Abstract
Activation of proteolytic enzymes, including cysteine proteases of the ced-3/ICE family, is a characteristic feature of the apoptotic program. In contrast, the role of the proteasome as the major nonlysosomal machinery to degrade or process proteins by ATP/ubiquitin-dependent proteolysis in this process is less clear. In human leukemic HL60 cells, inhibition of proteasome-mediated proteolysis by specific proteasomal inhibitors leads to the rapid induction of apoptosis as judged by morphological changes as well as by nuclear condensation and DNA fragmentation. HL60 apoptosis is due to activation of CPP32, a member of the ced-3/ICE family of cysteine proteases, and appears to occur independently from ICE activity. HL60 apoptosis is accompanied by an increase in the concentration of the cyclin-dependent kinase inhibitor p27Kip1. Labeling of the cells by the TUNEL technique demonstrates that HL60 cells undergoing apoptosis are primarily in the G1 phase of the cell cycle. Proteasomal activity therefore appears to be required in proliferating, but not in quiescent, HL60 cells for cell survival as well as normal progression through the cell cycle.
Apoptosis has been recognized as a distinct form of cell death that has an essential function in the regulation of cell turnover during development, tissue homeostasis, and cancer (1, 2). For a long time the characteristic cleavage of DNA into oligonucleosomal fragments has been regarded as a hallmark of apoptosis and was the only biochemical marker available. Recently the focus of interest has shifted toward proteolytic events during apoptotic cell death, and it has become apparent that activation of proteolytic enzymes, culminating in the disintegration of the cell, is a characteristic feature of apoptosis. In particular, cysteine proteases of the ced-3/ICE family have been implicated as central components of this proteolytic machinery (3). However, in contrast to the intense research efforts spent on the ced-3/ICE family of proteases, much less attention has been paid so far to the multicatalytic protease complex (MCP) or proteasome, which represents the cell’s major nonlysosomal tool to rapidly degrade or process proteins by ATP/ubiquitin-dependent proteolysis and its potential role in apoptotic cell death. In higher eukaryotic cells the MCP is involved in the degradation of most of the cytosolic proteins and in particular of short-lived proteins critical for cell proliferation and cell cycle regulation. Examples include the tumor suppressor protein p53 (4) and various cyclins (5), as well as the cyclin-dependent kinase inhibitor p27Kip1 (6). The proteasome in addition has a direct impact on transcriptional regulation by processing and degradation of NFκB and IκB respectively, as well as by proteolysis of transcription factors such as c-Fos (7, 8) and c-Jun (9). Finally, studies performed in two developmental systems, regression of the intersegmental muscles in the hawkmoth Manduca sexta and thyroxin-induced apoptosis in the tadpole tail, suggest a link between proteasome function and programmed cell death (10–12). In three recent in vitro studies the question of a potential involvement of proteasomes in apoptotic cell death was addressed, with a rather controversial outcome (13–15). On the basis of the known properties of the proteasome in combination with the fact that activation of a proteolytic cascade or of a proteolytic network occurs during apoptosis, it was therefore intriguing to study the potential involvement of the proteasome in the regulation of programmed cell death in more detail. Here it is reported that proteasomal inhibitors are capable of inducing apoptosis in proliferating HL60 cells, but not in quiescent, differentiated cells. Thus it appears that proteasome-mediated proteolysis is essential for cell survival and cell cycle progression of actively dividing cells and that the two events may be tightly coupled to each other by the proteasome.
MATERIALS AND METHODS
Materials.
N-Carbobenzoxy-l-leucyl-l-leucyl-l-norvalinal (LLnV), N-carbobenzoxy-l-isoleucyl-l-γ-t-butyl-l-glutamyl-l-alanyl-l-leucinal (PSI), and N-acetyl-l-aspartyl-l-glutamyl-l-valyl-l-aspartal (Ac-DEVD-cho) were obtained from the Peptide Institute, Osaka, Japan; the calpain inhibitors N-acetylleucylleucylnorleucinal (LLnL) and N-acetylleucylleucylnormethioninal (LLnM) were purchased from Calbiochem; N-acetyl-l-tyrosyl-l-valyl-l-alanyl-l-aspartyl chloromethyl ketone (Ac-YVAD-cmk) was a gift from R. Heumann (University of Bochum), and all other protease inhibitors were from Sigma. Compounds were dissolved in the amount of dimethyl sulfoxide (DMSO) required to etablish stock solutions of 50 mM. No more than 0.1% solvent was present in the assays unless otherwise stated. Antibodies used in Western blot analysis were obtained from Transduction Laboratories (p27Kip1; CPP32), Santa Cruz Biotechnology (ICE p10), Enzyme System Products [poly(ADP-ribose) polymerase (PARP)], and ICN (β-actin). The monoclonal anti-c-Myc antibody (clone 9E10) was kindly provided by S. Rose-John (University of Mainz).
Cell Culture.
