Abstract
To investigate the molecular basis of β-globin gene activation, we analyzed factor recruitment and histone modification at the adult β-globin gene in wild-type (WT)/locus control region knockout (ΔLCR) heterozygous mice and in murine erythroleukemia (MEL) cells. Although histone acetylation and methylation (Lys 4) are high before and after MEL differentiation, recruitment of the erythroid-specific activator NF-E2 to the promoter and preinitiation complex (PIC) assembly occur only after differentiation. We reported previously that targeted deletion of the LCR reduces β-globin gene expression to 1%–4% of WT without affecting promoter histone acetylation. Here, we report that NF-E2 is recruited equally efficiently to the adult β-globin promoters of the ΔLCR and WT alleles. Moreover, the LCR deletion reduces PIC assembly only twofold, but has a dramatic effect on Ser 5 phosphorylation of RNA polymerase II and transcriptional elongation. Our results suggest at least three distinct stages in β-globin gene activation: (1) an LCR-independent chromatin opening stage prior to NF-E2 recruitment to the promoter and PIC assembly; (2) an intermediate stage in which NF-E2 binding (LCR-independent) and PIC assembly (partially LCR-dependent) occur; and (3) an LCR-dependent fully active stage characterized by efficient pol II elongation. Thus, in its native location the LCR functions primarily downstream of activator recruitment and PIC assembly.
Keywords: Allele-specific chromatin immunoprecipitation analysis, locus control region, β-globin locus, transcription elongation, NF-E2, CTD-phosphorylation
Supplemental material is available at http://www.genesdev.org.
In higher eukaryotes, gene activation involves several events including chromatin opening, activator binding to regulatory regions, recruitment of basal transcription factors (TFIIA to TFIIJ) and RNA polymerase II (pol II) to the promoter [preinitiation complex (PIC) assembly], and transcription elongation. A general model has emerged in which activators function to stabilize or modulate transcription through interactions with histone acetyltransferases, as well as components of the basal transcription machinery, such as TATA binding protein (TBP), TBP-associated factors (TAFs), and TFIIB (for review, see Lemon and Tjian 2000). PIC assembly is followed by initiation and elongation, during which the C-terminal domain (CTD) of the RPB1 subunit of pol II is phosphorylated (for review, see Dahmus 1996).
These steps in gene activation are often regulated by distal elements called enhancers (for review, see Blackwood and Kadonaga 1998; Martin 2001). Enhancers increase either the rate of transcription, the number of the templates engaged in transcription, or both. Studies using transgenes or in vitro templates have linked enhancer activities to various events such as PIC assembly (Kim et al. 1998; Yie et al. 1999), histone acetylation at the promoter (Madisen et al. 1998), and nuclear localization (Francastel et al. 1999). However, it is poorly understood which events in transactivation are the actual targets of long-range enhancer function at native gene loci.
The murine β-globin gene locus is a model system for studying the molecular mechanisms of enhancer-dependent gene activation at a native locus. The locus contains embryonic and adult β-like globin genes that are ordered as they are expressed during development. High-level expression of the β-globin gene cluster is regulated by the locus control region (LCR), which consists of several DNase I hypersensitive sites (HSs) distributed in the region 30–60 kb upstream of the adult βmajglobin gene. Transient and stable transfection assays as well as studies in transgenic mice have suggested that the LCR not only enhances transcription but also has dominant chromatin opening activity (for review, see Bulger and Groudine 1999).
Surprisingly, studies of the targeted deletion of the LCR from the endogenous β-globin gene locus (ΔLCR) have revealed that the LCR is not required for the generalized DNaseI sensitive conformation of the locus or chromatin remodeling and histone hyperacetylation of the β-globin promoter (Epner et al. 1998; Bender et al. 2000; Schübeler et al. 2001); however, transcription from the ΔLCR allele is decreased to 1%–4% of wild-type (WT) levels (Bender et al. 2000; Schübeler et al. 2001). These results suggest that the LCR stimulates transcription at an event downstream of promoter remodeling, although the molecular mechanism of LCR function has not been determined.
Several recent chromatin immunoprecipitation (ChIP) analyses of factor binding in the β-globin locus of cultured erythroid cell lines have provided clues to understanding the regulation of β-globin gene expression (for review, see Bulger et al. 2002). NF-E2, an erythroid-specific activator, appears to play an important role in β-globin expression by stimulating PIC assembly. NF-E2 is composed of two subunits: p45 NF-E2 and p18 NF-E2 (Andrews et al. 1993a, 1993b; Chan et al. 1993; Ney et al. 1993), the latter of which is a member of the small Maf protein family, MafK (Igarashi et al. 1994; for review, see Motohashi et al. 1997). p45 NF-E2, which contains a transactivation domain, interacts with TAFII130 (Amrolia et al. 1997) and CBP in vitro (Cheng et al. 1997; Hung et al. 2001) and is required for pol II recruitment to the βmajglobin gene promoter in mouse erythroleukemia (MEL) cells (Johnson et al. 2001). Recently, we reported that NF-E2 binds to both the LCR and the β-globin promoter region, even though no canonical NF-E2 binding sites are present in the promoter (Sawado et al. 2001). This observation has reinforced speculation that the LCR functions through interactions with the promoter.
To gain insight into which transcription events are influenced by the LCR, we have performed ChIP analyses of histone modifications and factor recruitment at the adult β-globin gene in two model systems: (1) differentiation of 745A MEL cells, a process accompanied by a greater than 100-fold increase in β-globin gene expression (Sawado et al. 2001); and (2) primary erythroid cells derived from ΔLCR/WT heterozygous mice.
We find a high level of histone acetylation of the β-globin gene in uninduced MEL cells, and this does not change significantly upon differentiation. In contrast, significant binding of NF-E2, TFIIB, and pol II to the promoter, as well as efficient pol II elongation, is observed only in differentiated cells, consistent with the large increase in β-globin gene transcription. Analysis of wild-type and ΔLCR β-globin alleles in the ΔLCR/WT heterozygous mice suggests that NF-E2 binding to the β-globin promoter region occurs independent of the LCR, and that the LCR has only a small effect on PIC assembly. In contrast, deletion of the LCR dramatically decreases phosphorylation of the C-terminal domain (CTD) of pol II and pol II elongation, consistent with the large decrease in transcription observed. Our results suggest that the LCR plays multiple roles in the transition from an intermediate to a fully active stage, including a major effect on the transition from transcription initiation to elongation.
