Abstract
Caenorhabditis elegans contains a family of putative insulin-like genes proposed to regulate dauer arrest and senescence. These sequences often lack characteristic sequence features of human insulin essential for its folding, structure, and function. Here, we describe the structure and receptor-binding properties of INS-6, a single-chain polypeptide expressed in specific neurons. Despite multiple nonconservative changes in sequence, INS-6 recapitulates an insulin-like fold. Although lacking classical receptor-binding determinants, INS-6 binds to and activates the human insulin receptor. Its activity is greater than that of an analogous single-chain human insulin analog.
Keywords: Senescence, hormone, metabolism, protein structure, protein evolution, NMR
Development of Caenorhabditis elegans can arrest to form a specialized third-stage larva. Such dauer (“enduring”) larvae can survive up to eight times longer than the organism's normal 2-wk lifespan (Blaxter and Bird 1997). An adaption to limited food supply, increased population density, or elevated temperature (Riddle and Albert 1997), the dauer stage is triggered by chemosensory cues under the joint control of insulin-related and TGF-β-related signaling pathways (Ren et al. 1996; Kimura et al. 1997). On sensing favorable conditions for growth and reproduction, the dauer resumes development into adulthood. Genetic screens for dauer arrest have led to identification of C. elegans orthologs of the insulin receptor (daf-2) and post-receptor signaling pathway, including the catalytic subunit of PI-3-kinase (age-1), serine-threonine kinases Akt/PKB (akt-1 and akt-2) and a forkhead transcription factor (daf-16; Guarente and Kenyon 2000; Wolkow et al. 2000). Mutations in daf-2 and age-1 delay senescence of the adult independently of effects on dauer arrest (for review, see Finch and Ruvkun 2001).
Although genetic screens for diapause or long-lived phenotypes did not reveal DAF-2 ligands, search of the C. elegans genome by sequence- and structure-based algorithms has uncovered a family of 37 putative insulin-related (ins) genes (Pierce et al. 2001). These sequences are classified into three classes, designated α, β, and γ, based on pattern of cysteines (Pierce et al. 2001). α-Class proteins lack an A6–A11 disulfide bridge, an invariant feature of vertebrate insulins and insulin-like growth factors (Baker et al. 1988). β-Class proteins appear to retain insulin's three canonical disulfide bridges, plus a fourth, whose location is variable. γ-Class proteins contain three canonical (or pseudo-canonical) disulfide bridges. Because these sequences diverge from mammalian insulins at conserved sites required for biological activity, their classification as insulin-like has been presumptive. Genetic evidence is provided elsewhere in this issue that an ins gene (daf-28) functions in the daf-2 pathway of dauer arrest (Li et al. 2003).
We describe here the chemical synthesis and structure of a β-class INS protein. Because it lacks dibasic processing sites (as utilized in cleavage of proinsulin; Steiner 1998), the protein was prepared as a single chain. The polypeptide folds to form a well-defined structure with four disulfide bridges. Despite marked sequence divergence, an insulin-like fold is stabilized by reorganization of the hydrophobic core. Although the topography of the surface differs radically from that of insulin (Baker et al. 1988), the C. elegans protein can bind to and activate the human insulin receptor (hIR). As a seeming paradox, the affinity of INS-6 for the hIR is at least 10-fold greater than that of the corresponding single-chain analog of human insulin (“mini-proinsulin”; Markussen et al. 1985; Derewenda et al. 1991; Hua et al. 1998). Our results validate assignment of this ins gene to an insulin-like superfamily and broaden the scope of structure-function relationships in design of human insulin agonists (De Meyts and Whittaker 2002).
