Abstract
We investigated the translational regulation of SCL protein expression and its role in hematopoietic lineage choice. We show that the expression of different SCL protein isoforms is regulated by signal transduction pathways that modulate translation initiation factor (eIF) function. A conserved small upstream open reading frame (uORF) in SCL transcripts acts as a cis-regulatory element for isoform expression. At the onset of erythroid differentiation, truncated SCL protein isoforms arise by alternative translation initiation and favor the erythroid lineage. In comparison, full-length SCL proteins are more efficient at enhancing the megakaryocyte lineage. Together, our studies unravel translational control as a novel mechanism regulating hematopoietic outcome.
Keywords: Hematopoiesis, lineage commitment, translation initiation, uORF, eIF4E, eIF2α
The basic helix–loop–helix (bHLH) transcription factor SCL is essential for vertebrate hematopoiesis and vasculogenesis (Porcher et al. 1996). Deregulated expression of the scl gene is among the most common molecular abnormalities found in human T-cell acute lymphoblastic leukemia (T-ALL; for review, see Robb and Begley 1997). Intriguingly, full-length and a number of smaller SCL protein isoforms are expressed from mRNAs that contain the entire SCL reading frame (Pulford et al. 1995). Results presented here show that SCL protein isoforms with various portions of the N terminus are differentially expressed by a translation control mechanism. In addition, we show that distinct SCL isoforms mediate commitment and differentiation towards alternative hematopoietic lineages. It is intriguing that a similar situation exists with translationally controlled expression of C/EBPα and C/EBPβ isoforms that were shown to differentially recruit various chromatin remodeling and basic transcription factors to decisively affect proliferation and differentiation (Kowenz-Leutz and Leutz 1999; Calkhoven et al. 2000; Pedersen et al. 2001). Our data show that the expression of major hematopoietic transcription factors is translationally controlled and that this type of regulation plays an important role in hematopoiesis.
Results and Discussion
Expression of truncated SCL protein isoforms is induced upon erythroid differentiation
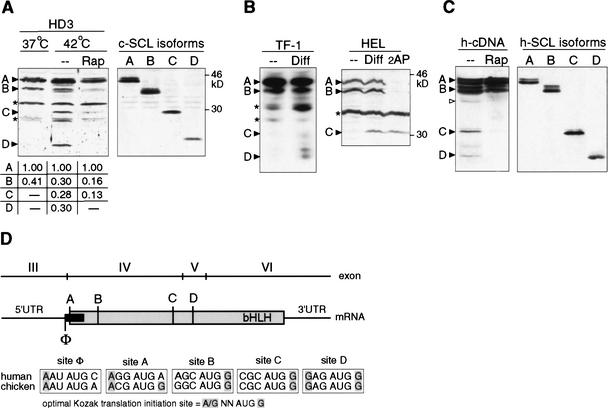
Erythroid differentiation is induced by inactivation of a temperature sensitive (ts) v-erbB oncogene kinase in the chicken HD3 erythroblast cell line (Beug et al. 1982). Similarly, in the human erythroid leukemia cell lines HEL or TF-1 erythroid differentiation is induced by DMSO and erythropoietin, respectively. In all three cases, we observed the appearance of faster migrating SCL peptides in addition to the full-length proteins after induction of differentiation, as shown in Figure 1A and B. The appearance of different SCL isoforms was altered by drugs that interfere with different signaling pathways that regulate translation initiation (De Benedetti and Baglioni 1983; Gingras et al. 2001). Rapamycin (Rap, inhibitor of mTOR) inhibits the appearance of short isoforms in erythroid cells (Fig. 1A), whereas 2-amino purine (2AP, inhibitor of eIF2α-kinases) represses long isoforms (Fig. 1B). Moreover, transient expression of full-length human SCL cDNA in COS1 cells yields a similar isoform pattern that was also rapamycin sensitive (Fig. 1C). The notion that SCL isoforms are generated by differential translation initiation was further supported by the observation that 5′-truncated cDNAs, which permit initiation from consecutive downstream AUG-codons, comigrate with the isoforms observed in HD3 or HEL cells (Fig. 1A,C). These results indicate that translational up-regulation of truncated SCL isoforms occurs early in erythroid differentiation, and that translational control of SCL-isoform expression is conserved in vertebrates.
Figure 1.