Human leukemic HL60 cells (ATCC no. CCL-240) were cultured in RPMI medium 1640 containing 10% fetal bovine serum (FBS). The cell density of the cultures was routinely maintained between 2 × 105 and 1 × 106 cells per ml. For experiments involving quantitation of apoptotic cell death approximately 2 × 107 cells were centrifuged through a Histopaque 1119 cushion, to separate viable cells from dead cells and debris. Viable cells were collected from the interphase, washed in an excess volume of fresh medium, and plated in 12-well plates at 1 × 106 cells in a final volume of 1 ml. Protease and proteasome inhibitors dissolved in DMSO were added to the cells from stock solutions at the final concentrations given in Table 1, and incubation was continued for 6 h. Differentiation of HL60 cells into adherent macrophage-like cells was achieved by addition of 4α-phorbol 12-myristate 13-acetate (PMA; 10 ng/ml) to the culture medium for 2–5 days.
Table 1.
Extent of DNA fragmentation induced by exposure of HL60 cells to protease inhibitors
| Treatment | DNA fragmentation, % |
|---|---|
| DMSO (0.1%) | 4.3 ± 1.4 |
| PMSF (1 mM) | 6.2 ± 2.0 |
| TLCK (25 μM) | 6.3 ± 2.7 |
| E64 (50 μM) | 5.6 ± 1.6 |
| Leupeptin (25 μg/ml) | 6.0 ± 1.7 |
| LLnM (50 μM) | 6.0 ± 2.5 |
| TPCK (25 μM) | 19.5 ± 4.0* |
| DCI (50 μM) | 44.0 ± 8.7* |
| LLnL (50 μM) | 27.4 ± 6.1* |
| LLnV (50 μM) | 45.1 ± 8.3* |
| PSI (50 μM) | 34.9 ± 4.9* |
Cells were plated in 12-well plates in medium containing 10% fetal bovine serum, and protease inhibitors were added to the cells at the final concentrations given above. After 6 h of incubation the cells were lysed and the percentage of DNA fragmentation was assessed by the diphenylamine assay. Data are means ± SD from three independent experiments; ∗, P < 0.01 relative to 0.1% DMSO-treated control HL60 cells (by Student’s t test).
Determination of DNA Fragmentation.
For qualitative analysis of DNA fragmentation, cells were harvested at the indicated times by centrifugation and lysed by the addition of 250 μl of lysis buffer consisting of 10 mM Tris·HCl (pH 7.5), 10 mM EDTA, and 0.1% Triton X-100. After centrifugation the soluble DNA fragments released into the supernatant were precipitated by addition of 0.5 vol of 7.5 M ammonium acetate and 2.5 vol of ethanol. DNA pellets were incubated in TE containing 20 μg/ml RNase A (30 min, 37°C), then loaded onto a 1.7% agarose gel and separated at 100 V for 3 h. DNA fragments were visualized after staining with ethidium bromide by translumination with UV light. DNA fragmentation was quantified as described in ref. 16. For flow cytometry, cells were subjected to terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end-labeling (TUNEL) as previously reported (17) and counterstained with propidium iodide. Contour plots were obtained by using a Becton Dickinson FACScan flow cytometer and CellQuest software.
Loading of Cells with ICE/CPP32 Inhibitor Peptides.
The osmotic lysis of pinosome method as described by Moore et al. (18) was used to enhance introduction of the inhibitor peptides into the cells. Briefly, for each sample 1 × 106 cells were pelleted in a conical tube and incubated for 8–10 min at 37°C with 500 μl of a prewarmed hypertonic solution of 0.5 M sucrose, 10% (vol/vol) polyethyleneglycol 1000 (Baker), and 10 mM Hepes (pH 7.0) in RPMI medium containing N-acetyl-l-tyrosyl-l-valyl-l-alanyl-l-aspartyl chloromethyl ketone (Ac-YVAD-cmk; 250 μM), N-acetyl-l-aspartyl-l-glutamyl-l-valyl-l-aspartal (Ac-DEVD-cho; 250 μM), or DMSO (0.5%). Hypotonic medium (60% RPMI 1640/40% distilled water) was added to a final volume of 15 ml, and incubation at 37°C was continued for 3 min. Cells were harvested by centrifugation at room temperature for 5 min at 200 × g and resuspended in RPMI 1640/10%FBS at 1 × 106 cells in a final volume of 1 ml. LLnV (50 μM) or DMSO (0.1%) was added to the cells, and the mixture was incubated for 6 h, followed by the diphenylamine assay to determine the extent of DNA fragmentation.
SDS/PAGE and Western Blotting.
After incubation with 50 μM LLnV for the indicated times (see Fig. 3), cells were lysed in 1% SDS/10 mM Tris·HCl, pH 7.5, proteins were denatured at 95°C for 10 min, and protein concentrations were determined by using the BCA assay (Pierce). Aliquots of each protein lysate (30 μg) were subjected to SDS/PAGE (19). After electrophoresis proteins were transferred to nitrocellulose membranes and blocked overnight at 4°C with TBST buffer (50 mM Tris·HCl, pH 7.4/150 mM NaCl/0.05%Tween-20) containing 5% nonfat dry milk powder. Primary antibodies were added in TBST/5% nonfat dry milk powder and incubated for 45 min at 37°C. Incubation with secondary peroxidase-coupled goat anti-mouse antibody was performed under the same conditions. Blots were developed by using the ECL system (Amersham) or the SuperSignal reagent (Pierce).