Results
Transcription factor recruitment to the promoter correlates with the conversion from basal to high level transcription during MEL cell induction
To gain insight into the pathway of β-globin gene activation, we analyzed recruitment of activators and components of the basal transcription machinery to the βmajglobin gene in MEL cells. MEL cells are blocked at an early stage of erythropoiesis and morphologically resemble proerythroblasts. In MEL cell clone 745A, βmajglobin gene transcripts are detectable in the uninduced state only by reverse transcriptase PCR (RT–PCR), and expression increases by >100-fold upon DMSO-mediated induction of terminal differentiation (Sawado et al. 2001). We performed ChIP with cross-linked chromatin from uninduced and differentiated MEL cells, using antibodies that recognize NF-E2, members of the PIC and histone modifications that are known markers for euchromatic genes. Using primer sets for the promoter region or exon 3 of the βmajglobin gene, the enrichment of these factors/histone modifications at the βmajglobin gene relative to myoD1 was determined by duplex PCR under conditions of linear amplification (Sawado et al. 2001). The myoD1 gene, which like the β-globin gene locus is located on murine chromosome 7, is expressed only in skeletal muscle and its precursors (Weintraub et al. 1991); thus, it serves as a negative control. The ratio of the two PCR products was determined for the antibody-bound fraction and normalized to the ratio of the control IgG-bound fraction.
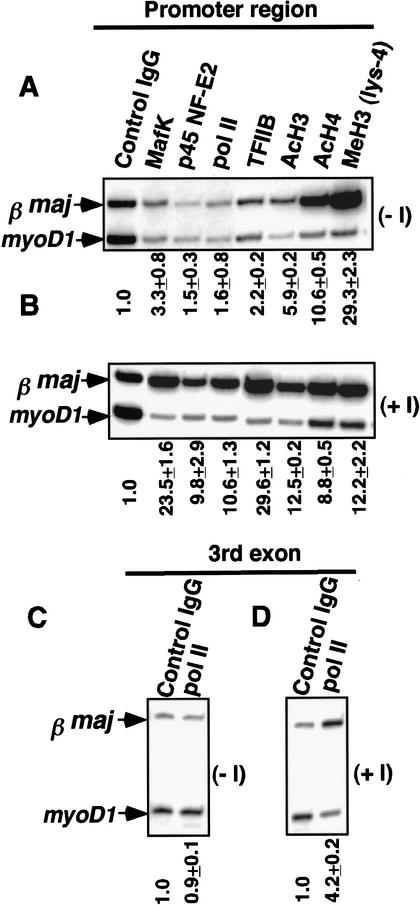
High levels of histone modifications such as acetylation of H3 (Lys 9 and/or Lys 14) and H4 (Lys 5, Lys 8, Lys 12, and/or Lys 16), dimethylation of H3 (Lys 4), which are known markers for euchromatic genes, are present at the promoter before induction and remain high throughout differentiation (Fig. 1; Sawado et al. 2001). In contrast, p45 NF-E2 recruitment is detectable only after induction (Fig. 1A,B), and the threefold enrichment in MafK binding at the βmajglobin promoter before induction increases to >20-fold upon differentiation, as previously reported (Fig. 1A,B; Sawado et al. 2001). Consistent with the observation that NF-E2 is required for pol II recruitment (Johnson et al. 2001), high levels of pol II enrichment, as measured with an antibody that detects both phosphorylated and unphosphorylated forms, are detected only after induction. TFIIB, a component of the PIC, is enriched only 2.2-fold before induction, and this enrichment increases to 29.6-fold after induction (Fig. 1A,B). Pol II enrichment at the third exon of the βmajglobin gene is also detectable after induction, indicating efficient transcription elongation (Fig. 1C). No enrichment of these factors or histone modifications is observed at the βmajglobin gene in a pro-B cell line (data not shown). These results suggest that, in MEL cells, an “open” chromatin conformation is present prior to induction, and that ensuing steps such as NF-E2 complex formation, PIC assembly, and transcription elongation at the β-globin gene are distinct from chromatin opening and are correlated with high levels of expression.
Figure 1.
Factor recruitment and histone modifications at the βmajglobin gene during MEL cell differentiation. Chromatin samples for immunoprecipitations were obtained from uninduced MEL cells (A,C; −I) and induced MEL cells (B,D; +I). Lanes are labeled with the antibodies used for ChIP. Duplex PCRs were performed with a primer set for myoD1 and a primer set for the promoter region (A,B) or the third exon of the βmajglobin gene (C,D). Enrichments relative to myoD1 are shown below the gels.
Allele-specific ChIP analysis for ΔLCR/WT heterozygous mice
Analysis of targeted deletions of the endogenous β-globin LCR in mice and in cultured cells have revealed that the LCR is not required for initiation or maintenance of general DNase I sensitivity of the locus, hyperacetylation of histones, or low-level transcription, but it is essential for achieving high levels of transcription (Epner et al. 1998; Bender et al. 2000; Schübeler et al. 2001). Given the similar chromatin structure and expression of the β-globin loci in uninduced MEL cells and the ΔLCR cells, we wished to determine if the differentiation-dependent events such as NF-E2 binding, PIC assembly, and elongation were LCR-dependent steps. Thus, we compared factor recruitment and histone modification at the ΔLCR with WT β-globin promoters in vivo.
