Abstract
The tobacco protein kinase NPK1 is a MAPKKK that regulates formation of the cell plate during cytokinesis. In the present study, we have identified tobacco NQK1/NtMEK1 and NRK1 as a MAPKK and a MAPK, respectively, downstream of NPK1. NQK1/NtMEK1 complements the mutation in the PBS2 MAPKK gene of yeast in a manner that depends on both NPK1 and its activator, NACK1, a kinesin-like protein. Active NPK1 and NQK1/NtMEK1 phosphorylate and activate NQK1/NtMEK1 and NRK1, respectively. Both NQK1/NtMEK1 and NRK1, as well as NPK1, are activated at the late M phase of the cell cycle in tobacco cells, and they are rapidly inactivated by depolymerization of phragmoplast microtubules. These results suggest the existence of a MAPK cascade that consists of NPK1, NQK1/NtMEK1, and NRK1 and functions in a process related to the architecture of phragmoplasts at the late M phase of the cell cycle. Overexpression of kinase-negative NQK1/NtMEK1 in tobacco cells generates multinucleate cells with incomplete cross-walls. Arabidopsis plants with a mutation in the ANQ1 gene, an ortholog of NQK1/NtMEK1, display a dwarf phenotype, with unusually large cells that contain multiple nuclei and cell-wall stubs in various organs. In addition, anq1 homozygotes set fewer flowers and produce large and malformed pollen grains with a tetrad structure. Thus, NQK1/NtMEK1 (ANQ1) MAPKK appears to be a positive regulator of plant cytokinesis during meiosis as well as mitosis.
Keywords: NPK1 MAPKKK, NQK1 MAPKK, NRK1 MAPK, plant cytokinesis, cell plate, Arabidopsis knockout
Mitogen-activated protein kinase (MAPK) cascades play a central role in the transduction of various extra- and intracellular stimuli. They function in the control of developmental and physiological processes, such as the growth and differentiation of cells and adaptation to stress (Widmann et al. 1999; Chen et al. 2001). They are conserved throughout eukaryotes, and individual organisms have multiple MAPK cascades. In general, each cascade depends on a MAPK kinase kinase (MAPKKK), which phosphorylates and activates a MAPK kinase (MAPKK), which, in turn, activates a MAPK by phosphorylation (Chen et al. 2001). In higher plants, various homologs of components that might participate in MAPK cascades have been identified (Ichimura et al. 2002), and a wide variety of abiotic and biotic stimuli have been shown to activate MAPKs (Jonak et al. 2002). In addition, the alfalfa SIMK MAPK has been shown to be involved in formation of root hairs (Samaj et al. 2002).
MAPK cascades also influence various aspects of cell division (Pages et al. 1993; Minshull et al. 1994; Wang et al. 1997; Takenaka et al. 1997, 1998; Wright et al. 1999; Graves et al. 2000). Recent studies on MAPKs of both animals and plants suggest that cytokinesis, in which complex processes separate a mother cell into two compartments, might be controlled by a MAPK cascade (Calderini et al. 1998; Shapiro et al. 1998; Zecevic et al. 1998; Bögre et al. 1999; Nishihama and Machida 2001; Nishihama et al. 2001, 2002). However, evidence for such a cascade was, until recently, circumstantial, being based on the activation of MAPKs at late M phase and the subcellular localization of MAPKs to the spindle midzone and the midbody (in animal cells) or the cell plate (the developing cross-wall of plant cells).
The NPK1 (nucleus- and phragmoplast-localized protein kinase 1) gene from tobacco encodes a member of the MAPKKK family, and its kinase domain can replace the functions of several yeast MAPKKKs (Banno et al. 1993; Machida et al. 1998). NPK1 and its orthologs in Arabidopsis, ANP1, ANP2, and ANP3, are transcribed in proliferating and division-competent cells (Banno et al. 1993; Nishihama et al. 1997; Nakashima et al. 1998). Recent studies have provided several lines of evidence for a requirement of the NPK1 group of MAPKKKs in cytokinesis (Nishihama et al. 2001; Jin et al. 2002; Krysan et al. 2002). The activity of NPK1 increases during late M phase. Overexpression of a kinase-negative mutant of NPK1 in tobacco cells results in the generation of multinucleate cells with incomplete cell plates. In these cells, phragmoplasts are formed but fail to expand all the way to the cell cortex (Nishihama et al. 2001). Such overexpression or silencing of the NPK1 gene in tobacco plants results in the generation of multinucleate guard cells (Nishihama et al. 2001; Jin et al. 2002). Moreover, anp2 anp3 double mutants of Arabidopsis exhibit defects in cytokinesis, forming unusually large multinucleate cells with cell-wall stubs (Krysan et al. 2002).