Results and Discussion
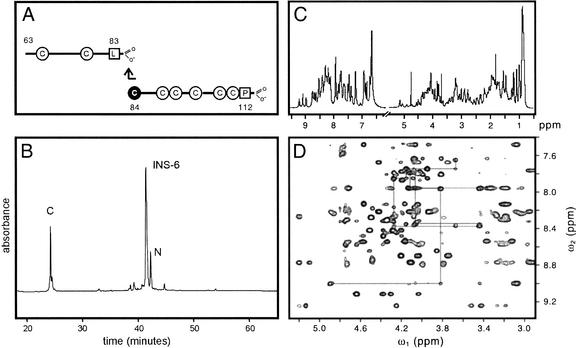
The sequence of INS-6 is shown in Figure 1 (top). Whereas insulin contains six cysteines (forming cystines A6–A11, A7–B7, and A20–B19), INS-6 contains eight; the additional cysteines are indicated by arrows. Exclusive of canonical cysteines (bold in Fig. 1), INS-6 and human insulin contain seven identical residues. A 50-residue polypeptide comprising the A and B domains of INS-6 was synthesized in two fragments (Fig. 2A) and joined by native ligation (Fig. 2B). Quantitative folding was observed in a redox buffer (see Materials and Methods). Under these conditions, the polypeptide is in equilibrium between a reduced, unfolded species and a single, predominant folded form. 1H-NMR spectra of the folded form at 600 MHz are well resolved (Fig. 2C,D); resonance line widths at pH 3.6–7.4 are consistent with a protein monomer. Chemical-shift dispersion and density of inter-residue nuclear Overhauser enhancements (NOEs) provide evidence of an ordered structure. A single set of spin systems was observed, indicating a single predominant conformation. Sequential assignment was obtained by standard methods (Wuthrich 1986).
Figure 1.
Sequence of INS-6 (residues 63–112; top) in relation to mammalian insulins (middle) and insulin-like growth factors (IGF1 and IGF2; bottom). The B domain is shown at left and the A domain at right. Color code: green, side chains in INS-6 core in accord with insulin structure; red, side chains in INS-6 core not in accord with insulin structure; purple, INS-6 exposed side chains not in accord with insulin structure. Canonical cysteines are in bold; arrows indicate noncanonical cysteines (residues 84 and 111). Boxes a–d indicate invariant sequence elements discussed in text.
Figure 2.
Synthesis of INS-6 and NMR Spectra. (A) Native ligation of N- and C-terminal segments. The eight cysteines are shown, including cysteine that participates in thioester ligation. Peptide sequences refer to intact INS-6. (B) Reverse-phase HPLC purification of reduced INS-6 polypeptide from ligation mixture. N- and C-terminal segments are labeled N and C, respectively. (C) One-dimensional 1H-NMR spectrum of folded INS-6 in H2O at pH 3.7 (left) and in D2O at pD 3.4 (right) at 25°C. (D) Fingerprint region of NOESY spectrum (mixing time 200 msec).
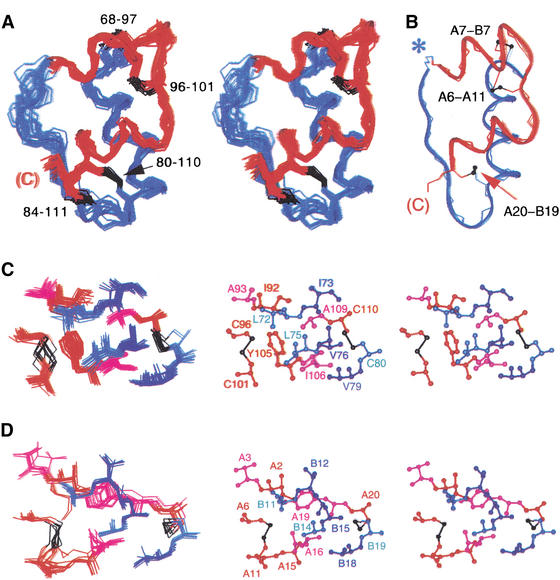
Analysis of secondary structure demonstrates three insulin-like α-helices. An ensemble of structures was calculated by distance geometry and simulated annealing (DG/SA; Fig. 3A). Input restraints include 621 NOEs (381 inter-residue and 240 intra-residue), 38 dihedral restraints (23 φ and 15 χ1), and 16 hydrogen-bond-related restraints. No main-chain dihedral angles are in disallowed Ramachandran regions. Root-mean-square deviations (RMSD) are 0.39 Å (main-chain atoms) and 0.70 Å (side-chain atoms) relative to the mean structure. Structures calculated in the absence of assumed disulfide bridges are essentially identical but less precise. Comparison with the crystal structure of mini-proinsulin (T-state protomer shown in Fig. 3B; Derewenda et al. 1991) demonstrates that an overall insulin-like fold is maintained. A fourth disulfide bridge (cystine 84–111) tethers residues whose homologs in insulin (Asn A21 and Gly B23) are in close proximity (Baker et al. 1988). Despite their overall similarities, superposition of INS-6 and mini-proinsulin demonstrates small changes in interhelical angles. An RMSD of 2.3 ± 0.1 Å is obtained on pairwise alignment of mini-proinsulin and the ordered INS-6 moiety (residues 68–111). This difference is significant given the precision of the DG/SA ensemble; the corresponding RMSD within the INS-6 ensemble is 0.6 ± 0.1 Å. Alignment of the three helical segments yields a main-chain RMSD of 1.7 ± 0.1 Å (RMSD within INS-6 0.4 ± 0.1 Å). Similarly, alignment of the B domain α-helices yields a segmental RMSD of 0.7 ± 0.1 Å (RMSD within INS-6 is <0.2 Å), whereas alignment of the A domains (INS-6 residues 92–111) yields a regional RMSD of 3.0 ± 0.1 Å (RMSD within INS-6 is 0.4 ± 0.1 Å).