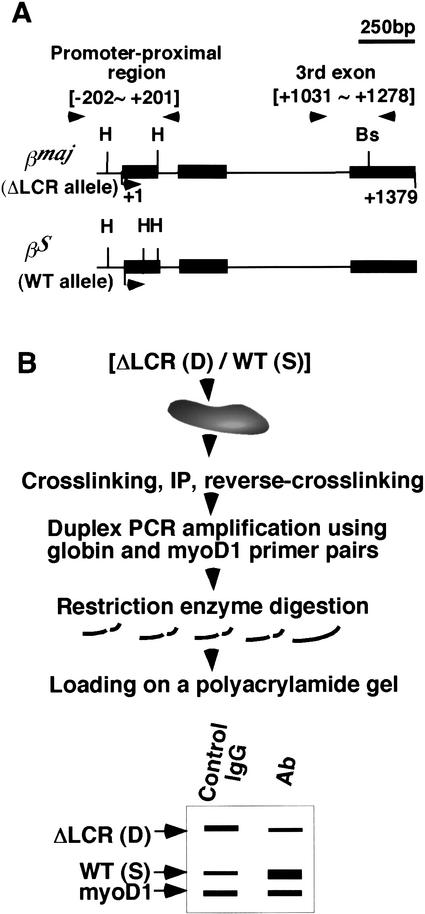
We developed a novel ChIP assay that allows simultaneous determination of factor enrichment and histone modification in each β-globin allele in cells derived from ΔLCR/WT heterozygous mice (Fig. 2). There are two advantages in analyzing heterozygous mice. First, using allele specific ChIP analysis, the ΔLCR and WT alleles are examined in the same ChIP reaction, allowing quantification of potentially subtle differences in factor recruitment or histone modification state between the WT and ΔLCR alleles. Second, by assessing factor recruitment or histone modification state in the same nuclear environment, we can exclude the possibility that differences in erythroid factor concentrations or the spectrum of erythroid cells present in the two homozygous mouse lines might bias assessment of factor enrichment or histone modification at one allele relative to the other.
Figure 2.
Allele-specific ChIP analysis. (A) Structures of the βmajglobin and βSglobin genes with polymorphisms for HaeIII (H) and the BstXI sites (Bs) are shown. PCR primer pairs for allele-specific ChIP analysis were designed to amplify both the βmajglobin and βSglobin genes and are indicated by arrowheads. Positions are relative to the Cap site (+1) of the βmajglobin gene. (B) An allele-specific ChIP analysis of ΔLCR (D)/WT (S) heterozygous mice is shown. Further details are presented in Materials and Methods.
To distinguish between binding at WT and ΔLCR alleles of heterozygous mice in ChIPs, we used mice containing two different murine β-globin haplotypes, HbbS (S) and HbbD (D). βmaj and βS, which are the primary adult β-globin genes of the D and S alleles, respectively, are highly homologous; for example, the nucleotide sequence in the upstream promoter region of the two genes (−249∼−1) is identical (data not shown). However, a βS-specific HaeIII site is located ∼100 bp downstream of the Cap site, and a βmaj-specific BstXI site is located in the third exon (Fig. 2A; Fiering et al. 1995). ChIPs were performed using spleen cells from ΔLCR (D)/WT (S) heterozygous mice. After coamplification of both alleles with a single set of primers, the PCR products from the WT (S) and the ΔLCR (D) alleles were digested with restriction enzymes recognizing these polymorphisms (Fig. 2B). The amount of a particular sequence in the antibody-bound chromatin was determined by duplex PCR and standardization to the amount of the myoD1 gene (not expressed in erythroid cells) in the bound fraction, as described previously (Sawado et al. 2001). Enrichment of signals from the WT (EWT) and ΔLCR (EΔLCR) alleles relative to myoD1 and the ratio of EΔLCR to EWT (EΔLCR/EWT) were determined as described in Materials and Methods.
NF-E2 and pol II recruitment to the β-globin promoter are LCR-independent
NF-E2 interacts not only with the LCR (Daftari et al. 1999; Forsberg et al. 2000; Sawado et al. 2001), but also with the promoter region, even though no canonical NF-E2 binding sites are present in the promoter (Sawado et al. 2001). Previously, we proposed two models for sequence-independent NF-E2 recruitment to the promoter: NF-E2 may be recruited to the promoter via interaction between the LCR and the promoter; alternatively, NF-E2 may reside in two independent protein complexes that bind to the LCR and the active promoters (Sawado et al. 2001). ChIP analyses of the WT and ΔLCR alleles in primary erythroid cells allow us to distinguish between these models.
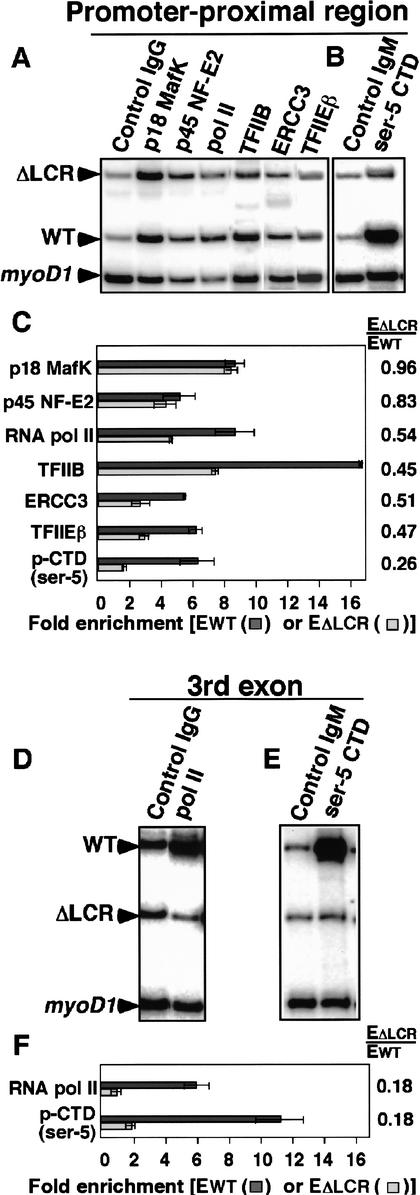
Using allele specific ChIP analyses, we find that NF-E2 is recruited to the adult βmajglobin promoter of the ΔLCR allele as strongly as to the βSglobin promoter of the WT allele (Fig. 3A). The ratio of enrichment on the ΔLCR allele to WT (EΔLCR/EWT) for MafK and p45 NF-E2 are 0.96 and 0.83, respectively (Fig. 3C). Therefore, NF-E2 recruitment to the promoter is minimally affected by deletion of the LCR, and NF-E2 recruitment to the promoter is not dependent on an interaction between the LCR and the promoter. We also found that pol II is significantly enriched at the promoter-proximal region even in the absence of the LCR (Fig. 3A), although this enrichment is reduced. EΔLCR/EWT for pol II is 0.54 (Fig. 3C), indicating that the number of pol II molecules in the promoter-proximal region of the ΔLCR allele is ∼50% of WT.
