper Runx protein function can result in leukemias, cleidocranial dysplasia, defective neuronal connectivity, and stomach cancer (Castilla et al. 1996; Mundlos et al. 1997; Inoue et al. 2002; Levanon et al. 2002; Li et al. 2002). It is therefore particularly important to understand how Runx proteins function in transcriptionally active and repressive roles.
The Drosophila Runx protein Lozenge (Lz) directly binds to DNA to activate transcription of target genes (Flores et al. 2000; Xu et al. 2000). For example, Lz and downstream effectors of the EGFR and Notch signaling pathways converge on the D-Pax2 enhancer to activate its expression in the nonneuronal cone cells in the developing eye (Flores et al. 2000). Consequently, D-Pax2 expression is lost in lz mutant eyes. In contrast, genetic evidence has shown that other transcription factors, such as Seven-up, are up-regulated in these same cells in lz mutants (Daga et al. 1996; Crew et al. 1997). The mode of this negative regulation by Lz was unknown, however, including whether Lz acts as a direct transcriptional repressor. In this study, we use the developing cone cell as an in vivo model system to understand how a Runx protein can accomplish both direct activation and repression in the same cell.
The developing Drosophila visual system is an excellent model system in which to study the problems of transcriptional regulation during cell fate specification. The eye contains ∼800 ommatidia, each of which contains an identical arrangement of cells, including eight neuronal photoreceptors (R cells), four nonneuronal cone cells which secrete lens, and several pigment and bristle cells. These cells all express distinct transcription factors that are required for their individual development (Kumar and Moses 1997), yet they all arise from a pool of undifferentiated, equipotent precursors. It is in this precursor population where the turning on and off of genes is crucial for establishing the expression pattern of cell-specific factors and creating diversity out of an equivalent group of cells. The Runx protein Lz has a pivotal role in this process (Flores et al. 1998).
Results and Discussion
Lozenge directly represses deadpan in cone cells
To understand negative regulation by the Lz protein, we investigated regulation of the deadpan (dpn) gene (Bier et al. 1992). In wild-type eyes, we found Dpn to be expressed in photoreceptors R3/R4 and R7 (Fig. 1A). In lz mutants, dpn is also ectopically activated in cone cells (Fig. 1B), suggesting that Lz either directly or indirectly represses dpn in these cells. We therefore used Dpn as a marker to investigate negative regulation by Lz.
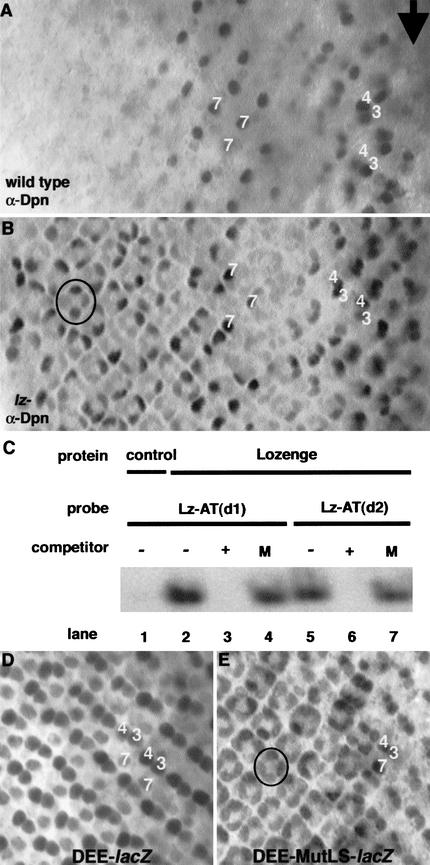
Figure 1.