Translational controlled differential expression of SCL protein isoforms during erythroid differentiation. (A) Erythroid differentiation in HD3 erythroblasts was induced by temperature shift (37°C–42°C for 12 h) to inactivate ts-v-erbB. (Left) Rapamycin was added 6 h after temperature shift where indicated. (Right) Expression of individual SCL isoforms from sequential translation initiation codons revealed by 5′ deletion of chicken SCL cDNA (termed A–D) in COS-1 cells. SCL-isoform expression was quantified by densitometry of X-ray films and normalized values are depicted below. For immunodetection, specific antiserum against C terminus of chicken SCL was used. (B) Human TF-1 or HEL cells were treated for 4–48 h with Epo or DMSO, respectively, to induce erythroid differentiation. 2-Amino purine (2AP) was added where indicated. Immunodetection of SCL in total cell extracts of TF-1 (data shown for 0 and 4 h postinduction) or HEL cells using a human SCL C terminus-specific antiserum is shown. In HEL cells, the SCL-D isoform was obscured by a nonspecific band and was therefore omitted. (C) Transiently transfected COS-1 cells with HA-tagged human SCL cDNA (left panel), and N-terminal-SCL deletion constructs (A–D, right panel). Cells were treated with rapamycin (Rap), or mock (− −) treated for 24 h where indicated. For immunodetection of SCL, an HA-immunotag-specific antiserum was used. SCL protein bands (▸), an alternative CUG (▹), and nonspecific/unidentified bands (*) are indicated. (D) Representation of the conserved vertebrate scl structure and translation initiation sites. Numbering of exons follows the nomenclature for the human scl gene. The SCL coding region is depicted in gray, the upstream open reading frame (uORF) is depicted in black. Potential translation initiation AUG-sites are designated A–D for SCL and Φ for the uORF. Shading in the initiation sites comparison indicates critical nucleotides at position −3 and +4 according to the optimal Kozak translation initiation consensus sequence (Kozak 1991).
The eukaryotic translation initiation factors eIF2α and eIF4E are rate limiting and decisive for regulated initiation of translation of a subset of mRNAs that have cis-regulatory small upstream open reading frames (uORFs; Calkhoven et al. 2000, 2002; Morris and Geballe 2000; Dever 2002). To further examine the effect of translation initiation factor activity on SCL-isoform expression, a kinase-inactive and dominant-negative form of the eIF2α-kinase PKR (PKRΔ6) that results in hypophosphorylation and constitutive activation of eIF2 (Koromilas et al. 1992), or the cap-binding eIF4E were cotransfected with HA-tagged SCL in HEK293A cells. Figure 2A shows that activation of eIF2 by PKRΔ6 or overexpression of eIF4E both enhance expression of truncated SCL isoforms C and D and an isoform that initiates at an alternative CUG-codon (Fig. 2A; ▹). Similarly, retroviral transfer of PKRΔ6 or eIF4E into HD3 cells enhanced expression of the endogenous SCL isoforms C and D (Fig. 2B). Expression of PKRΔ6 in HEL cells results in a shift in SCL-isoform expression towards more truncated isoforms (Fig. 2C). Analysis of polyA+ RNA by Northern blotting showed that the scl-splicing pattern was not altered by either PKRΔ6 or eIF4E expression in HD3 or HEL cells. Hence, activation of translation initiation by enhancing eIF2 or eIF4E function favors expression of truncated SCL isoforms.
Figure 2.
The SCL protein isoform ratio is modulated by translation initiation factor activity. (A) HA-tagged SCL was cotransfected with PKRΔ6 (to activate eIF2α), eIF4E, or control empty vector (−) into HEK293A. SCL expression was analyzed by immunoblotting using a HA-epitope-specific antiserum. The schematic representations of the SCL mRNA on the left indicate SCL initiation sites in relation to the protein bands. An alternative CUG initiation codon is depicted with an arrowhead (▹). (B) HD3 erythroblasts were infected with retrovirus encoding PKRΔ6, eIF4E, or a control vector (−) and endogenous SCL was examined by immunoblotting using chicken SCL-C terminus-specific antiserum. (C) HEL cells were infected with PKRΔ6 or control virus (−) and endogenous SCL expression was examined by immunoblotting using human SCL-C terminus-specific antiserum. (Right) Expression of eIF4E transgene was controlled by immunoblotting. eIF2α-specific and eIF2α phosphorylation-specific antibodies (eIF2α-P) were used to determine the effect of PKRΔ6 on eIF2α. Because the antibody raised against human eIF2α fails to detect the chicken homolog, a nonspecific band (*) detected by the eIF2α-P antibody serves as an internal control for the loading of HD3 extracts. Northern blots of polyA+ RNA revealed no alterations in scl RNA splicing pattern between control (−) and cells expressing PKRΔ6 or eIF2α.