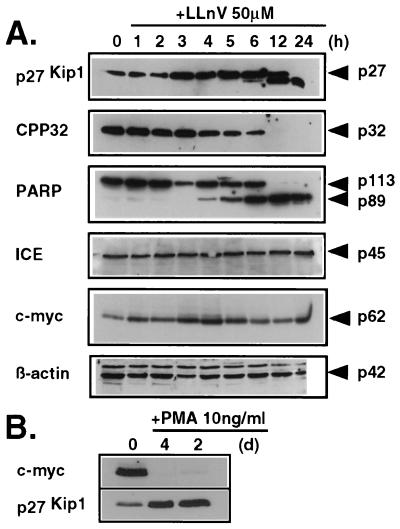
Figure 3.
(A) Relative changes in protein levels after treatment with the proteasomal inhibitor LLnV. (B) Relative changes of c-Myc and p27Kip1 during PMA-induced differentiation. Lysates of cells incubated with 50 μM LLnV (A) or 10 ng/ml PMA (B) for the indicated times were subjected to SDS/PAGE followed by Western blotting.
RESULTS
Proteasomal Inhibitors Induce Apoptosis in HL60 Cells.
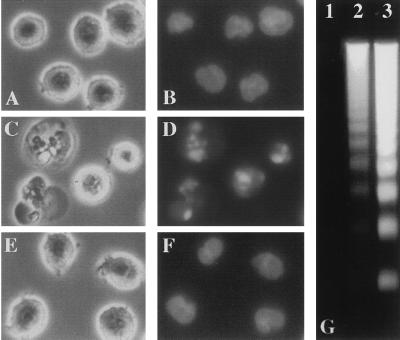
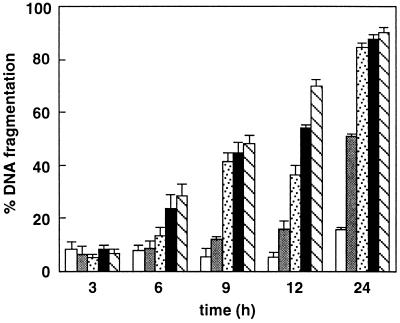
In unsynchronized HL60 cells, treated with N-tosylphenylalanyl chloromethyl ketone (TPCK), 3,4-dichloroisocoumarin (DCI), LLnL, or the specific proteasome inhibitors LLnV or PSI a fraction of the cells began to undergo apoptosis as early as 3–4 h after addition of the compounds. Membrane blebbing and cytoplasmic shrinkage were observed and the cells formed large protrusions. From 4 to 6 h after addition of proteasomal inhibitors cell nuclei became progressively pyknotic and were extensively fragmented (Fig. 1 C and D). The extent of DNA fragmentation, a hallmark of apoptotic cell death, was determined quantitatively using the diphenylamine assay, as described in Materials and Methods. The proportion of soluble DNA fragments which were extractable from apoptotic cells reached values of up to 45% for LLnV, 44% for DCI, and 34.9% for PSI of the total chromatin content after 6 h (Table 1). In contrast, cells treated with phenylmethylsulfonyl fluoride (PMSF), N-tosyllysyl chloromethyl ketone (TLCK), trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane (E64), N-acetylleucylleucylargininal (leupeptin), or the type II calpain inhibitor LLnM did not show any signs of apoptosis as assessed by morphological changes or by the extent of DNA fragmentation (Fig. 1 E and F; Table 1). DNA extracted from PSI-treated cells (Fig. 1G), as well as from TPCK-, DCI-, LLnL-, and LLnV-treated cells (not shown) displayed the oligonucleosomal laddering typically associated with apoptotic cells, when analyzed by agarose gel electrophoresis. The induction of DNA fragmentation by blocking proteasomal activity occurred in a dose-dependent fashion, as is shown for LLnV (Fig. 2) and was not detectable in cells treated with DMSO, PMSF, TLCK, E64, or LLnM (Fig. 2; data not shown).
Figure 1.
(A–F) Phase-contrast photomicrographs (A, C, and E) of HL60 cells and photomicrographs of the same cells after nuclear staining with Hoechst 33324 (B, D, and F) viewed 6 h after the following treatments: 0.1% DMSO (A and B), 50 μM PSI (C and D), or 50 μM E64 (E and F). (G) Oligonucleosomal DNA fragmentation from PSI-treated cells: 0.1% DMSO (lane 1), 10 μM PSI (lane 2), or 50 μM PSI (lane 3). Soluble DNA fragments were extracted, separated on a 1.7% agarose gel, and stained with ethidium bromide. The DNA fragmentation patterns were similar in cells treated with TPCK, DCI, LLnL, or LLnV (not shown).
Figure 2.
Dose–response relationship of LLnV-induced apoptosis. HL60 cells were maintained in medium containing 10% FBS, and treatments were as follows: 0.2% DMSO (empty bars); 10 μM LLnV (shaded bars); 20 μM LLnV (stippled bars); 50 μM LLnV (black bars); and 100 μM LLnV (hatched bars).
Inhibition of Proteasomal Function Activates CPP32, a Member of the ced-3/ICE Family of Proteases.