Lz directly represses dpn transcription in cone cells. (A) In wild type, Dpn is expressed in R3/R4 cells for four to five columns posterior to the furrow (arrow), then in R7 cells for five to six columns. (B) In lz mutants, Dpn is also ectopically activated in cone cells (one cluster circled). (C) Lz binds to the two Runx consensus binding sites in the dpn enhancer. Nuclear extracts were made from S2 cells transfected with vector alone (control) or a Lz-expressing vector. For the sequences of all probes used, see Table 1. Lz binds both sites (lanes 2,5), and this binding is specific as it is competed away with cold competitor oligo (+; lanes 3,6), but not by oligo containing a mutation (M) in the Runx-binding site (lanes 4,7). The portion of the gel shown represents the only shifted bands above the free probe. (D) The dpn eye enhancer (DEE) drives expression of a lacZ reporter in R3/R4 and R7, like the wild-type Dpn expression pattern, except that the expression of β-Gal perdures to the back of the eye disc. (E) When the two Lz-binding sites (LBS) are mutated in the DEE, expression is also seen in cone cells (one cluster circled), indicating that Lz directly represses the DEE in those cells.
The presence of two perfect consensus Runx protein-binding sites (5′-RACCRCA-3′) upstream of the dpn-coding region suggested possible direct negative regulation by Lz. Gel-shift experiments showed that Lz specifically binds to both sites (Fig. 1C). To determine whether these sequences are required for proper dpn regulation, we made lacZ reporter constructs driven by dpn upstream and intronic fragments, and transformed these into flies. A 4667-bp upstream fragment plus intron I (227 bp) caused expression of lacZ in R3/R4 and R7 (Fig. 1D) faithfully recapitulating the pattern of wild-type dpn expression in the eye. We therefore refer to this as the dpn eye enhancer (DEE). When the two Lz-binding sites (LBS) in the DEE were mutated (to 5′-RAAARCA-3′; DEE–MutLBS), lacZ expression was also seen in cone cells (Fig. 1E). Therefore, lack of Lz binding to this enhancer will cause its derepression in cone cells, establishing that Lz directly represses transcription of dpn in cone cells.
Lz-mediated repression requires the corepressor Groucho
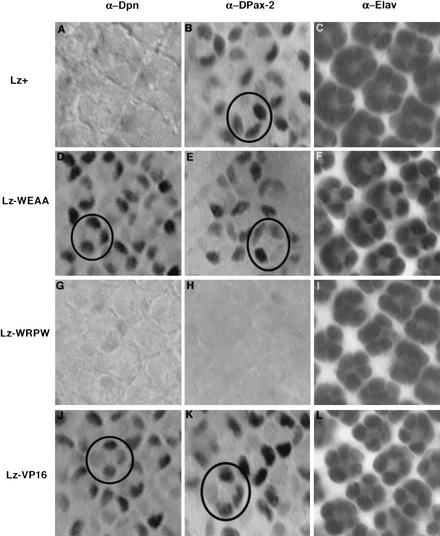
Like all Runx proteins, Lz contains the conserved C-terminal pentapeptide motif VWRPY, which binds the global corepressor Groucho (Gro; Aronson et al. 1997; Levanon et al. 1998). Gro does not bind DNA on its own, but functions as a repressor for sequence-specific DNA-binding factors (Fisher and Caudy 1998). Gro is expressed ubiquitously and has early pleiotropic roles in eye development, such as mediating repression by bHLH proteins (Paroush et al. 1994; Chanut et al. 2000), making it difficult to study possible involvement of Gro in cone cell development in loss-of-function mutant clones in the eye. We therefore altered the Gro-interaction domain at the C terminus of Lz from VWRPY to VWEAA, a change that abrogates Gro binding to bHLH proteins (Goldstein et al. 1999). Lz–EAA protein was then expressed under the control of the endogenous eye-specific lz enhancer (Flores et al. 1998) and tested for its ability to repress dpn in vivo. Whereas a wild-type lz+ transgene efficiently repressed dpn in cone cells (Fig. 2A), Lz–EAA was unable to keep dpn off in these same cells (Fig. 2D). Neuronal differentiation occurred normally in both cases as determined by the neural marker Elav (Fig. 2C,F). This showed that the C terminus of Lz, a known Gro-interaction domain (Aronson et al. 1997; Levanon et al. 1998), is required for Lz-mediated repression of dpn. The activation function of Lz-EAA, as determined by its ability to activate D-Pax2 expression, remained intact (Fig. 2E). Therefore, Gro mediates repression by Lz as it does for other Runx proteins. It still remained unclear, however, why in the same cell Lz represses dpn transcription while it directly activates D-Pax2. Clearly, the presence of Gro alone does not cause Lz to become a dedicated repressor in the cone cell.