Figure 3.
Allele-specific analysis of factor binding in the promoter-proximal region and third exon in ΔLCR(D)/WT(S) heterozygous mice. Lanes are labeled with the antibody used. The regions tested are promoter-proximal (A,B) and third exon (D,E) of the βmajglobin and βSglobin genes. Enrichment of the β-globin gene relative to myoD1 in WT (EWT; indicated as dark-gray bars) or ΔLCR (EΔLCR; indicated as light-gray bars) alleles in sample materials relative to those in control IgG or IgM bound materials are shown in C and F. The ratios between the WT and ΔLCR allele PCR products (EΔLCR/Ewt) are shown at the right of graphs. A table showing these data is included in Supplemental Material.
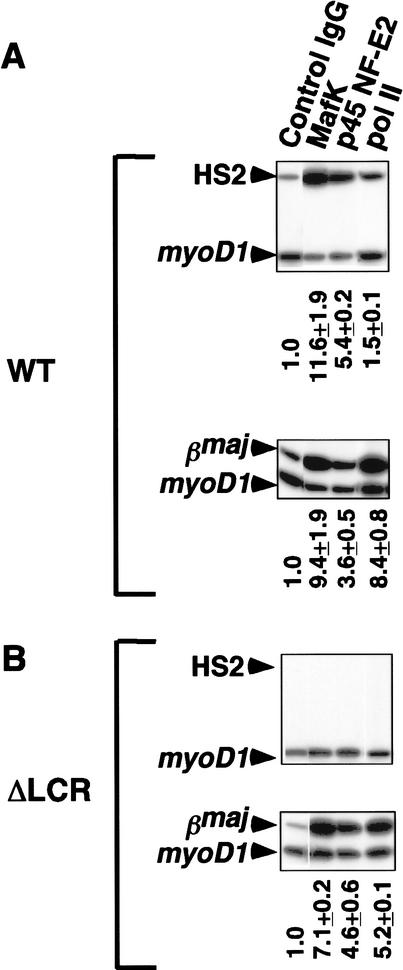
To examine the possibility that the LCR in the WT allele promotes factor recruitment to the βmajglobin promoter of the ΔLCR allele in trans, we also performed the ChIP assays using WT or ΔLCR homozygous mice (Fig. 4). As shown, MafK, p45 NF-E2, and pol II were all enriched relative to myoD1 at the βmajglobin promoter-proximal region in both WT and ΔLCR loci (Fig. 4). In addition, and consistent with our previous analysis in MEL cells, both components of NF-E2 were enriched relative to myoD1 at HS2 in the WT locus (Fig. 4A).
Figure 4.
ChIP analysis of occupancy of NF-E2 and pol II at HS2 and the βmajglobin gene promoter in WT (A) and ΔLCR (B) homozygous mice. Chromatin were immunoprecipitated with control IgG, anti-p45 NF-E2, anti-MafK, and anti-pol II. Enrichment relative to myoD1 is shown below the gels.
The LCR has only a twofold effect on PIC assembly
Because the size of the chromatin used in these ChIP analyses is 300–800 bp, the 50% reduction in pol II enrichment in the ΔLCR allele could occur at the level of pol II recruitment to the promoter and/or pol II elongation. To determine the effect of deletion of the LCR on PIC assembly, we determined the levels of general transcription factor (GTF) recruitment to the promoter-proximal regions in the WT and ΔLCR alleles. As shown in Figure 3A, TFIIB, a component of the PIC, is enriched in both WT and ΔLCR alleles, although enrichment is reduced in the ΔLCR allele (the EΔLCR/EWT for TFIIB is 0.45; Fig. 3C). TFIIE and TFIIH stimulate promoter melting and promote the initial stage of transcription elongation (for review, see Hampsey 1998). Both TFIIEβ, a subunit of TFIIE, and ERCC3, a helicase subunit of TFIIH, are enriched in both alleles (Fig. 3A). The EΔLCR/EWT for TFIIEβ is 0.47 and the EΔLCR/EWT for ERCC3 is 0.51 (Fig. 3C). We obtained similar results for TFIIEα, another subunit of TFIIE (data not shown). These results suggest that the LCR promotes PIC assembly approximately twofold. Importantly, despite dramatically decreased expression from the ΔLCR allele (1%–4% of WT; Schübeler et al. 2001), recruitment of GTFs and pol II remains significant. This suggests that the LCR functions primarily at a step downstream of PIC assembly. Therefore, we investigated the possibility that transition from initiation to elongation is affected by the LCR deletion.
The LCR acts primarily at the transition from initiation to elongation
The high homology between the 5′ portions of the four adult β-globin genes in the D and S alleles has precluded the use of conventional run-on assays to compare elongation efficiency in the β-globin locus of heterozygous mice. Therefore, we used allele-specific ChIP assays to compare pol II elongation efficiency in the βmaj and βSglobin genes on ΔLCR (D) and WT (S) alleles, respectively. In contrast to the nearly twofold higher enrichment of pol II in the promoter-proximal region in the WT compared to the ΔLCR allele, we detected an at least fivefold higher enrichment of pol II in the third exon (∼1 kb downstream of the promoter) in the WT compared to the ΔLCR allele. As shown in Figure 3D and F, pol II is enriched 6.1-fold in exon 3 in the WT allele, but no enrichment is observed in exon 3 in the ΔLCR allele. This result suggests that despite significant pol II recruitment to the promoter-proximal region of the ΔLCR allele (Fig. 3A,C), no pol II enrichment is detectable at the third exon. Similarly, we detected significant enrichment of pol II ∼600 bp downstream of the poly-A site in the WT, but not the ΔLCR allele (data not shown). These results suggest that pol II elongation is either blocked or inefficient in the absence of the LCR.