Figure 3.
Structure of INS-6 in relation to insulin. (A) Stereo pair showing INS-6 DG/SA ensemble (20 structures). B domain (residues 67–90; B6–B29 in insulin) and A domain (residues 91–112; A1–A22) are shown in blue and red, respectively. Disulfide bridges are shown in black. (B) Crystal structure of mini-proinsulin (T state protomer; PDB identifier 6INS) in similar orientation. The A domain is shown in red, the B domain in blue, and disulfide bridges in black. Despite similarities, differences occur in interhelical angles; a 2–4 Å translation in position of the A domain is also observed. Asterisk indicates B29–A1 peptide bond, not present in insulin. (C) Packing of side chains in core of INS-6. (Left) INS-6 ensemble showing 10 internal side chains and cystines 80–110 and 96–101. Color scheme is as in A. (Right) Stereo view of representative structure. (D) Packing of corresponding side chains in collection of insulin crystal structures (PDB identifiers 4INS, 1PID, 1APH, 1TRZ, 1TYL, 1TYM, 1ZNI, 3INS, and 6INS). Residues are labeled as in insulin. Molecules were aligned according to main-chain atoms of residues B9–B23, A2–A8, and A13–A19.
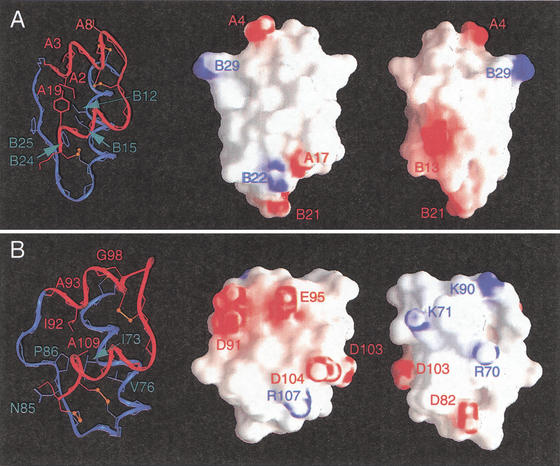
The core of INS-6, like that of insulin (Baker et al. 1988), contains nonpolar side chains (color coded in Fig. 1) and two disulfide bridges (80–110 and 96–101, corresponding to B19–A20 and A6–A11, respectively; Fig. 3A,B). Key side chains include Leu 72, Leu 75, Val 76, Val 79, Ile 92, Ala 93, Tyr 105, and Ala 109 (Fig. 3C). Although some of these residues (bold face) are conserved among mammalian sequences (Leu B11, Ala B14, Leu B15, Val B18, Ile A2, Val A3, Gln A15, and Ala A19; Fig. 1), others are substituted in INS-6 by side chains of smaller (underlined) or larger (italics) volumes. The substitution Tyr A19 → Ala is compensated in part by swiveling of a surface side chain in insulin (Gln A15) into the core of INS-6 (Tyr 105; highlighted in purple in Fig. 1). The latter packs against Leu 72 and Leu 75, interactions not observed among mammalian insulins. Insulin's invariant aromatic residues at B24–B26 (Fig. 1, box b) align with Asn-Pro-Gln in INS-6. The absence of Phe B24, which seals one edge of insulin's core, is compensated in part by the novel 84–111 disulfide bridge. Divergence at positions Tyr B26 and Pro B28, which pack against the A chain (Baker et al. 1988), is associated with an altered trajectory of the corresponding strand in INS-6. Because insulin's C-terminal B-chain β strand forms an anti-parallel β-sheet in the dimer, divergence at this and other self-assembly surfaces rationalizes impaired self-association. The surface of INS-6 thus differs in topography and electrostatic potential from that of insulin (Fig. 4).
Figure 4.