Differential expression of SCL isoforms depends on conserved mRNA features
The structure of the SCL gene with respect to composition of coding exons and their boundaries is highly conserved (Göttgens et al. 2000). As shown in Figure 1D, three in-frame translation initiation sites designated A, B, and C lie on the same exon IV, whereas initiation site D lies on exon V (Fig. 1D; human exon nomenclature). All but one mRNA splice variants converge on the noncoding exon III. Exon III carries a highly conserved uORF in front of the first SCL initiation codon on exon IV (Aplan et al. 1990). The uORF is out-of-frame with respect to the SCL reading frame (initiation site Φ), overlaps the first SCL initiation A-site, and terminates upstream of the second initiation B-site (Fig. 1D).
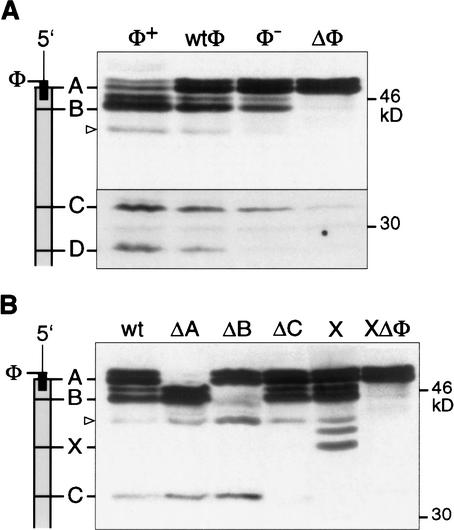
To analyze a potential regulatory function of the uORF in SCL-isoform generation and to rule out a proteolytic origin of SCL isoforms, we used a mutagenesis approach as described previously for the analysis of the translational control of C/EBPα and C/EBPβ (Calkhoven et al. 2000). First, we determined whether the small SCL-uORF serves as a cis-regulatory element in SCL protein isoform expression (Fig. 3A). The uORF was either destroyed by deleting its initiation site (ΔΦ), or converted into a weaker initiation site (Φ−) or an optimal initiation site (Φ+) and compared to the wild-type uORF (wtΦ). As shown in Figure 3A, alteration of uORF initiation site strength drastically alters the distribution of SCL isoforms: Increasing the initiation strength at the uORF (Φ+) enhances expression of truncated SCL isoforms. In contrast, reducing initiation strength (Φ−), or removal of the uORF initiation site (ΔΦ), diminishes or nearly abolishes expression of truncated isoforms, respectively. In addition, removal of the uORF also abrogates rapamycin and 2-amino purine sensitivity (data not shown). Mutations of the initiation sites of SCL isoforms into noninitiation sites (Fig. 3B, ΔA, ΔB, and ΔC) abolished expression of the corresponding SCL isoforms. Notably, mutations that abrogated expression from upstream initiation sites (ΔA, ΔB) simultaneously enhanced expression from downstream initiation sites. Introduction of a novel initiation codon between sites B and C, designated site X (Fig. 3B), generated a novel peptide at the expense of the isoform initiated at site C. Importantly, also expression from the engineered site X was abolished when the uORF was destroyed (Fig. 3B). These results are in accordance with differential translation initiation by a ribosomal scanning and reinitiation mechanism but are not compatible with a proteolytic mechanism of SCL-isoform generation. In summary, these data show that the SCL uORF directs expression of SCL isoforms.
Figure 3.
The uORF of SCL controls translation initiation at alternative initiation sites. COS-1 cells were transiently transfected with SCL constructs and analyzed by immunoblotting using HA-epitope-specific antiserum. (A) The wild-type uORF initiation site (wtΦ) was compared to an optimized initiation context (Φ+), a weakened context (Φ−), or a noninitiation site (ΔΦ). (B) Mutation of alternative translation initiation sites affects SCL-isoform expression. Null (Δ) mutations of the individual initiation sites (A, B, or C) are shown. X indicates an additional initiation site engineered between sites B and C with the uORF initiation codon in wild-type context (X) or removed (XΔΦ). Schematic representations of the SCL mRNA with initiation sites indicated in relation to the protein bands is shown at the left. An alternative CUG initiation codon is depicted with an arrowhead (▹).