CPP32 precursor protein was detected in the lysate from untreated HL60 cells, but it gradually declined between 3 and 4 h after addition of LLnV and was barely detectable after 12 h (Fig. 3A). When the cleavage of PARP was used as an indicator of CPP32-like activity (20–22), it was observed that the decrease in the levels of the CPP32 precursor molecule was accompanied by the generation of a C-terminal 89-kDa PARP fragment (Fig. 3A). The cleavage of PARP also correlated with the appearance of cells in tissue culture showing characteristic apoptotic features such as cytoplasmic bleb formation and nuclear condensation or fragmentation (not shown). An inverse relationship was observed between levels of p27Kip1 and CPP32 precursor protein (Fig. 3A). In addition, the increase in p27Kip1 protein appears to precede the decrease in CPP32 precursor protein by roughly 1 h (Fig. 3A), suggesting that p27Kip1 may act upstream of CPP32 activation. In contrast, the amounts of ICE precursor remain unchanged even during the later stages of apoptosis (12 h, 24 h; Fig. 3A), when the extent of apoptosis approached nearly 90% as assessed by DNA fragmentation (Fig. 2). This also holds true for β-actin, which was used as an internal control to monitor either induction of general proteolysis or protein accumulation upon inhibition of proteasomal function (Fig. 3A).
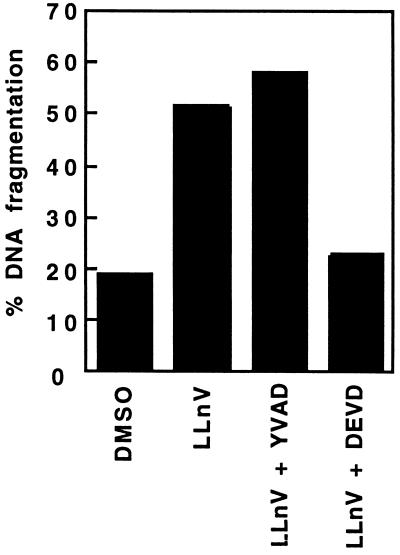
Two tetrapeptide derivatives, Ac-YVAD-cmk and Ac-DEVD-cho, which are selective inhibitors of ICE-like and CPP32-like proteases, respectively (23, 24), were used to preferentially block one or the other group of cysteine proteases. Since penetration of Ac-YVAD-cmk and Ac-DEVD-cho into HL60 cells was very poor when the inhibitors were added directly to the culture medium without further manipulation of the cells, peptides were loaded into the cells by using the technique of Moore et al. (18). The subsequent induction of cell death with LLnV could thus be completely blocked by the Ac-DEVD-cho peptide but not by the Ac-YVAD-cmk peptide (Fig. 4), confirming the involvement of CPP32 or CPP32-like proteases. It also suggests that LLnV-induced cell death proceeds independently from ICE or closely related family members, a suggestion that is supported by the lack of ICE precursor processing (Fig. 3A).
Figure 4.
Effect of Ac-DEVD-cho or Ac-YVAD-cmk on LLnV-induced apoptosis in HL60 cells. The tetrapeptide protease inhibitors Ac-DEVD-cho and Ac-YVAD-cmk were loaded into HL60 cells by osmotic lysis of pinosomes as described. Cell death was then induced by incubation with LLnV for 6 h, and the extent of DNA fragmentation was determined. Data shown are a representative sample of four independent experiments with similar results.
Inhibition of Proteasomal Activity Is Accompanied by Accumulation of the Cdk Inhibitor p27Kip1 and Apoptosis in G1.
Examination by Western blot analysis of the p27Kip1 levels in lysates of HL60 cells after incubation with LLnV demonstrated a clear accumulation of the p27Kip1 protein starting at 2–3 h after addition of inhibitor (Fig. 3A), indicating that proteasomal function is in fact blocked by LLnV (6). In lysates from cells incubated for more than 6 h with LLnV, an additional immunoreactive band at a lower molecular mass was detected which may represent a specific cleavage product of p27Kip1 due to an as-yet-unidentified proteolytic activity present in HL60 cells. Proteasomal degradation, however, is unlikely to account for the appearance of this band, as p27Kip1 is conjugated to ubiquitin prior to proteasome-mediated hydrolysis and hence migrates at higher relative molecular mass during SDS/PAGE, before it is finally degraded by the proteasome (6).
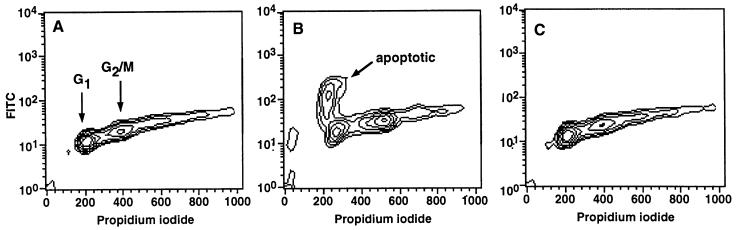
To examine whether the accumulation of p27Kip1 is also reflected in the distribution of the cells during cell cycle, cells were processed for flow cytometry by propidium iodide staining. To simultaneously assess, whether inhibition of proteasomal activity would render the cells of a particular cell cycle compartment more sensitive to execution of the cell death program, cells were additionally labeled by the TUNEL technique. FACS analysis of LLnV- or PSI-treated cells revealed that the majority of the apoptotic cells are in the G1 phase of the cell cycle (Fig. 5) which is consistent with the postulated function of p27Kip1 to control transit through the restriction point between G1 and S phase.