We have identified NACK1 and NACK2, novel members of the kinesin-like protein (KLP) family, as factors that increase the activity of NPK1 via formation of a complex (Nishihama et al. 2002). NACK1 accumulates at late M phase and is consistently colocalized with NPK1 at the equatorial zone of the phragmoplast during the phragmoplast expansion from anaphase to late telophase. Overexpression of motor domain-truncated NACK1 eliminates the accumulation of endogenous NPK1 at the equatorial zone of the phragmoplast and also interrupts lateral expansion of the cell plate (Nishihama et al. 2002). These results suggest that both the activity and the proper positioning of NPK1 are critical to cytokinesis. Mutations in the AtNACK1 (HINKEL) gene of Arabidopsis, which is the ortholog of NACK1, also result in the generation of multinucleate cells with incomplete cell walls (Nishihama et al. 2002; Strompen et al. 2002). Thus, it appears that formation of a protein complex that consists of NACK1 and NPK1 controls the formation of the cell plate at cytokinesis.
In the present study, we isolated a cDNA for a tobacco MAPKK, designated NQK1, that acts downstream of NPK1 and a cDNA for a tobacco MAPK, designated NRK1, that acts downstream of NQK1. These protein kinases were activated at late M phase during the cell cycle. Overexpression of kinase-negative NQK1 in tobacco cells resulted in the generation of multinucleate cells with aberrant cell plates, and this phenotype was similar to the phenotype produced by overexpression of kinase-negative NPK1 (Nishihama et al. 2001). Moreover, observations of Arabidopsis plants with mutations in ANQ1, an ortholog of NQK1, revealed that ANQ1 is essential for cytokinesis. A discussion is presented of the role of the NACK1/NPK1/NQK1/NRK1 signaling pathway in plant cells.
Results
Molecular cloning of a cDNA for a MAPKK that acts downstream of NPK1
To isolate a cDNA(s) for a MAPKK homolog(s) that acts downstream of NPK1, we took advantage of Saccharomyces cerevisiae cells (pbs2Δ) in which there is a mutation in the gene for the Pbs2p MAPKK, which functions in the osmosensing MAPK cascade (Brewster et al. 1993; Maeda et al. 1994, 1995). Yeast pbs2Δ cells cannot grow under high osmolarity (Fig. 1A; Brewster et al. 1993). The osmosensitivity of pbs2Δ cells was not suppressed by expression of NACK1 and NPK1cDNAs (data not shown). If a tobacco MAPKK that might be activated by NPK1 could activate Hog1p, then pbs2Δ cells that contain NACK1, NPK1, and such a MAPKK would be expected to grow. To isolate such a MAPKK, we made a cDNA library using mRNA prepared from tobacco BY-2 cells. The cDNAs were under the control of a galactose-inducible promoter, and they were introduced into pbs2Δ cells in which NACK1 and NPK1 cDNAs were expressed. We screened resultant transformants for cells that could form colonies under high-osmolarity conditions in the presence of galactose. We obtained five positive clones from 2.3 × 105 independent transformants. Plasmids containing the cDNAs of interest, derived from two clones (designated 6A and 17A), suppressed the osmosensitivity of pbs2Δ cells in a manner that depended on the expression of both NACK1 cDNA and NPK1 cDNA together (Fig. 1B; data for 17A not shown). We characterized the two cDNAs as described below.
Figure 1.