Differential SCL-isoform expression determines megakaryocytic versus erythroid lineage outcome
Regulated expression of various SCL isoforms during erythroid differentiation suggested distinct biological functions of individual isoforms. Indeed, retroviral transfer of SCL-A isoform in the HEL cell line results in a strong increase of cells that express the megakaryocytic cell surface marker CD41a, whereas the truncated SCL-D isoform failed to do so (Fig. 4A). These results indicated that the long SCL isoforms preferentially supports megakaryopoiesis.
Figure 4.
Functional differences between SCL protein isoforms in hematopoietic lineage outcome. (A) HEL cells were infected with the empty pMSCVpuro vector (−), or vectors encoding the SCL-A or SCL-D isoform. After puromycin selection cells were analyzed for expression of the megakaryocytic surface marker CD41a by flow cytometry. Data shown are the mean ± S.D. of two independent infections with analysis performed in duplicates. All HEL cells expressed similar amounts of recombinant proteins (data not shown). (B) Mouse bone marrow cells were infected with the empty pMSCV vector (−), or vectors encoding SCL-A (p42) or SCL-D (p22) isoforms. Recombinant SCL was detected in nuclear extracts by immunoblotting with a monoclonal antibody against human SCL. The blot was stripped and reprobed with an anti-E12 antibody as a control. (C) Increase of megakaryocyte colonies by full-length SCL-A. Mouse bone marrow cells were infected with the indicated retroviral constructs, antibiotic (G418)-selected, and plated in methylcellulose culture under selective pressure. Colonies were scored on day 9 of culture. Three independent experiments performed in duplicates are shown for each: control (open circles), SCL-A (gray circles), and SCL-D (black circles); the mean is indicated by a bar. The differences between control, SCL-A, and SCL-D are significant (p < 0.05). (D) Increased numbers of erythroid progenitors in multipotent colonies by SCL-D. Clonal analysis of erythroid development from multipotent progenitors. Mouse bone marrow cells were infected with the indicated retroviral constructs, antibiotic (G418)-selected, and plated in methylcellulose culture supplemented with erythropoietin. On day 10, pluripotent colonies were individually picked and assayed for their contents in erythroid progenitors (CFU-Es). The pluripotent colonies were ranked according to their capacities to give rise to increasing numbers of CFU-Es as shown. Note that SCL-D colonies show the highest incident of CFU-Es. Data shown are typical of two independent experiments.
Differential functions of long versus short isoforms in hematopoiesis were further assessed by introducing the full-length SCL-A isoform, the truncated SCL-D isoform, or a negative control into mouse bone marrow cells by retroviral gene transfer (Fig. 4B). As shown in Figure 4C, the frequency of megakaryocyte colonies was increased threefold by SCL-A, whereas SCL-D was not efficient in inducing megakaryocyte colony formation (Fig. 4C). In comparison, the effect of SCL-A and SCL-D on multipotent colonies was more modest (1.7- and 1.4-fold increase over control, respectively). Alteration in erythroid differentiation was examined by scoring for secondary erythroid colonies (CFU-E) derived from individual multipotent progenitor cells. As shown in Figure 4D, a strong enhancement of CFU-E formation was observed with SCL-D. Expression of the full-length SCL-A isoform increased the number of CFU-Es only to some extent (Fig. 4D). Consistent with CFU-E colony assays, molecular probing of cDNA samples derived from individual colonies displayed highest β-globin levels in SCL-D-isoform-expressing cells (data not shown). Taken together, these observations show that differential expression of alternative SCL isoforms determines cell fate during hematopoietic differentiation: SCL-D favors erythroid differentiation, whereas the SCL-A isoform favors megakaryocyte differentiation.