Figure 2.
The C terminus of Lz, a Gro-interaction domain, influences the mode of Lz regulation. lz transgenes were tested for their ability to regulate Lz targets dpn and D-Pax2. Transgenes and antibodies are as indicated. Stainings are of eye discs expressing one copy of the transgene in a lz77a7/Y mutant background. Circles mark one cluster of cone cells. Wild-type Lz+ represses dpn (A), but activates D-Pax2 (B) in cone cells. Lz–WEAA is unable to repress dpn in cone cells (D), but can activate D-Pax2 expression (E). Lz–WRPW represses dpn in cone cells (G), but is unable to activate D-Pax2 (H). Therefore, Lz–WRPW functions as a dedicated repressor. Lz–VP16 cannot repress dpn in cone cells (J), but activates D-Pax2 (K), and therefore functions as a constitutive activator. Expression of the neuronal differentiation marker Elav is unchanged in all the above genetic backgrounds (C,F,I,L).
Hairy-related proteins constitutively bind Gro through the conserved sequence WRPW, and function as dedicated repressors (Paroush et al. 1994; Jimenez et al. 1997). To further address the significance of the C terminus of Lz, we changed the C-terminal amino acids of Lz from WRPY to WRPW to resemble Hairy-related repressors. As a correlate, we also made a Lz–VP16 fusion, with the potent activation domain of VP16 fused onto the C terminus of Lz. We then tested the ability of Lz–WRPW and Lz–VP16 to regulate Lz targets in vivo. Lz–WRPW efficiently repressed dpn in cone cells (Fig. 2G) like the wild-type Lz+ (Fig. 2A) but was unable to activate expression of D-Pax2 (Fig. 2H). In contrast, Lz–VP16 failed to repress dpn in cone cells (Fig. 2J) but effectively activated D-Pax2 in cone cells (Fig. 2K). Therefore, Lz–WRPW functions as a dedicated repressor, and Lz–VP16 as a constitutive activator. These results suggested that Runx–Gro interactions are regulated, as wild-type Runx proteins function as both activators and repressors.
Sequences adjacent to Lz sites influence mode of Lz regulation
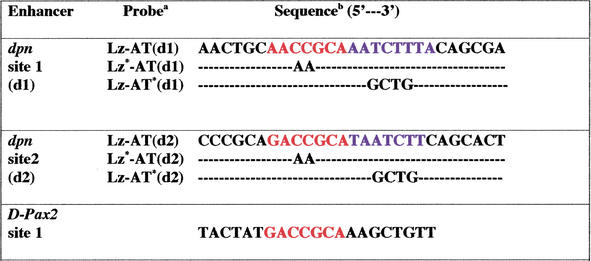
We compared the Lz-binding sites in the dpn and D-Pax2 enhancers and found distinct differences in the neighboring sequences. In the dpn enhancer, each Lz-binding site is followed by AT-rich sequences that are similar to each other (5′-AATCTTT-3′ and 5′-TAATCTT-3′; Table 1). In contrast, sequences near the three Lz-binding sites in the D-Pax2 enhancer, a positively regulated enhancer, are dissimilar and are not as rich in AT sequences (Table 1; Flores et al. 2000). To determine if the difference in these sequences influences the mode of Lz regulation, we replaced both AT-rich sequences in the DEE with the corresponding sequence (GCTG; Table 1) from the D-Pax2 enhancer. When transformed into flies, the resulting DEE–MutAT enhancer could not support repression of the reporter gene in cone cells (Fig. 3A). This was the same phenotype that was seen when the Lz-binding sites were mutated in the DEE (Fig. 1E). In this case, however, alteration of the AT-rich sites had no effect on Lz binding (Fig. 3B). Therefore, disruption of the AT-rich sequences in the DEE prevents repression of this enhancer by a mechanism that is independent of Lz binding. We conclude that a cofactor binds to the AT-rich regions next to the Lz-binding sites and is essential for mediating repression of dpn in cone cells.