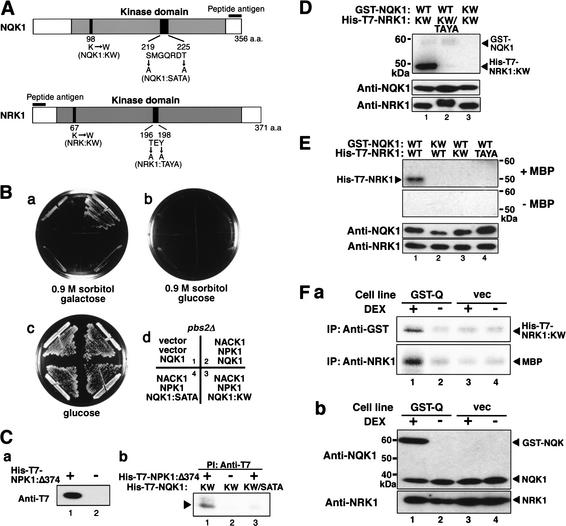
Molecular cloning of the cDNA for a tobacco MAPKK that acts downstream of NPK1. (A) Schematic diagram of the screening system, which exploited the osmoregulatory pathway in Saccharomyces cerevisiae. Osmotic signals, received by the osmosensor proteins Sln1p and Sho1p, activate the Ssk2/22p and Ste11p MAPKKKs, respectively, which, in turn, activate Hog1p MAPK via Pbs2p MAPKK (Brewster et al. 1993; Maeda et al. 1994, 1995; Posas and Saito 1997). We isolated cDNA that complemented the pbs2Δ mutation in a manner that depended on both NPK1 and NACK1. (B) Complementation of the pbs2Δ mutation by cDNA 6A. The pbs2Δ mutant was transformed with different combinations of vectors that encoded cDNAs for NACK1 and NPK1 and cDNA 6A (NQK1 cDNA), and the corresponding empty vectors, as indicated in panel d. Transformants were grown on plates that contained either galactose plus 0.9 M sorbitol (panel a), glucose plus 0.9 M sorbitol (panel b), or glucose (panel c) at 30°C for 4 d. Expression of cDNA 6A was driven by a galactose-inducible promoter. (C) Phosphorylation in vitro of NQK1 by NPK1 activated by NACK1. Immunocomplexes were prepared from lysates of yeast cells that expressed both NACK1 and NPK1 (lane 1), or either NPK1 (lane 2) or NACK1 (lane 3) with NPK1-specific (Anti-NPK1) antibodies and incubated with His-T7-NQK1:KW and [γ-32P]ATP (upper panel). Lysates of yeast cells were immunoblotted with NACK1-specific (Anti-NACK1) and NPK1-specific (Anti-NPK1) antibodies (bottom panel).
Sequence analysis showed that the two cDNAs included identical sequences but differed in length. They encoded a putative polypeptide that was strongly homologous to members of the MAPKK family. We named the putative protein NQK1 (Nicotiana kinase next to NPK1; GenBank accession no. BAB32405). The predicted kinase domain of NQK1 was 38% identical in terms of amino acid sequence to that of Pbs2p (Boguslawski and Polazzi 1987). The entire amino acid sequence of NQK1 was almost identical to that of NtMEK1, a recently identified tobacco MAPKK (the proteins differ at two positions in the kinase domain; Calderini et al. 2001). Although NQK1 and NtMEK1 seem to be encoded by the same gene, for convenience we refer to it as NQK1 in the present paper.
We prepared a DNA construct for expression of a kinase-negative mutant of NQK1 (Fig. 4A, below), in which the lysine codon at the predicted ATP-binding site was replaced by a tryptophan codon (NQK1:KW). Expression of NQK1:KW did not complement the pbs2Δ mutation (Fig. 4B, below), even when NPK1 and NACK1 were coexpressed. Expression of NQK1 cDNA did not suppress the osmosensitive phenotype caused by a mutation in the HOG1 gene (data not shown).
Figure 4.
Biochemical relationship between NPK1 and NQK1 and between NQK1 and NRK1. (A) Positions of the amino acid substitutions in various mutants of NQK1 and of NRK1, and the regions corresponding to the peptide antigens used for raising NQK1- and NRK1-specific antibodies. See text for details. (B) Suppression of the pbs2Δ mutation by NQK1 required its kinase activity and phosphorylation by NPK1. pbs2Δ mutant cells, expressing both NACK1 and NPK1 cDNAs, or neither cDNA, were transformed with vectors that encoded cDNAs for NQK1, NQK1:KW, or NQK1:SATA, as indicated in panel d. Cells were grown on plates that contained galactose plus 0.9 M sorbitol (panel a), glucose plus 0.9 M sorbitol (panel b), or glucose (panel c) at 30°C for 4 d. Expression of NQK1 cDNA and its derivatives was driven by a galactose-inducible promoter. (C) S219 and T225 of NQK1 were phosphorylated by NPK1. Lysates were prepared from yeast cells that expressed His-T7-NPK1Δ374 (panel a, lane 1; panel b, lanes 1,3) or did not express this construct (panel a, lane 2; panel b, lane 2). (Panel a) Immunoblotting of lysates with T7-specific antibodies. (Panel b) Immunocomplexes, prepared from lysates with T7-specific antibodies, were incubated with [γ-32P]ATP and either His-T7-NQK1:KW (lanes 1,2) or His-T7-NQK1:KW/SATA (lane 3). (D) Phosphorylation in vitro of NRK1 by NQK1. His-T7-NRK1:KW (lanes 1,3) and His-T7-NRK1:KW/SATA (lane 2) were incubated with [γ-32P]ATP and either GST-NQK1 (lanes 1,2) or GST-NQK1:KW (lane 3). The top panel shows the autoradiograph and the bottom panel shows results of immunoblotting of reaction mixtures with NQK1- and NRK1-specific antibodies. (E) Activation in vitro of NRK1 as a result of phosphorylation by NQK1. His-T7-NRK1 (lanes 1,2), His-T7-NRK1:KW (lane 3), and His-T7-NRK1:TAYA (lane 4) were incubated with unlabeled ATP and either GST-NQK1 (lanes 1,3,4) or GST-NQK1:KW (lane 2). Reaction products were subjected to the kinase assay within polyacrylamide gels prepared with (+) or without (−) MBP. After renaturation of proteins, the gels were incubated with [γ-32P]ATP. The top panel shows the autoradiograph and the bottom panel shows the results of immunoblotting of the reaction mixtures with the indicated antibodies. (F) Expression of GST-NQK1 in BY-2 cells resulted in activation of NRK1. GST-Q cells (lanes 1,2) and Vec cells (lanes 3,4), at the logarithmic phase of growth, were incubated in medium with (+, lanes 1,3) or without (−, lanes 2,4) 1 μM DEX for 5 h and then protein extracts were prepared. (Panel a) Activities of GST-NQK1 (top) and NRK1 (bottom) were analyzed by immunocomplex kinase assays. (Panel b) Immunoblotting of the extracts with the indicated antibodies. Twenty micrograms of protein was loaded in each lane.