Figure 5.
LLnV-treated HL60 cells become apoptotic primarily in the G1 phase of the cell cycle. The cells were incubated for 5 h in RPMI 1640 medium containing 10% FBS supplemented with 0.1% DMSO (A), 50 μM LLnV (B), or 50 μM E64 (C). Apoptotic cells were labeled with fluorescein isothiocyanate (FITC) by the TUNEL technique, DNA was stained with propidium iodide, and the distribution of apoptotic cells within the cell cycle was analyzed by using a Becton Dickinson FACScan. Data shown are a representative example of three independent experiments with similar results.
Noncycling Differentiated HL60 Cells Are Unresponsive to Treatment with PSI.
The c-Myc protein can be readily detected in the lysate of DMSO-treated control HL60 cells (Fig. 3), and its relative contents remained unchanged even after incubation for 24 h with LLnV or PSI, which resulted in massive apoptosis of virtually all cells in culture (Figs. 2 and 3A). This indicated that the HL60 cells are not arrrested in G0 and are actively proliferating. To examine whether inhibition of proteasome activity could also affect the viability of nonproliferating, differentiated cells, HL60 cells were first induced to differentiate into adherent and quiescent macrophage-like cells by treatment with PMA and then treated with PSI. Differentiation was apparent by the expected morphological changes, the absence of bromodeoxyuridine incorporation, and cell proliferation (not shown), and, most significantly, by concomitant up-regulation of p27Kip1 and complete down-regulation of c-Myc (Fig. 3B). Differentiated cells treated with 50 μM PSI for 5 h showed an increase of 2.9% ± 1.1% in DNA fragmentation in comparison to an 13.7% ± 2.8% increase for undifferentiated cells, compared with DMSO-treated control cells. Thus, differentiated HL60 cells, which have withdrawn from the cell cycle, display a greatly reduced sensitivity toward proteasomal inhibition.
DISCUSSION
To determine whether compounds capable of interfering with proteasomal activity affect cell viability, human leukemic HL60 cells were incubated with various protease inhibitors that are known to have the capability to block proteasomal activity as well as with unrelated protease inhibitors. To this end all of the inhibitors that were capable of inducing apoptosis in HL60 have been described as inhibitors of proteasome function. LLnV and PSI are cell-permeant compounds capable of specifically inhibiting proteasomal activity, and this property has been exploited to study ligand-induced degradation of platelet-derived growth factor receptor (25), to block antigen processing in major histocompatibility complex class I antigen-presenting cells (26), or to study NFκB activation/IκB degradation (27). More specifically, PSI inhibits the chymotrypsin-like activities of the proteasome (IC50acid = 0.25 μM; IC50neutral = 6.5 μM), leaving the trypsin-like activity and the peptidyl-glutamyl peptide hydrolyzing (PGPH) activity mostly unaffected (28, 29).
DCI is another potent proteasomal inhibitor, and it blocks at least four of the five known peptide-hydrolyzing activities of the proteasome, including the chymotrypsin-like activity (30–32). Nevertheless, as blocking the chymotrypsin-like activity of the proteasome by PSI induced a level of DNA fragmentation similar to that obtained by incubation with DCI (Fig. 1G), it is suggested that the cytotoxic effect of DCI is primarily due to inhibition of the chymotrypsin-like activities of the proteasome. Preferential inhibition of the chymotrypsin-like activities of the proteasome may also account for the observed differences in the ability of TPCK and TLCK, two structurally closely related chloromethyl ketones, to induce apoptosis in HL60 cells. TPCK is an irreversible inhibitor of chymotrypsin and is widely used to inactivate chymotrypsin in trypsin preparations, whereas TLCK irreversibly inhibits trypsin-like serine proteases.
LLnL and LLnV have been shown to block proteasome activity as well, in contrast to LLnM, which exerts only a weak inhibitory activity toward proteasomal activity (26). All three aldehyde peptides, on the other hand, inhibit calpain I and II as well as cathepsin B with rather similar Ki values between 5 and 12 nM (26). If the cytotoxic effect of these inhibitors on HL60 cells were mediated by inhibition of calpains or cathepsin B, then LLnM should have elicited a cytotoxic response with similar potency, assuming that LLnM, LLnL, and LLnV are able to enter HL60 cells under identical conditions equally well. It is therefore concluded that the observed cytotoxic effect is due to blocking proteasomal activity and not to inhibition of calpains or lysosomal cathepsin B. This conclusion is also supported by the finding that treatment of HL60 cells with LLnV leads to increased concentrations of the Cdk inhibitor p27Kip1, a protein that has not been described as a substrate of calpain or cathepsin B. Instead, the decline in levels of p27Kip1 has been shown to be due to proteasomal degradation, when cells traverse the restriction point between G1 and S phase, following stimulation with specific mitogens (6, 33).