Table 1.
Upstream sequences containing Lz binding sites and adjacent regions from dpn and D-Pax2 enhancers, and probes used for DNA binding assays
The top probe for each site is the wild-type sequence. Asterisks (*) indicate mutations in either the Lz site, or the AT-rich stretch.
Runx consensus binding sites are in red, and AT-rich domains are in blue. Sequences for probes that contain mutations to sites show only those nucleotides that have been changed with respect to the wild-type probe.
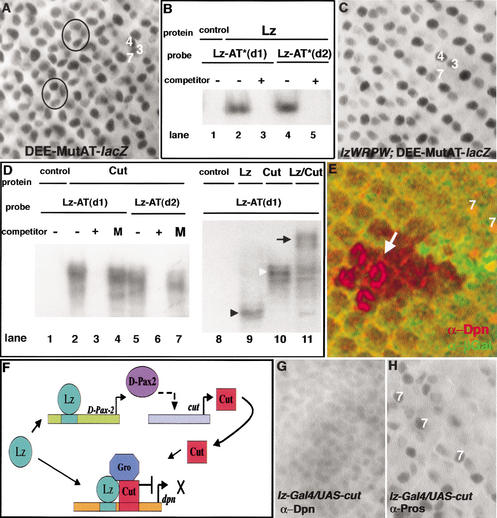
Figure 3.
Cut binding to the AT-rich sequences adjacent to the Lz-binding sites is required for dpn repression. (A) Mutation of both AT-rich sequences in the DEE causes derepression of this enhancer, as lacZ reporter gene expression is now also seen in cone cells (two clusters circled; cf. with Fig. 1D). (B) Lz binding is not affected by mutations in the AT-rich sequences. Nuclear extracts were made from S2 cells transfected with vector alone (control) or with a Lz-expressing vector. For probe sequences, see Table 1. Lz binds to both of its binding sites (lanes 2,4) even when the adjacent AT-sequences are altered. This binding is specific as it is competed away with cold competitor (lanes 3,5). (C) Lz–WRPW is able to repress the DEE in cone cells despite the mutated AT-rich sites. The lacZ reporter gene is expressed in R3/R4 and R7, but not in the cone cells. (D) Cut binds to the AT-rich sequences in the dpn enhancer. Nuclear extracts were made from cells expressing proteins as indicated. Probes are defined in Table 1. Left and right panels represent different gels. (Left panel) Cut binds to the two AT-rich sequences in the DEE (lanes 2,5), and this binding is specific as it is competed away with cold oligo containing known Cut-binding sites (lanes 3,6), but not by cold oligo that contains a mutation (M) in the AT-rich sites (lanes 4,7). (Right panel) A probe containing a Lz-binding site and adjacent AT sequences from the dpn enhancer [Lz–AT(d1); Table 1] can bind Lz (black arrowhead, lane 9) and Cut (white arrowhead, lane 10). Extracts from cells transfected with both lz and cut cause a supershifted band (arrow, lane 11) indicating that Lz and Cut can together bind the same probe. (E) Dpn is ectopically expressed in cone cells in clones of cut mutant cells in the eye. Arrow indicates cone cells within the mutant clone (nongreen) expressing Dpn (red). The counter marker β-Gal (green) marks wild-type tissue. (F) Model for a developmental circuit for positive and negative regulation by Lz in cone cells. Lz directly activates D-Pax2, which is required for activation of cut. Cut protein then binds and forms a repressive complex with Lz and Gro that represses dpn in cone cells. (G) Misexpression of Cut in R7 cells using the lz–Gal4 driver causes repression of dpn in those cells. (H) The R7-cell-specific marker Prospero is unchanged in this genetic background.