As shown in Figure 1C, histidine- and T7-epitope-tagged NQK1:KW (His-T7-NQK1:KW) was phosphorylated by the immunocomplex prepared with NPK1-specific antibodies from a lysate of yeast cells that expressed both NACK1 and NPK1 (Fig. 1C, lane 1). A very low level of phosphorylation was detected when His-T7-NQK1:KW was incubated with the immunocomplex isolated from yeast cells that expressed either NACK1 or NPK1 (Fig. 1C, lanes 2,3).
Molecular cloning of a cDNA for a MAPK that acts downstream of NQK1
We attempted to use yeast two-hybrid screening to isolate a MAPK that might act downstream of NQK1. Using NQK1:KW as “bait,” we cloned several cDNAs for proteins that were able to interact with NQK1. Two of the isolated cDNAs encoded a putative homolog of MAPK, which was designated NRK1 (Nicotiana kinase next to NQK1). An interaction between recombinant NQK1 and NRK1 proteins was also detected by examination of binding in vitro (data not shown). The predicted amino acid sequence of NRK1 was almost identical to that of NTF6 (there were different amino acids at three positions, one in the noncatalytic region at the N terminus and two in the kinase domain; Wilson et al. 1995). It has been reported that NTF6 might be a MAPK that functions downstream of NtMEK1 (Calderini et al. 2001). However, the nucleotide sequence of NRK1 cDNA (GenBank accession no. BAB32406) was significantly different from that of the NTF6 cDNA: the coding, 5′-untranslated and 3′-untranslated regions were 98%, 98%, and 80%, respectively, identical to each other. Therefore, NRK1 cDNA was apparently derived from a gene different from the NTF6 gene.
We examined two-hybrid interactions among NACK1, NPK1, NQK1, and NRK1; our results are summarized in Table 1. When NQK1 was used as “bait,” it interacted with NPK1 in addition to NRK1, but not with NACK1:ST, a truncated form of NACK1 that lacked a motor domain but included the NPK1-binding region (Nishihama et al. 2002). Interaction of NPK1 with NQK1 required the kinase domain of NPK1 (NPK1:KD), whereas it did not interact with the regulatory domain at the C-terminal half of NPK1 (NPK1:RD). When NRK1 was used as the “bait,” we detected interactions with NQK1 and with NRK1 itself.
Table 1.
Summary of the two-hybrid interactions among NACK1, NPK1, NQK1, and NRK1
|
|
LexA-NQK1
|
LexA-NRK1
|
LexA
|
|---|---|---|---|
| VP16-NACK1:ST | − | − | − |
| VP16-NPK1 | + | − | − |
| VP16-NPK1:KD | + | ND | − |
| VP16-NPK1:RD | − | ND | − |
| VP16-NQK1 | − | ++ | − |
| VP16-NRK1 | ++ | + | − |
| VP16 | − | − | − |
Interactions (++, strong; +, weak; −, none; ND, not done) were estimated by monitoring the growth of yeast cells on histidine-free medium, which was supplemented with 20 mM and 40 mM 3-aminotriazol, respectively, when NQK1 and NRK1 were used as “bait.”