One prominent event during the transition from initiation to elongation is phosphorylation of the CTD of the RPB1 subunit of pol II (for review, see Dahmus 1996). Two residues in the CTD, Ser 2 and Ser 5, are known targets for phosphorylation during transcription enhanced by the HIV-1 long terminal repeat (Barboric et al. 2001), as well as transcription of several mammalian (Nissen and Yamamoto 2000; Soutoglou and Talianidis 2002) and yeast genes (Komarnitsky et al. 2000; Cho et al. 2001). In the β-globin locus, the Ser 5 phosphorylated form of CTD was shown to be enriched at the βmajglobin promoter after MEL cell induction (Johnson et al. 2001). To determine if CTD phosphorylation is affected by the LCR, we performed an allele-specific ChIP analysis using an antibody against the phosphorylated CTD. As shown in Figure 3B and C, Ser 5 phosphorylation of the CTD is present at the promoter-proximal region in the WT allele, but is significantly (fourfold) reduced in the ΔLCR allele. These results suggest that even though pol II is enriched at the ΔLCR promoter-proximal region (Fig. 3A), the CTD is not highly phosphorylated in the absence of the LCR.
The difference in CTD phosphorylation between the WT and the ΔLCR alleles becomes even more prominent at the third exon (Fig. 3E,F), which is due in part to the relative inefficiency of pol II elongation in the ΔLCR allele (Fig. 3D). Considering the slight reduction in pol II enrichment at the third exon in the WT allele, the increase of CTD phosphorylation of Ser 5 in the third exon is most likely due to hyperphosphorylation of the CTD (see Discussion).
Discussion
Mechanisms of NF-E2 recruitment to the promoter-proximal region
Previously we reported that subunits of NF-E2 are recruited to both the LCR and the promoter upon MEL cell induction (Sawado et al. 2001). Although there are multiple NF-E2 binding sites in the LCR, the promoter does not contain any canonical NF-E2 binding sites. Thus, NF-E2 may be recruited to the promoter via communication with the LCR, or NF-E2 may interact with both the LCR and the promoter independently (Sawado et al. 2001). Here, we find that NF-E2 recruitment to the promoter does not require the LCR. This result suggests that NF-E2 recruitment to the promoter may be mediated by interactions with proteins associated with the promoter-proximal region, for example, CBP (Cheng et al. 1997; Hung et al. 2001) and/or TafII130 (Amrolia et al. 1997).
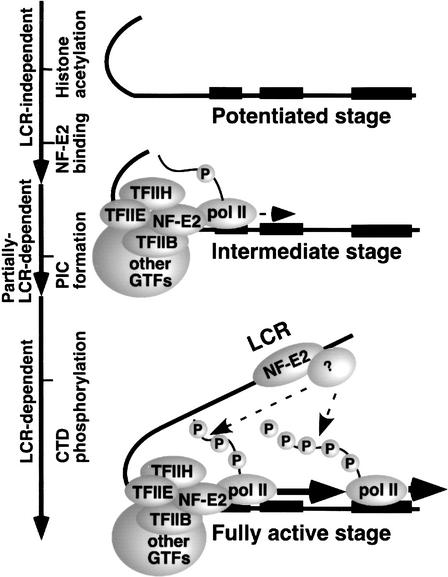
Three distinct stages of β-globin gene activation
Together with our previous observations (Epner et al. 1998; Bender et al. 2000; Sawado et al. 2001; Schübeler et al. 2001), the results presented here suggest a three-step model for β-globin gene activation (Fig. 5). The first step is represented by uninduced MEL cells. Histone H3 and H4 acetylation, and H3 dimethylation (Lys 4) at the βmajglobin promoter occur prior to formation of a functional NF-E2 complex and PIC. Interestingly, we find a twofold reduction in dimethylation of histone H3 at Lys 4 in the promoter region. It is not clear whether this modest decrease is due to conversion of the dimethyl to the trimethyl form of H3 upon high level of transcription, as recently suggested (Santos-Rosa et al. 2002) or caused by demethylation of H3. Regardless, in uninduced MEL cells the βmajglobin promoter is in a “potentiated” state, but not active state. Previously we reported that neither histone acetylation (Schübeler et al. 2001) nor remodeling of the adult β-globin promoter (Epner et al. 1998; Bender et al. 2000) require the LCR. Using ΔLCR/S heterozygous mice, we find that NF-E2, pol II, and general transcription factors are recruited to the promoter-proximal region in the absence of the LCR. Thus, the status of the promoter on the ΔLCR allele appears to represent an intermediate stage in the pathway that leads to fully activated transcription. The third step (fully active stage), characterized by high levels of CTD phosphorylation and efficient elongation, occurs in the WT but not the ΔLCR allele, and is thus LCR-dependent. In this model, the LCR functions primarily to promote the transition from the intermediate to fully active stage.
Figure 5.
β-globin gene activation can be divided into three distinct steps. An LCR-independent potentiated stage is characterized by histone acetylation and methylation (Lys 4) in the absence of NF-E2 complex and PIC assembly at the promoter. An intermediate stage is characterized by LCR-independent NF-E2 binding and partially LCR-dependent PIC assembly at the promoter. An LCR-dependent, fully active stage is characterized by CTD phosphorylation and efficient pol II elongation. The unknown factor (indicated by question mark) shown binding to the LCR may play a role in the transition to full activation (see Discussion). Each exon of adult β-globin is shown by a black bar.