Electrostatic surface of human insulin (A) and INS-6 (B). GRASP representations of “front” and “back” protein surfaces (middle and right images). Regions of positive or negative potential are indicated in red or blue. Charged side chains are labeled to orient surface features. In each panel, a ribbon structure at same orientation is shown at left; disulfide bridges are shown in orange.
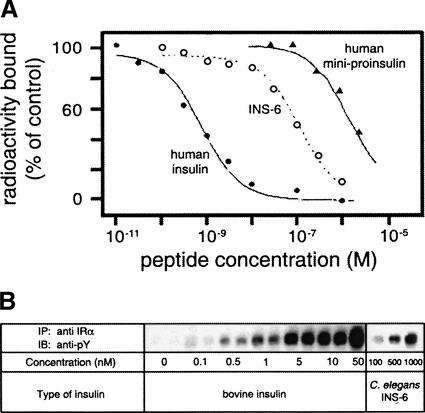
Binding of INS-6 to the hIR was evaluated by competitive displacement of 125I-labeled human insulin (Fig. 5A). Its relative affinity is 0.6% (Fig. 5A, open circles). Although low relative to insulin (Fig. 5A, closed circles), such binding is at least 10-fold tighter than that of mini-proinsulin (Fig. 5A, triangles). Competition between insulin and INS-6 suggests that the two ligands bind to overlapping or structurally linked sites. In independent assays of transmembrane signaling, INS-6 stimulates autophosphorylation of the cytoplasmic domain of the hIR to the same extent as insulin at corresponding levels of ectodomain occupancy (i.e., INS-6 functions as an agonist). Radioimmunoassays demonstrated that such activity is not the result of contamination by insulin. Although these observations are broadly consistent with the biological activity of human proinsulin in transgenic worms (presumably mediated by binding to DAF-2; Pierce et al. 2001), the activity of INS-6 would not be expected on the basis of its divergent surface. The following structure-function relationships in insulin predict that INS-6 should be essentially inactive. (1) The B29–A1 peptide bond, retained in INS-6, causes a 3000-fold reduction in insulin's activity (Hua et al. 1998). (2) Insulin residues Phe B24, Phe B25, Val A3, and Tyr A19 are critical to its activity (Baker et al. 1988; De Meyts and Whittaker 2002). Substitution of Phe B24 or Phe B25 (Fig. 1, box b) by nonaromatic amino acids impairs receptor binding (Mirmira et al. 1991) and can cause a monogenic form of diabetes mellitus (Shoelson et al. 1983). Respective substitutions of Tyr A19 (Fig. 1, box d) by Phe and Ala impair activity by 8- and 1000-fold (Kristensen et al. 1997; Du and Tang 1998). Respective substitution of Leu B6 and Val A3 (Fig. 1, boxes a and c) by Ala likewise impairs activity by 40- and 70-fold (Nakagawa and Tager 1991, 1992). Accordingly, divergence at these positions of INS-6, if considered one at a time, would be expected to block binding to the insulin receptor. We suggest an analogy between protein folding and receptor binding; just as multiple compensating interactions within INS-6 permit maintenance of an insulin-like fold, perhaps the extensive differences between the surfaces of insulin and INS-6 permit recapitulation of an analogous signaling conformation. Understanding the molecular basis of INS-6 activity will require crystal structures of insulin and INS-6/receptor complexes.
Figure 5.
INS-6 binds to and triggers the hIR. (A) Competitive displacement of 125I-human insulin by INS-6 (○), unlabeled human insulin (●), or human mini-proinsulin (▴). (B) Relative activity of bovine insulin and INS-6 in stimulating autophosphorylation of the hIR (tyrosine phosphorylation).
In summary, we have demonstrated conservation of an insulin-like fold in a C. elegans INS protein. Such conservation raises the possibility that the human genome may likewise contain unrecognized insulin-like genes. The relationship of sequence to structure exhibits combinatorial interactions of intricate complexity. Binding of INS-6 to the hIR—a seeming paradox in light of classical structure-function relationships—suggests that INS proteins may provide novel starting points for design of agonists and antagonists of therapeutic interest.