NQK1 and NRK1 are activated in a parallel fashion at late M phase during the cell cycle
Rabbit polyclonal NQK1-specific and NRK1-specific antibodies were raised against synthetic oligopeptides that corresponded to amino acid residues 337–354 of NQK1 and 1–18 of NRK1, respectively. As shown in Figure 2A, the antibodies recognized proteins of 38 kD and 45 kD, which corresponded to the estimated molecular masses of NQK1 and NRK1, respectively, in protein extracts prepared from tobacco BY-2 cells (Fig. 2A, lanes 1,3). When these antibodies were preincubated with the respective antigenic peptides, no signals indicative of an immunoreaction were detected (Fig. 2A, lanes 2,4). Therefore, we concluded that the respective antibodies recognized NQK1 and NRK1 specifically.
Figure 2.
Activation of NQK1 and NRK1 during the cell cycle in tobacco BY-2 cells. (A) Specificity of NQK1-specific antibodies and NRK1-specific antibodies. An extract of BY-2 cells at logarithmic phase was analyzed by immunoblotting with affinity-purified NQK1-specific (lanes 1,2) and NRK1-specific (lanes 3,4) antibodies, which had been preincubated in the presence (+, lanes 2,4) or absence (−, lanes 1,3) of the respective peptide antigens. (B) Activation of NQK1 and NRK1 during the cell cycle. (Panel a) Relative proportions of mitotic cells at different times (in hours) after the removal of aphidicolin. (Panel b) Activities of NQK1, NRK1, and CDKs during the cell cycle. Proteins were extracted from cells that had been harvested at the indicated times (in hours). Immunocomplex kinase assays were performed with NQK1- (top) and NRK1-specific (middle) antibodies using His-T7-NRK1:KW and MBP, respectively, as substrate. NQK1- and NRK1-specific antibodies that had been incubated with the respective peptide antigens (+peptide) were used to confirm that phosphorylation of substrates, as detected here, was due to the activities of NQK1 and NRK1. CDKs were precipitated with p13SUC1 beads, and then CDK-bead complexes were examined for their ability to phosphorylate histone H1 (bottom). (Panel c) Extracts used for the immunocomplex kinase assay were immunoblotted with the indicated antibodies. Twenty micrograms of proteins were loaded in each lane. (C) Activities of NQK1 and NRK1 during the M-to-G1 transition. (Panel a) Mitotic indices (bars) and relative kinase activities of NQK1 (open circles) and NRK1 (filled circles), as determined in panel b, at the indicated times (in hours) after removal of propyzamide. The patterns in bars indicate populations of BY-2 cells at different phases of the M phase, as indicated. (Panel b) Activities of NQK1 (top), NRK1 (middle), and CDKs (bottom) analyzed by the immunocomplex kinase assay or the p13Suc1 bead-complex kinase assay. (Panel c) Immunoblotting of extracts with the indicated antibodies.
We measured the activities of NQK1 and NRK1 in synchronized populations of BY-2 cells by immunocomplex kinase assays. His-T7-tagged kinase-negative NRK1 (His-T7-NRK1:KW), in which the lysine residue at the predicted ATP-binding site had been replaced by a tryptophan residue (Fig. 4A, below), was used as the substrate for NQK1, and myelin basic protein (MBP) was used as the substrate for NRK1 in these assays. The cell cycle of BY-2 cells was arrested at the G1/S boundary by treatment of cells with aphidicolin, and cells were released from the blockage by washing out the drug. Analysis of mitotic indices showed that cells that were in M phase appeared from 6 to 10 h after the removal of aphidicolin and that the highest proportion of M-phase cells was found at 8 h (Fig. 2B, panel a). The highest activities of both NQK1 and NRK1 were detected at 8 and 10 h (Fig. 2B, panel b). Although changes in the activities of NQK1 and NRK1 occurred almost in parallel to changes in mitotic indices, high levels of activities of both protein kinases were still detected even when the mitotic index was reduced at 10 h. These results suggested that NQK1 and NRK1 were activated relatively late in M phase. Immunoblot analysis showed that both NQK1 and NRK1 themselves were present consistently throughout the cell cycle (Fig. 2B, panel c). We also measured the activities of cyclin-dependent protein kinases (CDKs), which were activated prior to activation of NQK1 and NRK1 (2 h), although the maximum activities of these three types of protein kinase were found roughly at the same time (8 h).