Relationship between expression levels and PIC assembly
In numerous genes in yeast, it has been shown that TBP and TFIIB binding are strictly correlated with transcription levels (Kuras and Struhl 1999; Li et al. 1999); however, we find that removal of the LCR leads to only a twofold reduction in recruitment of GTFs and pol II to the promoter, while the rate of transcription is reduced 25- to 100-fold (Schübeler et al. 2001). Therefore, PIC assembly does not reflect the level of expression in the β-globin gene locus. Similar inconsistencies between expression level and PIC assembly were observed in the HMRa1 and HSP82 genes in yeast, where PIC assembly takes place even in silent chromatin (Sekinger and Gross 2001). In a small subset of yeast promoters, TBP binding is high even when gene activity is low and increases only modestly with high expression levels (Kuras and Struhl 1999). Also, GTFs bind to Drosophila promoters that are repressed by Polycomb group proteins (Breiling et al. 2001). It has been shown that chromatin remodeling of the α1-antitrypsin gene promoter occurs after PIC assembly (Soutoglou and Talianidis 2002), and that elongation is the rate-limiting step in the regulation of many genes, including c-myc (Bentley and Groudine 1986; Krumm et al. 1992), c-fos (Plet et al. 1995), heat-shock protein (Brown et al. 1996; Lis 1998), and transcription regulated by the HIV-1 LTR (Marciniak and Sharp 1991; for review, see Jones and Peterlin 1994). These observations demonstrate that events downstream of PIC assembly can be rate-limiting steps in transcription, and may represent common mechanisms for establishing an active state in many genes. Surprisingly, our results reveal that the β-globin LCR functions primarily at just such downstream steps, because it mediates the transition from a state defined by PIC assembly, low CTD phosphorylation and inefficient elongation to a fully active stage.
Our results implicating a primary function of the LCR downstream of PIC assembly contrast with a recent report showing that the endogenous 3′ TCRβ enhancer (Eβ) acts to modify chromatin and in the recruitment of pol II, TBP, and transactivators at the upstream pDβ1 promoter (Spicuglia et al. 2002). Clearly, these differences could be due to different technical approaches (e.g., ChIP procedures). However, the order of chromatin modification and factor recruitment is not the same at different gene loci (Cosma et al. 1999; Agalioti et al. 2000), and this may reflect different enhancer-mediated steps in gene activation.
LCR-dependent CTD phosphorylation
One major effect of the LCR is CTD phosphorylation at Ser 5, which is required for the transition from transcription initiation to elongation. We find that although basal levels of CTD phosphorylation can be detected at the ΔLCR allele, high-level CTD phosphorylation requires the LCR. Our observation that CTD hyperphosphorylation of Ser 5 is detected at the 3′ end of the gene does not correlate with previous analyses of several yeast genes (Komarnitsky et al. 2000; Cho et al. 2001) and the human α1-antitrypsin gene (Soutoglou and Talianidis 2002). In these genes, the Ser 5 phosphorylated form of the CTD is enriched only at the promoter-proximal region, and not at the 3′ end, whereas Ser 2 phosphorylated CTD is detected at the 3′ end. Despite using several different antibodies, we failed to detect the Ser 2 phosphorylated form of the CTD in the β-globin gene in murine erythroid cells, and Ser 2 phosphorylation of the CTD is not observed in the myoD1 gene in murine muscle cells (B. Penn and S. Tapscott, unpubl.). Moreover, hyperphosphorylation at Ser 5 of the CTD, in the absence of Ser 2 phosphorylation, is sufficient for optimal transcription of the Drosophila sgs genes (Kaplan et al. 2000).
Several CTD-kinases such as TFIIH-associated kinase (cdk7), cdk8, and positive transcription elongation factor b (P-TEFb) associated kinase (cdk9) are known to phosphorylate CTD at Ser 5 in vivo (for review, see Prelich 2002). Although it remains to be determined which CTD kinase(s) is/are responsible for activation of the β-globin gene, one possibility is that LCR-bound factors may stimulate CTD kinase activity in the protein complex at the β-globin gene. Alternatively, a CTD kinase associated with the LCR may be directly recruited to the βmajglobin gene and promote pol II elongation (Fig. 5).
Materials and Methods
Cell lines, culture conditions, and antibodies
The 745A MEL cell line was cultured and induced by DMSO as described previously (Sawado et al. 2001). Control IgG (normal IgG), anti-p45 NF-E2, anti-MafK (p18 NF-E2), anti-pol II (for both phosphorylated and unphosphorylated form), and anti-TFIIB, anti-ERCC3, and anti-TFIIEα, and anti-TFIIEβ were obtained from Santa Cruz Biotechnology. H14, an anti-CTD of RPB1 (phosphorylated at Ser 5) IgM was obtained from Covance. Antiacetylated H3 (Lys 9 and/or Lys 14), antiacetylated H4 (Lys 5, Lys 8, Lys 12, and/or Lys 16), antidimethylated H3 (Lys 4) were obtained from Upstate Biotech. Control IgM (clone K76) was a gift from Dr. M. Roth (Fred Hutchinson Cancer Research Center, Seattle, WA).
ChIP using MEL cells
Chromatin fixation and immunoprecipitation procedures were as described (Shang et al. 2000). Duplex PCR reactions and quantifications were performed as described (Sawado et al. 2001).