Materials and methods
Peptide synthesis
INS-6 segments (peptide 1–21 thioester and peptide 22–50) were synthesized by solid-phase BOC-chemistry. The methionine residue (peptide residue 15) was substituted by selenomethionine. The reduced INS-6 polypeptide was prepared by native ligation (Dawson et al. 1994) in 23% yield, purified by rp-HPLC, and characterized by mass spectrometry (observed molecular mass 5346.97 ± 0.87 D in agreement with calculated mass-average isotope composition, 5345.95 D). Folding was effected by stirring overnight in 100 mM Tris-HCl (pH 8.7), 8 mM cysteine, 1 mM cystine, and 1 M guanidine(HCl; guanidine was included to enhance the solubility of the reduced polypeptide without denaturing the folded (oxidized) form. The predominant folded product was isolated by rp-HPLC (observed mass 5339.37 ± 0.84 D in agreement with calculated value 5337.95 D). Folding yield was 65% relative to the reduced polypeptide.
Insulin receptor-binding assay
Assays were performed as described (Weiss et al. 2000) using human placental cell membranes. Membrane fragments (25 μg protein/tube) were incubated with 125I-labeled insulin (ca. 30,000 cpm) in the presence of selected concentrations of INS-6 for 18 h at 4°C in 0.25 mL of 0.05 M Tris-HCl and 0.25% (w/v) BSA at pH 8, diluted with 1 mL of ice-cold buffer, and centrifuged (10,000g) for 5 min at 4°C. The supernatant was removed by aspiration and membrane pellet counted for radioactivity. Data were corrected for nonspecific binding (membrane-associated radioactivity in presence of 1 μM human insulin). Four replicates were performed. Mini-proinsulin data are as described (Hua et al. 1998).
Tyrosine phosphorylation
Assays were performed as described (Bass et al. 1998) using human embryonal kidney cells expressing the hIR. Cells were grown to confluence on poly-L-lysine-treated 12-well plates, incubated for 18 h in DMEM containing 1% FBS, washed in PBS, and stimulated in serum-free medium containing 0.1% BSA with indicated concentrations of insulin at 37°C for 5 min, and then lysed in prechilled Triton X 100 lysis buffer containing phosphatase inhibitors (1% Triton X-100, HEPES, 100 mM NaF, 2 mM sodium orthovanadate, and 4 mM sodium pyrophosphate). Receptors were immunoprecipitated (Rouard et al. 1999), and samples processed by sodium dodecyl sulfate gel electrophoresis, followed by electroblotting. Phosphorylated proteins were detected with horseradish peroxidase-conjugated monoclonal anti-phophotyrosine antibody (Upstate Biotechnology, Inc.) and visualized by chemiluminescence (Amersham). Blots were stripped (63 mM Tris at pH 6.8, 10% SDS, and 0.7% v/v β-mercaptoethanol) and reprobed with anti-insulin receptor β-subunit antibodies (Upstate Biotechnology, Inc.). Percentage of tyrosine-phosphorylated hIR to total receptor recovered was compared by densitometry.
NMR spectroscopy
Spectra were obtained at 600 MHz at 25°C (pH 3.7 and 7.3) as described in Hua et al. (1996); the protein concentration was 1.5 mM. Resonance assignment was based on 2D NOESY (mixing times 100 and 200 msec), TOCSY, and DQF-COSY spectra. The ensemble was refined by inclusion of hydrogen-bond restraints derived from amide protection data and imposition of disulfide bridges based on sulfur positions in initial models.
Acknowledgments
We thank S.B. Kent and P. Katsoyannis for advice regarding peptide synthesis; P. Rue at the Diabetes Research and Training Center at the University of Chicago for radioimmunassays; W. Li and G. Ruvkun for communication of results prior to publication; and G.G. Dodson, B. Roth, E. Stavnezer, D.F. Steiner, and J. Whittaker for discussion. This work was supported by a grant from the National Institutes of Health to M.A.W. (R21 DK61651).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL weiss@biochemistry.cwru.edu; FAX (216) 368-3419.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1058003.