To examine more precisely the timing of activation of NQK1 and NRK1 during the M phase, we blocked the cell cycle of BY-2 cells that had been synchronized with aphidicolin at prometaphase using propyzamide, a microtubule-depolymerizing drug, and then we released cells from prometaphase by washing out the drug (Fig. 2C, panel a). No activity of either NQK1 or NRK1 was detected at prometaphase. Both activities increased transiently 0.5–2.0 h after removal of propyzamide: the peak of NRK1 activity was detected at 1 and 1.5 h, but the peak of NQK1 activity seemed to be broader than that of NRK1 activity (Fig. 2C, panel b), consistent with the observations in Figure 2B. It is noteworthy that the proportions of cells in anaphase and telophase were the highest at this time (Fig. 2C, panel a). These patterns of activation of NQK1 and NRK1 were essentially the same as that of activation of NPK1 (Nishihama et al. 2001) and that of the accumulation of NACK1 itself (Nishihama et al. 2002).
Simultaneous decreases in the activities of NPK1, NQK1, and NRK1 upon depolymerization of microtubules
We examined whether the activities of NPK1, NQK1, and NRK1 might be affected by the depolymerization of phragmoplast microtubules (MTs). Accordingly, 60 min after release from prometaphase arrest, when the majority of cells were in anaphase or telophase, a culture of the BY-2 cells was divided into two aliquots, one of which was again treated with propyzamide and one that was not. Treatment with the drug resulted in depolymerization of MTs within 10 min, as monitored by immunofluorescence microscopy with α-tubulin-specific antibodies (data not shown). We measured the activities of NPK1, NQK1, and NRK1 by immunocomplex kinase assays with the respective antibodies. As shown in Figure 3A, the activities of NPK1, NQK1, and NRK1 were detected 60 min after the release of cells from the prometaphase arrest and increased for up to 90 min in the absence of propyzamide (−propyzamide). However, these activities decreased rapidly within 10 min after reintroduction of propyzamide (+propyzamide).
Figure 3.
Rapid decreases in the activities of NPK1, NQK1, and NRK1 upon depolymerization of microtubules. Cultures of BY-2 cells were divided into two aliquots 60 min after the release from prometaphase-arrest (arrows), incubated with (+) or without (−) 6 μM propyzamide, and harvested at the indicated times (in minutes) after the release. (A) Activities of NPK1, NQK1, NRK1, and CDKs. (B) Immunoblotting with the indicated specific antibodies. Twenty micrograms of protein was loaded in each lane.
Levels of NPK1 and NACK1 proteins themselves decrease as cells complete mitosis (Nishihama et al. 2001, 2002). In contrast, levels of NQK1 and NRK1 did not (Fig. 2C). Reintroduction of propyzamide accelerated the decrease not only in the levels of NPK1 and NACK1 but also in the levels of NQK1 and NRK1 (Fig. 3B). However, the decreases in levels of NQK1 and NRK1 themselves occurred much more slowly than the decreases in the kinase activities of NPK1, NQK1, and NRK1. Therefore, decreases in activity might not have resulted solely from protein degradation. The activities of CDKs decreased after the release from propyzamide arrest, but reintroduction of propyzamide stabilized these activities (Fig. 3A). Thus, inhibition of the activities of NPK1, NQK1, and NRK1 was not caused by critical damage to cells by propyzamide.
NPK1 activates NQK1 through phosphorylation
In yeast and animal cells, MAPKKs are activated upon phosphorylation of serine and threonine residues in the S/TXXXS/T motif that is located between kinase subdomains VII and VIII. However, all the plant MAPKKs isolated to date contain five residues between putative sites of phosphorylation (S/TXXXXXS/T). We predicted that serine (S219) and threonine (T225) residues in the SMGQRDT sequence of NQK1 would be sites of phosphorylation by NPK1. To examine this possibility, we replaced S219 and T225 with alanine residues to generate NQK1:SATA (Fig. 4A). NQK1:SATA did not complement the pbs2Δ mutation even when NPK1 and NACK1 were coexpressed (Fig. 4B, panel a, sector 4).
We examined whether S219 and T225 could be phosphorylated by NPK1 by fusing His and T7 epitopes to a constitutively active form of NPK1 (His-T7-NPK1Δ374), in which the C-terminal regulatory domain of NPK1 had been removed (Banno et al. 1993). We prepared protein extracts from yeast cells that expressed this construct and performed immunocomplex kinase assays (Fig. 4C). As shown in Figure 4C, panel b, His-T7-NQK1:KW was phosphorylated by His-T7-NPK1Δ374 (lane 1). No phosphorylation was detected in the absence of His-T7-NPK1Δ374 (Fig. 4C). Recombinant His-T7-NQK1:KW/SATA protein was not phosphorylated by His-T7-NPK1Δ374 (lane 3).