Conventional or allele-specific ChIP analyses using mouse splenic cells
Two alleles of the murine β-globin gene, HbbD (D) and HbbS (S), were analyzed in this study. Previously we showed that the expression level of the adult β-globin from D and S alleles in WT(D)/WT(S) mice is quite similar (Fiering et al. 1995). In this study the D allele contains the ΔLCR mutation, while the WT (S) allele is used as a control. For ChIP assays using homozygous mice, we used WT and ΔLCR mice with D alleles. Both strains were also homozygous for a YAC expressing the human β-globin gene (H) as ΔLCR homozygous mice are not viable (Bender et al. 2000). For allele-specific analyses, heterozygous mutant mice ΔLCR (D)/WT (S) with or without one copy of the YAC (H) were used and gave identical results. To increase the erythroid cell population in spleens, ΔLCR (D)/WT (S)-H mice were treated with phenylhydrazine (PHZ) for 3 d and the spleens were harvested on day 6 (Reitman and Felsenfeld 1990). The spleens of ΔLCR (D)/WT (S) mice consist primarily of erythroid cells; thus, PHZ was not used. Spleens were harvested from mice and disrupted in RPMI media containing 2% fetal calf serum to prepare a single cell suspension. After passage through a 70-μm Nylon cell strainer (Falcon), cells were washed once with PBS containing 2% serum and were fixed with 1% formaldehyde for 5 min at room temperature in PBS containing 2% serum. The cross-linking reaction was terminated by adding 1/20 volume of 2.5 M glycine. Cells were washed twice with ice-cold PBS containing 2% serum and then lysed in buffer containing 1% SDS, 5 mM EDTA, and 50 mM Tris-HCl (pH 8.1). Chromatin samples were sonicated to obtain 300–800 bp DNA fragments. Samples were diluted 10-fold by adding buffer containing 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris-HCl (pH 8.1). For all the antibodies except IgMs, chromatin was incubated with normal serum (Jackson Immunoresearch), protein A/G beads in the presence of 20 μg/mL of salmon sperm DNA. Following centrifugation to remove A/G beads, antibodies were added to the supernatant and incubated overnight at 4°C. Samples were then incubated with preblocked beads for 1 h. To harvest immunoprecipitated chromatin, beads were sequentially washed with buffer I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.1, 150 mM NaCl), buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.1, and 500 mM NaCl), buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl at pH 8.1), and then twice with TE. Chromatin was eluted from the beads by washing with elution buffer containing 1% SDS, 0.1 M NaHCO3, and 0.1 mg/mL of proteinase K. Soluble chromatin was reverse-cross-linked overnight in elution buffer overnight at 65°C. DNA was purified using a PCR purification kit (Qiagen). All of the solutions, except TE washes and elution buffer, contain phosphatase inhibitors (microcystin, sodium fluoride, and sodium vanadate), a proteinase inhibitor cocktail (Roche), and histone deacetylase inhibitor (sodium butyrate).
For antiphosphorylated pol II IgM, we used another ChIP procedure described by Nissen and Yamamoto (2000) with minor modifications.
Semiquantitative PCR for allele-specific analyses
Duplex PCR reactions were performed in 1× buffer II (Applied Biosystems), 1.25 mM MgCl2, 0.2 mM dNTP, 5–10 pmole of each globin gene primer set, 5 pmole of the myoD1 gene primer set, 1 μCi [α-32P] dCTP, and 0.05 units of AmpliTaq Gold. The PCR reaction included one cycle of 10 min at 95°C, followed by 30 or 31 cycles of 96°C for 30 sec, 54°C for 30 sec, and 72°C for 30 sec. For allele specific analysis, PCR products were digested with HaeIII for the promoter-proximal region, or BstXI for the third exon of the β-globin gene. All the PCR reactions were performed within the range of linear amplification. The PCR products were separated on a 6% polyacrylamide gel. Quantification of the ChIP results using homozygous mice was done as described previously (Sawado et al. 2001; Schübeler et al. 2001). Quantification of allele-specific analysis was performed as follows.
Enrichment of the β-globin gene relative to myoD1 in WT (EWT) or ΔLCR (EΔLCR) alleles in sample materials relative to those in control IgG bound materials were calculated by:
 |
To account for differences in incorporation of [α-32P] dCTP, constant values for the WT (CWT) or ΔLCR (CΔLCR) alleles were determined based on GC contents of PCR products from each allele. For the adult β-globin promoters, CWT and CΔLCR are 1.25 and 1.0, respectively. For the third exon of the adult β-globin gene, CWT and CΔLCR are 1.00 and 1.26, respectively. ChIP experiments were perfomed at least twice and the standard deviation for each value was determined. When the enrichment from both alleles was significantly high (more than twofold, relative to myoD1), the ratio between the WT and ΔLCR alleles in PCR products (EΔLCR/Ewt) was determined to be significant.
Oligonucleotides for ChIP using homozygous mice and MEL cells were as described previously (Sawado et al. 2001). Oligonucleotide pairs for PCRs using the heterozygous mice were as follows: myoD1 gene (268 bp), md2176, TTCCAGTCTAGCA AGTCCTCAGTT; myoD-s−, TTAGGGATGCCCCCTCTGG CGGA. Digestion of the myoD1 gene with HaeIII or BstXI yields products of 148 and 74 bp, 47 bp, or 169 and 100 bp, respectively. The primer pairs for the βmaj and βSglobin promoter-proximal regions (403 bp) were, β-maj+, GACAAACAT TATTCAGAGGGAGTA; 114706–114728, TGTCTCCAAGCA CCCAACTTCTT.
Digestion of the D allele with HaeIII yields products of 211, 127, and 65 bp, while the S-allele yields 170, 127, 41, and 65 bp. Primer pairs for the 3-exon of βmaj and βSglobin genes (270/273 bp) were: 115558–15583, TAATTTGTCAGTAGTTTAAGGTT GCA; 115827–15805, CATTGTTCACAGGCAAGAGCAGG.
Digestion of the D allele with BstXI yields products of 217 and 53 bp, while the S-allele yields a 273-bp product. Linear amplification in duplex PCR reaction was confirmed for each primer pair. All globin primer pairs were designed to amplify only the βmaj and βSglobin genes without coamplification of the two minor adult globin genes (βmin and βt) or the human β-globin genes in YAC.