We then investigated if it was possible for the dpn enhancer to be repressed independently of the AT-rich sequences. Earlier, we showed that Lz–WRPW functions as a dedicated repressor in cone cells (Fig. 2G,H). We therefore tested the ability of Lz–WRPW to regulate DEE–MutAT. Significantly, although wild-type Lz failed to repress DEE–MutAT (Fig. 3A), Lz–WRPW was effective in repressing this enhancer in cone cells (Fig. 3C). Therefore, Lz–WRPW is able to repress transcription of the DEE without a requirement for the nearby AT-rich sites.
Cut binding to AT-rich sequences is required for Lz-mediated repression
The homeodomain protein Cut is expressed specifically in the four cone cells in the eye (Blochlinger et al. 1988) and has been shown previously to bind AT-rich sequences (Valentine et al. 1998). We tested the ability of Cut to bind the AT-rich sequences next to the Lz sites in the DEE. Electromobility-shift assays were conducted using probes containing the Lz-binding sites and adjacent AT sequences from the dpn enhancer (Table 1). Nuclear extracts of cells transfected with a Cut-expressing vector bound the two AT-rich sequences, and this binding was specific as established by competition assays (Fig. 3D, left panel). Further, extracts from cells transfected with both lz and cut caused a supershifted band (Fig. 3D, right panel), indicating that Lz and Cut can bind together to the same probe.
To address the in vivo relevance of the above results, we made FLP/FRT-mediated clones in the eye that were mutant for the cut locus. Strikingly, Dpn was ectopically expressed in cone cells in the absence of Cut (Fig. 3E). This provides genetic proof that, in vivo, Cut represses dpn expression in cone cells. Cut is therefore required along with Lz for repression of dpn in these cells.
Interestingly, D-Pax2, which is directly activated by Lz, is needed to activate cut in cone cells (Fu and Noll 1997). Therefore, although indirectly, Lz positively regulates cut. This presents an interesting developmental circuit in which Lz, acting as a transcriptional activator, causes expression of a cofactor that then binds with Lz to convert it into a direct repressor of transcription (Fig. 3F). Both the presence of the cofactor and binding sites for this cofactor in the controlling regions of a Lz target gene are required for Lz-mediated repression.
We then tested this model in R7 cells where both Dpn and Lz are coexpressed. Here, Lz does not repress dpn, presumably because Cut is absent from R7. Consistent with this notion, mis-expression of Cut in R7 cells using lz–Gal4 causes repression of dpn in these cells (Fig. 3G). This is not a secondary result of a change in cell fate as the expression of the R7 cell-specific marker Prospero remains unchanged in this genetic background (Fig. 3H).
These results add another level of complexity to recent studies demonstrating a combinatorial code whereby a relatively small number of signaling pathways and activated transcription factors work together to generate unique cell fates (Flores et al. 2000). In cone cells, the Notch and EGFR pathways are required along with Lz to activate D-Pax2, and therefore cut. In contrast, the combination of these few inputs is not right for activation of cut in the R7 neurons, and therefore dpn is not repressed. The circuit described here demonstrates a higher order of sophistication necessary for a cell to choose between a neuronal and nonneuronal fate using a very limited number of inputs. Using a self-regulated circuit and just two signaling pathways, a single Runx protein is capable of causing opposing effects on different enhancers in the same cell, resulting in a unique fate.