Translational control as a regulatory event in hematopoietic cell fate determination
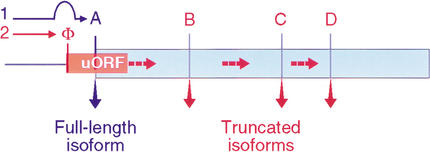
In addition to the bHLH domain, parts of the N-terminal SCL sequences, including those involved in transactivation, are highly conserved between vertebrates suggesting additional functions that have not yet been addressed. Our data show that differential expression of SCL protein isoforms that contain various portions of the N terminus modify lineage output in hematopoiesis. Since distinct SCL isoforms arise from alternative initiation sites, the data entail translational control as a regulatory event in hematopoietic cell fate determination. The translational regulation of SCL depends on its uORF. uORFs relay the activity of eukaryotic translation initiation factors (eIFs) to determine initiation from alternative sites (Fig. 5; Morris and Geballe 2000; Dever 2002). uORF-dependent initiation site selection by sensing activities of eukaryotic initiation factors, eIF2 and eIF4E, has also been observed with the “hematopoietic” transcription factors, C/EBPα and -β (Calkhoven et al. 2000), and ATF4 (Harding et al. 2000; Morris and Geballe 2000). Components of the signaling pathways that constitutively activate eIF4E and/or eIF2 display oncogenic activity which can be blocked by drugs that interfere with eIF functions (Aktas et al. 1998; Palakurthi et al. 2000; Aoki et al. 2001). It is therefore anticipated that signaling pathways involved in adjusting the activity of elFs, in particular the rate-limiting eIF4E and eIF-2α, are of major importance in hematopoiesis. Interestingly, in acute myeloid leukemia or Down's syndrome-related acute megakaryocytic leukemia, the translationally controlled transcription factors C/EBPα and GATA1, respectively, are targets for mutations that disrupt proper isoform expression (Pabst et al. 2001; Wechsler et al. 2002).
Figure 5.
Model of the SCL uORF as a ribosome relay. Translation initiation at site A occurs by leaky-scanning over the uORF initiation site Φ (path 1, blue). Initiation at the uORF relays ribosomes over the initiation site A. Subsequent reinitiation occurs at internal initiation sites B, C, and D (path 2, red, broken arrows). Efficiency of initiation at Φ and efficiency of reinitiation at the downstream sites are regulated by translation initiation factors. Accordingly, the activities of eIFs in conjunction with the uORF determine initiation site preference and thus the expression of various SCL protein isoforms.
Materials and methods
DNA constructs
SCL 5′UTR (exon III) and coding sequences were isolated by reverse transcriptase PCR (RT–PCR) from K562 cells. Constructs were C-terminally tagged with HA-epitope and cloned in pSG5. All mutations were generated on the SCLwt-HA-pSG5 by PCR or site-directed mutagenesis using oligonucleotide primers containing the following mutated sequences: A, ACCATGGCC; B, ACCATGGCC; C, ACCATGGTG; D, ACCATGTTC; E, ACCATGGAG; X, ACGGCCGAA → ACCATGGAA; ΔΑ, AGGATGACC → AGGGGTACC; ΔB, AGCATGGCC → AGCGGG GCC; ΔC, CGCATGGTG → CGCCACGTG; Φ+, AATATGCCC → ACC ATGGCC; Φ− −, AATATGCCC → TGCATGCCC; ΔΦ, AATATGCCC → AATATTCCC. Human SCL cDNAs, encoding A or D isoforms were cloned into the EcoRI site of pMSCV-neo (Hawley et al. 1994). The murine β-globin probe spans the 3′UTR and was a generous gift from Dr. Gordon Keller (Mount Sinai, New York; Robertson et al. 2000).
Cells and tissue culture
COS-1 and HEK293A cells (ATCC, CRL-1650) were propagated and transfected as described in (Calkhoven et al. 2000). Cells were transfected with 6 μg h-SCL-HA-pSG5, 2 μg PKRΔ6-pSG5, or eIF4E-pSG5 and 2 μg carrier pSG5 DNA using a calcium phosphate method. HD3 cells were propagated in DMEM/8% FCS/2% chicken serum/50 μM β-mercapto-ethanol/10 mM HEPES at pH 7.4 at 37°C or 42°C. HEL and TF-1 cells were propagated in RPMI/10% FCS (GIBCO) at 37°C. Erythroid differentiation was induced by 1.5% DMSO for 2 d. HD3 and HEL cells were infected with the retroviral PKRΔ6-pMSCV or eIF4E-pMSCV expression constructs as described in Calkhoven et al. (2000) using the GP2-293 virus producer cell line (Clontech). Rapamycin was used in a concentration of 1 μM, 2-amino purine was used in a concentration of 5 mM (Calbiochem).