References
- Baker EN, Blundell TL, Cutfield JF, Cutfield SM, Dodson EJ, Dodson GG, Hodgkin DM, Hubbard RE, Isaacs NW, Reynolds CD. The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Phil Trans Royal Soc London Ser. 1988;319:369–456. doi: 10.1098/rstb.1988.0058. [DOI] [PubMed] [Google Scholar]
- Bass J, Chiu G, Argon Y, Steiner DF. Folding of insulin receptor monomers is facilitated by the molecular chaperones calnexin and calreticulin and impaired by rapid dimerization. J Cell Biol. 1998;141:637–646. doi: 10.1083/jcb.141.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M, Bird D. Parasitic nematodes. In: In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. Elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 851–878. [PubMed] [Google Scholar]
- Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- De Meyts P, Whittaker J. Structural biology of insulin and IGF1 receptors: Implications for drug design. Nat Rev Drug Discov. 2002;1:769–783. doi: 10.1038/nrd917. [DOI] [PubMed] [Google Scholar]
- Derewenda U, Derewenda Z, Dodson EJ, Dodson GG, Bing X, Markussen J. X-ray analysis of the single chain B29-A1 peptide-linked insulin molecule. A completely inactive analogue. J Mol Biol. 1991;220:425–433. doi: 10.1016/0022-2836(91)90022-x. [DOI] [PubMed] [Google Scholar]
- Du X, Tang JG. Hydroxyl group of insulin A19Tyr is essential for receptor binding: Studies on (A19Phe)insulin. Biochem Mol Biol Int. 1998;45:255–260. doi: 10.1080/15216549800202622. [DOI] [PubMed] [Google Scholar]
- Finch CE, Ruvkun G. The genetics of aging. Annu Rev Genomics Hum Genet. 2001;2:435–462. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- Hua Q-X, Hu SQ, Frank BH, Jia W, Chu YC, Wang SH, Burke GT, Katsoyannis PG, Weiss MA. Mapping the functional surface of insulin by design: Structure and function of a novel A-chain analogue. J Mol Biol. 1996;264:390–403. doi: 10.1006/jmbi.1996.0648. [DOI] [PubMed] [Google Scholar]
- Hua Q-X, Hu SQ, Jia W, Chu Y-C, Burke GT, Wang SH, Wang RY, Katsoyannis PG, Weiss MA. Mini-proinsulin and mini-IGF-I: Homologous protein sequences encoding non-homologous structures. J Mol Biol. 1998;277:103–118. doi: 10.1006/jmbi.1997.1574. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kristensen C, Kjeldsen T, Wiberg FC, Schaffer L, Hach M, Havelund S, Bass J, Steiner DF, Andersen AS. Alanine scanning mutagenesis of insulin. J Biol Chem. 1997;272:12978–12983. doi: 10.1074/jbc.272.20.12978. [DOI] [PubMed] [Google Scholar]
- Li, W., Kennedy, S.G., and Ruvkun, G. 2003. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Markussen J, Jorgensen KH, Sorensen AR, Thim L. Single chain des-(B30) insulin. Intramolecular crosslinking of insulin by trypsin catalyzed transpeptidation. Int J Peptide Protein Res. 1985;26:70–77. [PubMed] [Google Scholar]
- Mirmira RG, Nakagawa SH, Tager HS. Importance of the character and configuration of residues B24, B25, and B26 in insulin-receptor interactions. J Biol Chem. 1991;266:1428–1436. [PubMed] [Google Scholar]
- Nakagawa SH, Tager HS. Implicatons of invariant residue LeuB6 in insulin-receptor interactions. J Biol Chem. 1991;266:11502–11509. [PubMed] [Google Scholar]
- ————— Importance of aliphatic side-chain structure at positions 2 and 3 of the insulin A chain in insulin-receptor interactions. Biochemistry. 1992;31:3204–3214. doi: 10.1021/bi00127a023. [DOI] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes & Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P, Lim C-S, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-β homolog. Science. 1996;274:1389–1392. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Albert PS. Genetic and environmental regulation of dauer larva development. In: In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. Elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 739–768. [PubMed] [Google Scholar]
- Rouard M, Bass J, Grigorescu F, Garrett TPJ, Ward CW, Lipkind G, Jaffiole C, Steiner DF, Bell GI. Congenital insulin resistance associated with a conformational alteration in a conserved β-sheet in the insulin receptor L1 domain. J Biol Chem. 1999;274:18487–18491. doi: 10.1074/jbc.274.26.18487. [DOI] [PubMed] [Google Scholar]
- Shoelson S, Haneda M, Blix P, Nanjo A, Sanke T, Inouye K, Steiner D, Rubenstein A, Tager H. Three mutant insulins in man. Nature. 1983;302:540–543. doi: 10.1038/302540a0. [DOI] [PubMed] [Google Scholar]
- Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- Weiss MA, Hua Q-X, Jia W, Chu Y-C, Wang R-Y, Katsoyannis PG. Hierarchiacal protein ‘un-design’: Insulin's intrachain disulfide bridge tethers a recognition a-helix. Biochemistry. 2000;39:15429–15440. doi: 10.1021/bi001905s. [DOI] [PubMed] [Google Scholar]
- Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulin-like signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- Wuthrich K. NMR of proteins and nucleic acids. New York, NY: John Wiley & Sons; 1986. [Google Scholar]