Runx–Gro interactions are regulated
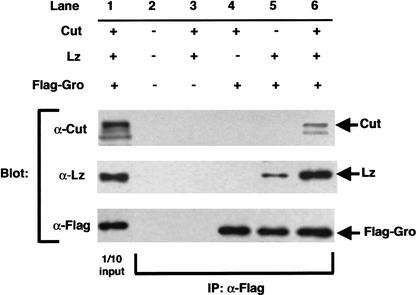
The above observations suggest that Gro binds proteins with a WRPW motif in a stable manner and causes constitutive repression as seen for both Lz–WRPW (Fig. 2G,H) and Hairy-related proteins that contain the WRPW motif (Paroush et al. 1994). In contrast, Gro interaction with the WRPY motif in Runx proteins requires a cofactor, such as Cut, for stabilization. Therefore, repression is regulated as Runx forms a functional repressor complex with Gro only in the presence of the cofactor Cut. We tested this hypothesis in immunoprecipitation (IP) experiments. On its own, Lz weakly interacts with Gro (Fig. 4, lane 5). In the presence of Cut, however, the Lz–Gro interaction is dramatically increased (Fig. 4, lane 6). As expected, Lz–WEAA did not coimmunoprecipitate with Gro, with or without Cut, and Lz–WRPW interacted strongly with Gro, in both the presence and absence of Cut (data not shown). These results are entirely consistent with all of our in vivo observations. First, Lz functions as a repressor only in the cells that express the Cut protein (Fig. 1A,B). Second, Lz–WRPW, which functions as a constitutive repressor, can repress DEE–MutAT (Fig. 3C), in spite of the mutant AT-sites and absence of Cut binding. Finally, wild-type Lz does not repress DEE–MutAT because Cut cannot bind, and therefore the Lz–Gro complex is not stabilized.
Figure 4.
Cut stabilizes an interaction between Lz and Gro. Nuclear extracts were made from S2 cells transfected with lz, cut, and Flag–gro as indicated, and used in immunoprecipitation experiments. Lane 1 shows blots of extracts (1/10 of input), and lanes 2–6 are blots of extracts immunoprecipitated with α-Flag antibody. Extracts in lane 2 are from cells transfected with vector alone. The control in lane 3 shows that α-Flag does not precipitate Lz or Cut in the absence of Gro. Also, Flag–Gro does not interact with Cut (lane 4). A small amount of Lz coimmunoprecipitates with Gro (lane 5), but this interaction is significantly increased in the presence of Cut (lane 6). The same results were obtained in the presence of DNA containing Lz- and Cut-binding sites (data not shown).
Runx proteins have been shown to act as positive and negative regulators. This study, however, is the first to demonstrate that a Runx protein can act as both a direct transcriptional activator and repressor in vivo in the same cell, and that the repressive role requires involvement of the cofactor Cut. The mechanism unraveled here for a Runx protein is similar to that described for a Rel protein (Valentine et al. 1998), suggesting a common strategy adopted by transcription factors that switch between positive and negative regulation. Furthermore, Cut is conserved in mammals (called CDP or Cux; Nepveu 2001) and has been implicated in the repression of several genes, including osteocalcin (OC; van Gurp et al. 1999). Interestingly, the OC gene is positively regulated by Runx2 (Javed et al. 1999; Gutierrez et al. 2002). These in vitro studies did not investigate a relationship between Runx and Cux. Our analysis of dpn repression by Lz and Cut raises the possibility that mammalian Runx proteins may also switch from activation to repression modes through involvement of Cux proteins. If confirmed, such correlations will prove to be important as the mammalian Runx protein AML-1 (Acute Myeloid Leukemia-1) is the most frequent site of translocations that cause leukemia (Okuda et al. 1996; Wang et al. 1996), and human CutL1 is located in a chromosomal region that is often rearranged in cancers, including myeloid leukemia (Ellis et al. 2001).