The members of the ICE family of cysteine proteases are present as inactive zymogens within the cell and are processed at the onset of apoptosis into enzymatically active heterodimer complexes (3, 34–39). In particular, activation of CPP32, one of the ICE family members, is markedly increased in cells undergoing apoptosis, and the cleavage of PARP has been widely used as an indicator for the activation of CPP32 or of a closely related enzyme with similar substrate specificity (20–22). CPP32 most likely is also involved in LLnV-induced apoptosis, as the disappearance of the CPP32 precursor protein starting at 3–4 h after addition of LLnV was directly correlated with the cleavage of PARP, suggesting that CPP32 is processed to its active form. Further evidence for this conclusion comes from the observation that Ac-DEVD-cho, a selective inhibitor of CPP32-like enzymes, could nearly entirely block LLnV-induced cell death. The changes induced in the relative levels of CPP32 precursor protein as well as of p27Kip1 appear to be rather selective and are apparently not due to random dysregulation of proteolysis, because several other proteins remain unaffected until late stages of apoptosis (ICE, β-actin, c-Myc).
ICE has been reported to act upstream of CPP32 in Fas-mediated cell death and to be directly involved in the processing and activation of CPP32 (40). However, during HL60 apoptosis no processing of ICE precusor could be detected in Western blot analysis using an antibody directed against the p10 subunit of mature ICE, either by the reduction in the relative levels of the precusor protein or by the detection of the cleavage product p10. In addition, Ac-YVAD-cmk, which selectively inhibits ICE-like family members, could not block the LLnV-induced cell death (Fig. 4), demonstrating that in HL60 cells ICE itself or closely related proteases probably do not play a central role in LLnV-induced CPP32 activation or the apoptotic cell death which ensues. This is in agreement with the finding that mice generated to be deficient in ICE are morphologically normal and show only minor defects regarding apoptosis induction (41). However, these results do not exclude the possibility that ICE or ICE-like proteases may be important for HL60 cell death induced by other stimuli, nor do they completely rule out the possibility that the observed cleavage of PARP could be due to the activition of other members of the ced-3/ICE family with substrate specificities similar to CPP32, for instance Mch-2, Mch-3, or Mch-4.
In contrast to CPP32 and p27Kip1, the levels of c-Myc protein remained constant over the course of the experiment. Given that at the same time levels of p27Kip1 are increased, the conclusion is supported that apoptotic cell death in HL60 cells may result from conflicting signaling events, as has been described for Myc-induced apoptosis in serum-deprived fibroblasts (42) or Myc-accelerated apoptosis in interleukin-3-deprived myeloid cells (43). In normally proliferating cells c-Myc is expressed continuously in a cell cycle-independent manner and is rapidly down-regulated upon mitogen withdrawal or by the action of cytostatic cytokines. Aberrant c-Myc expression however, in particular under unfavorable growth conditions, is associated with an increase in apoptotic cell death (42–44). Therefore, an increase in the levels of p27Kip1 protein induced by blocking proteasomal function along with persistent expression of c-Myc represents an analogous situation for the cell, and the apparent conflict promotes entry of the cell into the apoptosis pathway. This view is also supported by the observations made with PMA-differentiated HL60 cells, which are in a quiescent state. Their exit from the cell cycle is again accompanied by an increase in the levels of p27Kip1 protein, but at the same time, c-Myc protein declines to levels below the detection limit. Therefore the assumed signaling conflict is avoided and obviously renders the cells unresponsive to the otherwise apoptosis-inducing effect of proteasomal inhibition.
In the light of these results it is not surprising that in nonproliferating tissues like thymus, or in terminally differentiated cell populations like neuronal cells, inhibition of proteasomal function does not produce the same rapid effect of apoptosis induction and instead protects the cells from undergoing programmed cell death induced by various means (13, 15). Similar effects have been described earlier (45–48), although the protective effects of TPCK, DCI, or LLnL on cell survival noted in these studies were attributed to inhibition of an unidentified serine protease or of calpain. These observations seem at first glance to be contradictory to previous results (14) and to the findings reported here, but they can be explained on the basis of cell-specific differences in cell cycle regulation and the proliferative status of the cell, as manifested, for instance, by the expression of c-Myc.
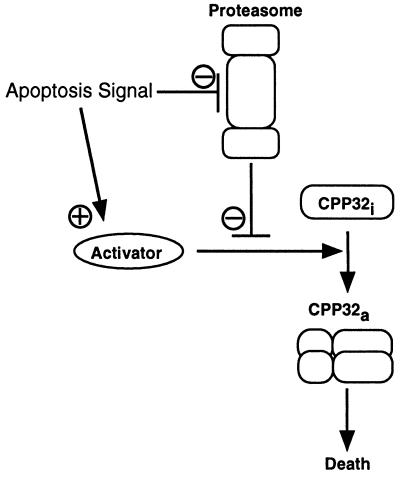
On the basis of the results presented here, I propose that the combination of a cell cycle block mediated by proteasomal inhibitors with the simultaneous deregulation of c-Myc expression as observed in various tumor cells will drive the cells into apoptosis. Apparently, it is one of functions of the proteasome to ensure the orderly progression of the cell through the cell cycle. At the same time, activation of CPP32 or of a closely related protease is prevented. It is tempting to speculate that this inhibition of CPP32 activation may also be accomplished by the proteasome through constant degradation or processing of a protein located upstream of CPP32 that is critical for CPP32 activation. Blocking cell cycle progression in such an actively dividing cell population may then lead to a net increase in activator protein, by reduction of proteasome activity (as mediated by the action of LLnV or PSI), by increased de novo synthesis of the activator, or by both (Fig. 6). Quiescent or terminally differentiated cells consequently would not be affected to the same extent by such a treatment. Thus a close relationship exists between the mechanisms controlling cell cycle progression and the mechanisms activating the cell death program, and it appears that the proteasome is in a prime position in which it functions as a switchboard to execute the decision of a cell either to proceed with proliferation or to undergo apoptosis.