NQK1 phosphorylates and activates NRK1 in vitro and in vivo
We examined whether NQK1 could phosphorylate and activate NRK1 in vitro using recombinant proteins. First, we examined the phosphorylation of His-T7-NRK1:KW by a chimeric recombinant NQK1 protein in which glutathione-S-transferase (GST) had been fused to the N terminus (GST-NQK1). The activity of this recombinant protein was 60-fold higher than that of His-T7-tagged NQK1 (data not shown). As shown in Figure 4D, GST-NQK1 efficiently phosphorylated His-T7-NRK1:KW (Fig. 4D, lane 1) but the kinase-negative mutant (GST-NQK1:KW) did not (Fig. 4D, lane 3). GST-NQK1 failed to phosphorylate His-T7-NRK1:KW/TAYA in which amino acids T196 and Y198 had been replaced by alanine residues (Fig. 4A,D). T196 and Y198 are amino acid residues in the TEY-sequence that are conserved in MAPKs as the motif for phosphorylation by their respective MAPKKs. To examine whether phosphorylation of NRK1 might lead to its activation, we incubated the recombinant NRK1 protein and mutant forms of this protein with GST-NQK1 or GST-NQK1:KW in buffer that contained unlabeled ATP and then NRK1 proteins were subjected to in gel kinase assay with [γ-32P]ATP and MBP as the substrate. His-T7-NRK1 that had been incubated with GST-NQK1, but not with GST-NQK1:KW, phosphorylated MBP within the gel (Fig. 4E, +MBP, lanes 1,2). His-T7-NRK1:KW and His-T7-NRK1:TAYA (Fig. 4A) did not phosphorylate MBP, even when incubated with GST-NQK1 (Fig. 4E, +MBP, lanes 3,4).
To examine whether NQK1 could activate NRK1 in vivo, we generated BY-2 cells that had been transformed with the GST∷NQK1 gene, whose expression was driven by a dexamethasone-inducible (DEX-inducible) promoter (Aoyama and Chua 1997), and with the empty binary vector, which had been used for construction of the DEX-inducible GST∷NQK1 gene, as the control. The former cell line is abbreviated as GST-Q and the latter as Vec. Upon treatment of GST-Q cells with DEX, GST-NQK1 proteins were produced and accumulated to about three times the level of endogenous NQK1 proteins (Fig. 4F, panel b). The GST-NQK1 proteins that were immunoprecipitated with the GST-specific antibodies phosphorylated His-T7-NRK1:KW in vitro (Fig. 4F, panel a, top). An immunocomplex kinase assay with NRK1-specific antibodies revealed that NRK1 proteins were activated in GST-Q cells that had been treated with DEX (Fig. 4F, panel a, bottom). In contrast, immunocomplexes prepared with NRK1-specific antibodies from DEX-untreated GST-Q cells and Vec cells phosphorylated MBP only slightly (Fig. 4F, panel a, bottom). Levels of endogenous NQK1 and NRK1 did not differ significantly among the various cell extracts (Fig. 4F, panel b). These results showed that expression of GST-NQK1 induced the activation of endogenous NRK1 in BY-2 cells.
Overexpression of kinase-negative NQK1 in tobacco BY-2 cells results in generation of multinucleate cells
We examined the effects of overexpression of NQK1:KW on cytokinesis of tobacco BY-2 cells. We transformed BY-2 cells with a DEX-inducible GST∷NQK1:KW construct. We selected two cell lines (GST-Q:KW cell lines) that grew normally without DEX but synthesized GST-NQK1:KW in the presence of DEX (maximum levels were obtained 18–24 h after the addition of 0.1–1.0 μM DEX) but not in the absence of DEX. Vec cell lines were used as controls. As shown in Figure 5A, when GST-Q:KW cells were incubated in the presence of 1 μM DEX, a significant proportion of cells (5%–10%) was binucleate (Fig. 5A, panel d) or multinucleate (Fig. 5A, panel e, eight nuclei) and usually large (Fig. 5A, panel e). Cells with other than two, four, or eight nuclei were rarely seen. Without DEX, all the cells had single nuclei (Fig. 5A, panel c). The Vec lines produced no multinucleate cells (Fig. 5A, panels a,b).
Figure 5.
Generation of multinucleate cells upon expression of kinase-negative NQK1. (A,B) GST-Q:KW cells (A, panels c–e; B) and Vec cells (A, panels a,b) were cultured in the presence (+DEX; A, panels b,d,e; B, panels b,c) or absence (−DEX; A, panels a,c; B, panel a) of 1 μM DEX for 5 d, and then stained with orcein (A), or Calcofluor (blue; B) and PI (red; B). N1 and N2 in A, panel d, and N1–N8 in A, panel e, indicate nuclei in binucleate or octanucleate cells. Arrowheads in B indicate incomplete cell plates. Note that a cell with two sets of chromosomes in B, panel c, is undergoing mitosis. (C,D) Seeds of GST-Q;KW tobacco plants were allowed to germinate on solid medium supplemented with (C, panel b; D, panels b–d) or without (C, panel a; D, panel a) 0.1 μM DEX. (C) Surface views of 10-day-old seedlings. (D) Nomarski images of orcein-stained guard cells (panels a–c) and pavement cells (panel d) of cotyledons. Bars: A,B,D, 20 μm; C, 0.5 cm.