Materials and methods
Fly stocks and genetics
The following strains were used: Oregon R (wild type), lz77a7 (Flores et al. 1998), lz-Gal4 (gift from J. Pollock), FRT18A cut145/FM7 and UAS-cut (gifts from R. Saint), and hsFLP;FRT18A/arm–lacZ (Bloomington Stock Center). Clones of homozygous cut145mutant cells were generated by expression of heatshock-FLP recombinase. A 1.5-h heatshock (37°C) was given in early second instar.
Immunohistochemistry
The following primary antibodies were used: rabbit α-Dpn (1:400; gift from H. Vaessin), mouse α-βGal (1:100; Promega), rat α-Elav and mouse α-Prospero (1:100; Iowa Hybridoma Bank), and α-D-Pax2 (1:50; gift from M. Noll). Secondary antibodies were HRP-α-mouse, HRP-α-rat, HRP-α-rabbit, FITC-α-mouse, and CY3–α-rabbit (1:100; Jackson Labs). α-Flag(M2)-conjugated beads were obtained from Sigma. Staining of eye discs was performed as described (Flores et al. 1998).
DNA constructs and P-element transformation
dpn upstream and intronic enhancer fragments were PCR-amplified from genomic BAC DNA (BAC No. 9N11) and cloned into PwHZ128, which contains a heatshock promoter and a lacZ gene (gift from A. Courey). lz transgenes were cloned into pCasper-hs, downstream of the lz eye-specific enhancer (Flores et al. 1998). PCR-mediated mutagenesis was performed to mutate the dpn enhancer and lz cDNA. Germ-line transformations were performed as described (Rubin and Spradling 1982).
Biochemistry
Schneider 2 (S2) cells were transiently transfected using FuGene (Roche) according to the manufacturer's protocol. Constructs for transfection were made in the pPac vector (Invitrogen). pPac–cut and pPac–Flag–gro were gifts from A. Courey. Nuclear extracts were obtained using NET lysis buffer. For electromobility-shift assays, extracts were incubated with P32-labeled, double-stranded DNA probes and resolved on a 5% native gel. Cold competitor oligos were used at 40×. In competition assays for Cut binding, the oligo contained a known Cut-binding site: 5′-ATGAACGAATATTGATTGGGTTTCT-3′ (Valentine et al. 1998). Immunoprecipitation experiments were performed using M2-Flag affinity resin (Sigma), and subjected to SDS-PAGE.
Acknowledgments
We thank A. Courey, L. Zipursky, and members of the Banerjee lab for helpful discussions. We acknowledge the work of G. Flores in early aspects of this project that established that the P82 enhancer trap line is caused by an insertion upstream of the dpn locus. We thank N. Ken for help with the S2 cell experiments. We thank M. Caudy and P. Gergen for useful discussions. We thank A. Courey, R. Saint, G. Rubin, H. Vaessin, M. Noll, J. Pollock, K. Matthews, and the Bloomington stock center for fly stocks and reagents. The Elav and Prospero antibodies were obtained from the University of Iowa Hybridoma Bank, developed under the auspices of the National Institute of Child Health and Human Development. This work was supported by National Institutes of Health grant EY08152 to U.B. and a USPHS National Research Service Award (GM07185) to J.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL banerjee@mbi.ucla.edu; FAX (310) 206-9062.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1064803.