Figure 6.
Suggested model of proteasome function in the control of apoptotic cell death. See Discussion for further explanations.
Acknowledgments
I thank Stefanie Pebler for performing the FACS analysis, M. Eilers and M. Clauss for helpful suggestions, C. Mitchell for critically reading the manuscript, and W. Risau for continuing generous support.
Footnotes
Abbreviations: LLnV, N-carbobenzoxy-l-leucyl-l-leucyl-l-norvalinal; PSI, N-carbobenzoxy-l-isoleucyl-l-γ-t-butyl-l-glutamyl-l-alanyl-l-leucinal; LLnL, N-acetylleucylleucylnorleucinal; LLnM, N-acetylleucylleucylnormethioninal; Ac-YVAD-cmk, N-acetyl-l-tyrosyl-l-valyl-l-alanyl-l-aspartyl chloromethyl ketone; Ac-DEVD-cho, N-acetyl-l-aspartyl-l-glutamyl-l-valyl-l-aspartal; PARP, poly(ADP-ribose) polymerase; TPCK, N-tosylphenylalanyl chloromethyl ketone; TLCK, N-tosyllysyl chloromethyl ketone; DCI, 3,4-dichloroisocoumarin; PMSF, phenylmethylsulfonyl fluoride; E64, trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane; leupeptin, N-acetylleucylleucylargininal; DMSO, dimethyl sulfoxide; PMA, 4α-phorbol 12-myristate 13-acetate.
References
- 1.Steller H. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 2.Ellis R E, Yuan J, Horvitz H R. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- 3.Yuan J Y, Shaham S, Ledoux S, Ellis H M, Horvitz H R. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 4.Maki C G, Huibregtse J M, Howley P M. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 5.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Del S G, Chau V, Yew P R, Draetta G F, Rolfe M. Science. 1995;286:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 7.Stancovski I, Gonen H, Orian A, Schwartz A L, Ciechanover A. Mol Cell Biol. 1995;15:7106–7116. doi: 10.1128/mcb.15.12.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsurumi C, Ishida N, Tamura T, Kakizuka A, Nishida E, Okumura E, Kishimoto T, Inagaki M, Okazaki K, Sagata N, Ichihara A, Tanaka K. Mol Cell Biol. 1995;15:5682–5687. doi: 10.1128/mcb.15.10.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jariel-Encontre I, Pariat M, Martin F, Carillo S, Salvat C, Piechaczyk M. J Biol Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- 10.Jones M E E, Haire M F, Kloetzel P M, Mykles D L, Schwartz L M. Dev Biol. 1995;169:436–447. doi: 10.1006/dbio.1995.1159. [DOI] [PubMed] [Google Scholar]
- 11.Dawson S P, Arnold J E, Mayer N J, Reynolds S E, Billett M A, Gordon C, Colleaux L, Kloetzel P M, Tanaka K, Mayer R J. J Biol Chem. 1995;270:1850–1858. doi: 10.1074/jbc.270.4.1850. [DOI] [PubMed] [Google Scholar]
- 12.Phillips M E, Platt P J. Gen Comp Endocrinol. 1994;95:409–415. doi: 10.1006/gcen.1994.1140. [DOI] [PubMed] [Google Scholar]
- 13.Grimm L M, Goldberg A L, Poirier G G, Schwartz L M, Osborne B A. EMBO J. 1996;15:3835–3844. [PMC free article] [PubMed] [Google Scholar]
- 14.Imajoh-Ohmi S, Kawaguchi T, Sugiyama S, Tanaka K, Omura S, Kikuchi H. Methods Enzymol. 1995;217:1070–1077. doi: 10.1006/bbrc.1995.2878. [DOI] [PubMed] [Google Scholar]
- 15.Sadoul R, Fernandez P-A, Quiquerez A-L, Martinou I, Maki M, Schröter M, Becherer J D, Irmler M, Tschopp J, Martinou J-C. EMBO J. 1996;15:3845–3852. [PMC free article] [PubMed] [Google Scholar]
- 16.Duke R C, Cohen J J. In: Current Protocols in Immunology. Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, editors. Vol. 1. New York: Wiley; 1995. pp. 3.17.4–3.17.5. [Google Scholar]