FACS analysis
For FACS analysis 2 × 105 cells were stained with FITC-conjugated antihuman CD41a antibody (R&D systems). Control cells were treated with IgGa-FITC. After staining with 1 μg/mL propidium iodide, cells were examined on a FACSCalibur (Beckton Dickinson). Viability gates were set by propidium iodide exclusion.
Bone marrow infection and colony assays
Bone marrow cells were collected from 5-fluorouracil-treated mice and infected as described previously (Krosl et al. 1998). Cells were plated in prestimulation medium (IMDM, supplemented with 15% fetal bovine serum, 5 ng/mL IL-3, 100 ng/mL Steel factor, and 10 ng/mL IL-6) for 2 d. Cells were then infected by cocultivation on confluent monolayers of GP + E-86 cells in the presence of polybrene (8 μg/mL) for 48 h (Krosl et al. 1998). Infection efficiency was in the range of 20%. Following infection, the cells were allowed to recover for 24 h, plated in semisolid IMDM (0.8% methylcellulose, Fluka), supplemented with 10% fetal calf serum (FCS, GIBCO), 50 ng/mL Steel factor, 100 ng/mL IL-11, 10 ng/mL IL-6, Erythropoietin (1 U/mL), 2.7 × 10−5 M α-monothioglycerol (Calbiochem), and 0.8 mg/mL Geneticin (G-418, GIBCO). Colonies were scored on day 9, and multipotent colonies were individually picked. Multipotent colonies were replated into methylcellulose culture, containing erythropoietin (1 U/mL) to assay for late erythroid progenitors (CFU-E). β-globin gene expression was assessed by global amplification of cDNA (Brady et al. 1995). Another batch of cells was taken after infection for Western blot analysis of nuclear extracts. For megakaryocyte colony assays, mouse bone marrow cells were isolated, infected, preselected in G418 for 5 d and plated in methylcellulose culture under selective pressure. Colonies were scored on day 9. For statistic analysis, each sample set (SCL-A or SCL-D) was compared to the control set (empty vector) using t-test (paired two samples for mean).
Western blot analysis
Cell extracts were prepared and Western blot analysis was performed as described (Calkhoven et al. 1994; Lecuyer 2002). The following antisera were used: HA.11, BabCO (1:1000); SCL, Geneka Biotechnology (1:250), human SCL, gift from Richard Baer (Department of Microbiology, Southwestern Medical School, Dallas, TX); monoclonal human SCl, gift from Daniele Mathieu-Mahul (Institut de Genetique Moleculaire, Montpellier, France; 1:1000). eIF2α (C-20), Santa Cruz Biotechnology (0.5 μg/mL); Phosphorylated-eIF2α, Cell Signaling Technology (1:1000); eIF4E, Transduction Laboratories (0.5 μg/mL). The chicken SCL antiserum was raised against the C-term peptide HHAILPVEGSAQR in rabbits (1:1000). Protein bands were quantified by densitometry of X-ray films using the FUJIFILM Science Lab/Image Gauge computer program.
Northern blot analysis
Northern analysis was performed as described in Kowenz-Leutz and Leutz (1999) using 3 μg poly-A+ RNA and 700-bp C-terminal SCL coding sequences as probes recognizing all known splice variants.
Acknowledgments
We thank Dr. Royer-Pokora for providing a hSCL clone, Dr. Richard Baer for human SCL antiserum, Dr. Daniele Mathieu-Mahul for monoclonal antihuman SCL, Eric Lecuyer for help with the Western blots, and Marina Scheller for help with FACS analysis. We thank Drs. T. Green and B. Göttgens for discussion. G.K. was supported by the Canadian Institute for Health Research by grants to T.H. (MT14430), R.M. by a studentship from the National Science and Engineering Research Council of Canada. C.M. was supported by the Deutsche Forschungsgemeinschaft (DFG) by grants to A.L. (LE770/2-3) and C.F.C. by a DFG grant (LE770/3-3) and by a Helmholtz Association fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL aleutz@mdc-berlin.de; FAX 49-30-9406-3298.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.251903.