References
- Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Younger-Shepherd S, Jan LY, Jan YN. deadpan, an essential pan-neural gene in Drosophila, encodes a helix–loop–helix protein similar to the hairy gene product. Genes & Dev. 1992;6:2137–2151. doi: 10.1101/gad.6.11.2137. [DOI] [PubMed] [Google Scholar]
- Blochlinger K, Bodmer R, Jack J, Jan LY, Jan YN. Primary structure and expression of a product from cut, a locus involved in specifying sensory organ identity in Drosophila. Nature. 1988;333:629–635. doi: 10.1038/333629a0. [DOI] [PubMed] [Google Scholar]
- Canon J, Banerjee U. Runt and Lozenge function in Drosophila development. Semin Cell Dev Biol. 2000;11:327–336. doi: 10.1006/scdb.2000.0185. [DOI] [PubMed] [Google Scholar]
- Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, Marin-Padilla M, Collins FS, Wynshaw-Boris A, Liu PP. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- Chanut F, Luk A, Heberlein U. A screen for dominant modifiers of ro(Dom), a mutation that disrupts morphogenetic furrow progression in Drosophila, identifies groucho and hairless as regulators of atonal expression. Genetics. 2000;156:1203–1217. doi: 10.1093/genetics/156.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MM., Jr RUNX genes, neoplasia, and cleidocranial dysplasia. Am J Med Genet. 2001;104:185–188. [PubMed] [Google Scholar]
- Crew JR, Batterham P, Pollock JA. Developing compound eye in lozenge mutants of Drosophila: Lozenge expression in the R7 equivalence group. Dev Genes Evol. 1997;206:481–493. doi: 10.1007/s004270050079. [DOI] [PubMed] [Google Scholar]
- Daga A, Karlovich CA, Dumstrei K, Banerjee U. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes & Dev. 1996;10:1194–1205. doi: 10.1101/gad.10.10.1194. [DOI] [PubMed] [Google Scholar]
- Ellis T, Gambardella L, Horcher M, Tschanz S, Capol J, Bertram P, Jochum W, Barrandon Y, Busslinger M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes & Dev. 2001;15:2307–2319. doi: 10.1101/gad.200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AL, Caudy M. Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes & Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- Flores GV, Daga A, Kalhor HR, Banerjee U. Lozenge is expressed in pluripotent precursor cells and patterns multiple cell types in the Drosophila eye through the control of cell-specific transcription factors. Development. 1998;125:3681–3687. doi: 10.1242/dev.125.18.3681. [DOI] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes & Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RE, Jimenez G, Cook O, Gur D, Paroush Z. Huckebein repressor activity in Drosophila terminal patterning is mediated by Groucho. Development. 1999;126:3747–3755. doi: 10.1242/dev.126.17.3747. [DOI] [PubMed] [Google Scholar]
- Gutierrez S, Javed A, Tennant DK, van Rees M, Montecino M, Stein GS, Stein JL, Lian JB. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem. 2002;277:1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ozaki S, Shiga T, Ito K, Masuda T, Okado N, Iseda T, Kawaguchi S, Ogawa M, Bae SC, et al. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat Neurosci. 2002;5:946–954. doi: 10.1038/nn925. [DOI] [PubMed] [Google Scholar]
- Javed A, Gutierrez S, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D-responsive transcription and contribute to chromatin organization. Mol Cell Biol. 1999;19:7491–7500. doi: 10.1128/mcb.19.11.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes & Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Moses K. Transcription factors in eye development: A gorgeous mosaic? Genes & Dev. 1997;11:2023–2028. doi: 10.1101/gad.11.16.2023. [DOI] [PubMed] [Google Scholar]
- Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, et al. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Valentine SA, Chen G, Shandala T, Fernandez J, Mische S, Saint R, Courey AJ. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol Cell Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gurp MF, Pratap J, Luong M, Javed A, Hoffmann H, Giordano A, Stein JL, Neufeld EJ, Lian JB, Stein GS, et al. The CCAAT displacement protein/cut homeodomain protein represses osteocalcin gene transcription and forms complexes with the retinoblastoma protein-related protein p107 and cyclin A. Cancer Res. 1999;59:5980–5988. [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JC, Shigesada K, Gergen JP, Ito Y. Mechanisms of transcriptional regulation by Runt domain proteins. Semin Cell Dev Biol. 2000;11:369–375. doi: 10.1006/scdb.2000.0184. [DOI] [PubMed] [Google Scholar]
- Xu C, Kauffmann RC, Zhang J, Kladny S, Carthew RW. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell. 2000;103:87–97. doi: 10.1016/s0092-8674(00)00107-0. [DOI] [PubMed] [Google Scholar]