Acknowledgments
This work was supported by a fellowship from the Uehara memorial foundation (T.S.), a grant from the Cooley's Anemia Foundation (M.A.B.), an American Society of Hematology Scholar Award (M.A.B.), and NIH Grants DK44746 and HL57620 (M.G.). We thank Drs. Myles Brown and Stephen Buratowski (Harvard Medical School) for technical advice and discussion; Dr. Anton Krumm (Seattle V.A. Hospital) for helpful discussions; Rachel Byron, Andrew Gingras, and Wendy Paulsene (FHCRC) for technical assistance; and Drs. Steve Hahn, Tobias Ragoczy, Mike Bulger, Dirk Schübeler, and Fred van Leeuwen (Fred Hutchinson Cancer Research Center) for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL markg@fhcrc.org; FAX (206) 667-5894.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1072303.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Amrolia PJ, Ramamurthy L, Saluja D, Tanese T, Jane SM, Cunningham JM. The activation domain of the enhancer binding protein p45NF-E2 interacts with TAFII130 and mediates long-range activation of the α- and β-globin gene loci in an erythroid cell line. Proc Natl Acad Sci. 1997;94:10051–10056. doi: 10.1073/pnas.94.19.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993a;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- Andrews NC, Kotkow KJ, Ney PA, Erdjument-Bromage H, Tempst P, Orkin SH. The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc Natl Acad Sci. 1993b;90:11488–11492. doi: 10.1073/pnas.90.24.11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Bender MA, Bulger M, Close J, Groudine M. β-globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT. Going the distance: A current view of enhancer action. Science. 1998;281:61–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes & Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: Toward a model for long-distance gene activation. Genes & Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Bulger M, Sawado T, Schübeler D, Groudine M. ChIPs of the β-globin locus: Unraveling gene regulation within an active domain. Curr Opin Genet Dev. 2002;12:170–177. doi: 10.1016/s0959-437x(02)00283-6. [DOI] [PubMed] [Google Scholar]
- Chan JY, Han XL, Kan YW. Isolation of cDNA encoding the human NF-E2 protein. Proc Natl Acad Sci. 1993;90:11366–11370. doi: 10.1073/pnas.90.23.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Reginato MJ, Andrews NC, Lazar MA. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Daftari P, Gavva NR, Shen CK. Distinction between AP1 and NF-E2 factor-binding at specific chromatin regions in mammalian cells. Oncogene. 1999;18:5482–5486. doi: 10.1038/sj.onc.1202916. [DOI] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Epner E, Reik A, Cimbora D, Telling A, Bender MA, Fiering S, Enver T, Martin DI, Kennedy M, Keller G, et al. The β-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse β-globin locus. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin DI, Enver T, Ley TJ, Groudine M. Targeted deletion of 5′HS2 of the murine β-globin LCR reveals that it is not essential for proper regulation of the β-globin locus. Genes & Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- Forsberg EC, Downs KM, Bresnick EH. Direct interaction of NF-E2 with hypersensitive site 2 of the β-globin locus control region in living cells. Blood. 2000;96:334–339. [PubMed] [Google Scholar]
- Francastel C, Walters MC, Groudine M, Martin DI. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung HL, Kim AY, Hong W, Rakowski C, Blobel GA. Stimulation of nf-e2 dna binding by creb-binding protein (cbp)-mediated acetylation. J Biol Chem. 2001;276:10715–10721. doi: 10.1074/jbc.M007846200. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature. 1994;367:568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- Johnson KD, Christensen HM, Zhao B, Bresnick EH. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- Jones KA, Peterlin BM. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes & Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Kim TH, Maniatis T. Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon-β enhanceosome in vitro. Proc Natl Acad Sci. 1998;95:12191–12196. doi: 10.1073/pnas.95.21.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes & Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Lemon B, Tjian R. Orchestrated response: A symphony of transcription factors for gene control. Genes & Dev. 2000;14:2551–2269. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- Li XY, Virbasius A, Zhu X, Green MR. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Madisen L, Krumm A, Hebbes TR, Groudine M. The immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. Mol Cell Biol. 1998;18:6281–6292. doi: 10.1128/mcb.18.11.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak RA, Sharp PA. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DI. Transcriptional enhancers—On/off gene regulation as an adaptation to silencing in higher eukaryotic nuclei. Trends Genet. 2001;17:444–448. doi: 10.1016/s0168-9525(01)02368-x. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Shavit JA, Igarashi K, Yamamoto M, Engel JD. The world according to Maf. Nucleic Acids Res. 1997;25:2953–2959. doi: 10.1093/nar/25.15.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ney PA, Andrews NC, Jane SM, Safer B, Purucker ME, Weremowicz S, Morton CC, Goff SC, Orkin SH, Nienhuis AW. Purification of the human NF-E2 complex: cDNA cloning of the hematopoietic cell-specific subunit and evidence for an associated partner. Mol Cell Biol. 1993;13:5604–5612. doi: 10.1128/mcb.13.9.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet A, Eick D, Blanchard JM. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene. 1995;10:319–328. [PubMed] [Google Scholar]
- Prelich G. RNA polymerase II carboxy-terminal domain kinases: Emerging clues to their function. Eukaryot Cell. 2002;1:153–162. doi: 10.1128/EC.1.2.153-162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M, Felsenfeld G. Developmental regulation of topoisomerase II sites and DNase I-hypersensitive sites in the chicken β-globin locus. Mol Cell Biol. 1990;10:2774–2786. doi: 10.1128/mcb.10.6.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Sawado T, Igarashi K, Groudine M. Activation of β-major globin gene transcription is associated with recruitment of NF-E2 to the β-globin LCR and gene promoter. Proc Natl Acad Sci. 2001;98:10226–10231. doi: 10.1073/pnas.181344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, Groudine M, Bender MA. The murine β-globin locus control region regulates the rate of transcription but not the hyperacetylation of histones at the active genes. Proc Natl Acad Sci. 2001;98:11432–11437. doi: 10.1073/pnas.201394698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Gross DS. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell. 2001;105:403–414. doi: 10.1016/s0092-8674(01)00329-4. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- Spicuglia S, Kumar S, Yeh JH, Vachez E, Chasson L, Gorbatch S, Cautres J, Ferrier P. Promoter activation by enhancer-dependent and -independent loading of activator and coactivator complexes. Mol Cell. 2002;10:1479–1487. doi: 10.1016/s1097-2765(02)00791-8. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell TK, Turner D, Rupp R, Hollenberg S, et al. The myoD gene family: Nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Yie J, Senger K, Thanos D. Mechanism by which the IFN-β enhanceosome activates transcription. Proc Natl Acad Sci. 1999;96:13108–13113. doi: 10.1073/pnas.96.23.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]