References
- Aktas H, Fluckiger R, Acosta JA, Savage JM, Palakurthi SS, Halperin JA. Depletion of intracellular Ca2+ stores, phosphorylation of eIF2α and sustained inhibition of translation initiation mediate the anticancer effects of clotrimazole. Proc Natl Acad Sci. 1998;95:8280–8285. doi: 10.1073/pnas.95.14.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki M, Blazek E, Vogt PK. A role of the kinase mTOR in cellular transformation induced by the oncoproteins P3k and Akt. Proc Natl Acad Sci. 2001;98:136–141. doi: 10.1073/pnas.011528498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplan PD, Begley CG, Bertness V, Nussmeier M, Ezquerra A, Coligan J, Kirsch IR. The SCL gene is formed from a transcriptionally complex locus. Mol Cell Biol. 1990;10:6426–6435. doi: 10.1128/mcb.10.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beug H, Doederlein G, Freudenstein C, Graf T. Erythroblast cell lines transformed by a temperature-sensitive mutant of avian erythroblastosis virus: A model system to study erythroid differentiation in vitro. J Cell Physiol. 1982;Suppl. 1:195–207. doi: 10.1002/jcp.1041130427. [DOI] [PubMed] [Google Scholar]
- Brady G, Billia F, Knox J, Hoang T, Kirsch IR, Voura EB, Hawley RG, Cumming R, Buchwald M, Siminovitch K. Analysis of gene expression in a complex differentiation hierarchy by global amplification of cDNA from single cells. Curr Biol. 1995;5:909–922. doi: 10.1016/S0960-9822(95)00181-3. [DOI] [PubMed] [Google Scholar]
- Calkhoven CF, Bouwman PR, Snippe L, Ab G. Translation start site multiplicity of the CCAAT/enhancer binding protein α mRNA is dictated by a small 5′ open reading frame. Nucleic Acids Res. 1994;22:5540–5547. doi: 10.1093/nar/22.25.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPα- and C/EBPβ-isoform expression. Genes & Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- Calkhoven CF, Muller C, Leutz A. Translational control of gene expression and disease. Trends Mol Med. 2002;8:577–583. doi: 10.1016/s1471-4914(02)02424-3. [DOI] [PubMed] [Google Scholar]
- De Benedetti A, Baglioni C. Phosphorylation of initiation factor eIF-2 α, binding of mRNA to 48 S complexes, and its reutilization in initiation of protein synthesis. J Biol Chem. 1983;258:14556–14562. [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- Gottgens B, Barton LM, Gilbert JG, Bench AJ, Sanchez MJ, Bahn S, Mistry S, Grafham D, McMurray A, Vaudin M, et al. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hawley RG, Lieu FH, Fong AZ, Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Leutz A. A C/EBP β isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: Intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosl G, He G, Lefrancois M, Charron F, Romeo PH, Jolicoeur P, Kirsch IR, Nemer M, Hoang T. Transcription factor SCL is required for c-kit expression and c-Kit function in hemopoietic cells. J Exp Med. 1998;188:439–450. doi: 10.1084/jem.188.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Herblot S, Saint-Denis M, Martin R, Begley CG, Porcher C, Orkin SH, Hoang T. The SCL-complex regulates c-kit expression in hematopoietic cells through functional interaction with SP1. Blood. 2002;100:2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, Behre G, Hiddemann W, Tenen DG. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- Palakurthi SS, Fluckiger R, Aktas H, Changolkar AK, Shahsafaei A, Harneit S, Kilic E, Halperin JA. Inhibition of translation initiation mediates the anticancer effect of the n-3 polyunsaturated fatty acid eicosapentaenoic acid. Cancer Res. 2000;60:2919–2925. [PubMed] [Google Scholar]
- Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C. Cooperation between C/EBPα TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes & Dev. 2001;15:3208–3216. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- Pulford K, Lecointe N, Leroy-Viard K, Jones M, Mathieu-Mahul D, Mason DY. Expression of TAL-1 proteins in human tissues. Blood. 1995;85:675–684. [PubMed] [Google Scholar]
- Robb L, Begley CG. The SCL/TAL1 gene: Roles in normal and malignant haematopoiesis. Bioessays. 1997;19:607–613. doi: 10.1002/bies.950190711. [DOI] [PubMed] [Google Scholar]
- Robertson SM, Kennedy M, Shannon JM, Keller G. A transitional stage in the commitment of mesoderm to hematopoiesis requiring the transcription factor SCL/tal-1. Development. 2000;127:2447–2459. doi: 10.1242/dev.127.11.2447. [DOI] [PubMed] [Google Scholar]
- Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]