- 17.Gavrieli Y, Sherman Y, Bensasson S A. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore M W, Carbone F R, Bevan M J. Cell. 1988;54:777–785. doi: 10.1016/s0092-8674(88)91043-4. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Nature (London) 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 21.Tewari M, Quan L T, O’Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann S H, Desnoyers S, Ottaviano Y, Davidson N E, Poirier G G. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 23.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, Munday N A, Raju S M, Smulson M E, Yamin T T, Yu V L, Miller D K. Nature (London) 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 24.Schlegel J, Peters I, Orrenius S, Miller D K, Thornberry N A, Yamin T T, Nicholson D W. J Biol Chem. 1996;271:1841–1844. doi: 10.1074/jbc.271.4.1841. [DOI] [PubMed] [Google Scholar]
- 25.Mori S, Tanaka K, Omura S, Saito Y. J Biol Chem. 1995;270:29447–29452. doi: 10.1074/jbc.270.49.29447. [DOI] [PubMed] [Google Scholar]
- 26.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 27.Traenckner E B, Wilk S, Baeuerle P A. EMBO J. 1994;13:5433–5441. doi: 10.1002/j.1460-2075.1994.tb06878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilk S, Figueiredo-Pereira M E. Enzyme Protein. 1993;47:306–313. doi: 10.1159/000468688. [DOI] [PubMed] [Google Scholar]
- 29.Figueiredo-Pereira M E, Chen W E, Yuan H M, Wilk S. Arch Biochem Biophys. 1995;317:69–78. doi: 10.1006/abbi.1995.1137. [DOI] [PubMed] [Google Scholar]
- 30.Orlowski M, Cardozo C, Michaud C. Biochemistry. 1993;32:1563–1572. doi: 10.1021/bi00057a022. [DOI] [PubMed] [Google Scholar]
- 31.Vinitsky A, Michaud C, Powers J C, Orlowski M. Biochemistry. 1992;31:9421–9428. doi: 10.1021/bi00154a014. [DOI] [PubMed] [Google Scholar]
- 32.Djaballah H, Harness J A, Savory P J, Rivett A J. Eur J Biochem. 1992;209:629–634. doi: 10.1111/j.1432-1033.1992.tb17329.x. [DOI] [PubMed] [Google Scholar]
- 33.Coats S, Flanagan W M, Nourse J, Roberts J M. Science. 1996;272:877–880. doi: 10.1126/science.272.5263.877. [DOI] [PubMed] [Google Scholar]
- 34.Fernandes-Alnemri T, Litwack G, Alnemri E S. J Biol Chem. 1994;269:30761–30764. [PubMed] [Google Scholar]
- 35.Fernandes-Alnemri T, Litwack G, Alnemri E S. Cancer Res. 1995;55:2737–2742. [PubMed] [Google Scholar]
- 36.Fernandes-Alnemri T, Takahashi A, Armstrong R, Krebs J, Fritz L, Tomaselli K J, Wang L, Yu Z, Croce C M, Salveson G, Earnshaw W C, Litwack G, Alnemri E S. Cancer Res. 1995;55:6045–6052. [PubMed] [Google Scholar]
- 37.Wang L, Miura M, Bergeron L, Zhu H, Yuan J Y. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 38.Kamens J, Paskind M, Hugunin M, Talanian R V, Allen H, Banach D, Bump N, Hackett M, Johnston C G, Li P, Mankovich J A, Terranova M, Ghayur T. J Biol Chem. 1995;270:15250–15256. doi: 10.1074/jbc.270.25.15250. [DOI] [PubMed] [Google Scholar]
- 39.Kumar S, Kinoshita M, Noda M, Copeland N G, Jenkins N A. Genes Dev. 1994;8:1613–1626. doi: 10.1101/gad.8.14.1613. [DOI] [PubMed] [Google Scholar]
- 40.Enari M, Talanian R V, Wong W W, Nagata S. Nature (London) 1996;380:723–726. doi: 10.1038/380723a0. [DOI] [PubMed] [Google Scholar]
- 41.Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C, Mcdowell J, Paskind M, Rodman L, Salfeld J, Towne E, Tracey D, Wardwell S, Wei F Y, Wong W, Kamen R, Seshadri T. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 42.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 43.Askew D, Ashmun R, Simmons B, Cleveland J. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 44.Harrington E A, Bennett M R, Fanidi A, Evan G I. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fearnhead H O, Rivett A J, Dinsdale D, Cohen G M. FEBS Lett. 1995;357:242–246. doi: 10.1016/0014-5793(94)01367-a. [DOI] [PubMed] [Google Scholar]
- 46.Cain K, Inayathussain S H, Kokileva L, Cohen G M. Biochem Cell Biol. 1994;72:631–638. doi: 10.1139/o94-083. [DOI] [PubMed] [Google Scholar]
- 47.Cain K, Inayathussain S H, Kokileva L, Cohen G M. FEBS Lett. 1995;358:255–261. doi: 10.1016/0014-5793(94)01436-5. [DOI] [PubMed] [Google Scholar]
- 48.Squier M K, Miller A C, Malkinson A M, Cohen J J. J Cell Physiol. 1994;159:229–237. doi: 10.1002/jcp.1041590206. [DOI] [PubMed] [Google Scholar]