We also examined the formation of cell plates in multinucleate cells by staining with Calcofluor, which detects β-glucans in cell walls. As shown in Figure 5B, almost all the multinucleate cells had incomplete cell plates, which seemed to have been terminated prematurely (Fig. 5B, panels b,c). In DEX-treated Vec cells and DEX-untreated cells, there were no such abnormal structures (Fig. 5B, panel a; data not shown).
We also generated transgenic tobacco plants that harbored the DEX-inducible GST∷NQK1:KW gene (GST-Q:KW plant lines). When seeds of the transgenic plants were allowed to germinate on DEX-containing medium, cotyledons with rough surfaces were formed and they were poorly developed (Fig. 5C, panel b). Analysis of these cotyledons revealed that 5% of guard cells and pavement cells were binucleate and cell plates in some cells were incomplete or undetectable (Fig. 5D, panels b–d). These features can be explained by the occurrence of nuclear division with arrested or delayed cytokinesis during the development of guard cells and pavement cells. We found similar abnormalities in the epidermal cells of cotyledons in which kinase-negative NPK1 was overexpressed (Nishihama et al. 2001).
Disruption of an Arabidopsis ortholog of NQK1 causes cytokinetic defects
A search of sequences in the Arabidopsis genome revealed that the NQK1 gene was closely related to a hypothetical gene designated At5g56580 (AtMKK6; Ichimura et al. 2002), which encoded a deduced amino acid sequence that was 84% identical to that of NQK1. We named this gene ANQ1 (Arabidopsis NQK1). Using a PCR-based screening system (Krysan et al. 1996), we identified a mutant allele of the ANQ1 gene with a T-DNA insert in the first intron (anq1; Fig. 6A). In addition, we found that the mutant had additional copies of part of the sequence of the ANQ1 gene, which might have been duplicated during insertion of the T-DNA. Transcripts that were synthesized from these mutated ANQ1 sequences were detected. Analysis of nucleotide sequences of cDNAs corresponding to the transcripts showed that the 5′-noncoding region and the first exon that includes the putative initiator methionine codon were deleted in all the cDNAs (data not shown), suggesting that no active ANQ1 protein would be synthesized from either transcript in the anq1 mutant even if translation takes place from the first available in-frame ATG codon, which corresponds to the 98th methionine in the kinase subdomain II of wild-type ANQ1.
Figure 6.
Effects of the knockout of the ANQ1 gene on the development of Arabidopsis and on cell division. (A) Schematic diagram of the T-DNA insert in the ANQ1 gene. Boxes represent exons. The T-DNA insert is not drawn to scale. (B–E) Phenotypes of various plants grown on Murashige and Skoog's basic medium. (B) From left to right, wild-type (WT; Wassiliewskija), anq1, and atnack1-1 plants (11 d after vernalization, DAV). (C) anq1 and atnack1-1 plants (11 DAV). (D) Dark-field views of roots of wild-type, anq1, and atnack1-1 plants (11 DAV). (E) Scanning electron micrographs of wild-type and anq1 mature pollen grains. (F) Sections of leaves and roots of plants (12 DAV) stained with toluidine blue. (Panel a) Transverse sections of the fourth rosette leaf of a wild-type plant and third rosette leaves of anq1 and atnack1-1 plants. (Panels b–d) Magnified views of boxed regions in panel a. (Panel e) Transverse sections of roots of wild-type and anq1 plants. Arrows indicate incomplete cell walls. Arrowheads indicate nuclei in multinucleate cells. Asterisks indicate chloroplasts. Bars: B, 1 cm; C, 1 mm; D, 0.2 mm; E,F, panels a,e, 50 μm; F, panelsb–d, 25 μm.
The anq1 homozygous mutant of Arabidopsis exhibited dwarfism, producing small leaves and short roots during vegetative growth (Fig. 6B). The cotyledons had rough surfaces (Fig. 6C) and outgrowths of cells were evident on the root epidermis (Fig. 6D). These features were observed in 26% (n = 339) of progeny of self-pollinated anq1 heterozygotes. A normal phenotype was restored by a transgene that contained the ANQ1 cDNA (data not shown). The phenotype of the anq1 mutant was similar to that of the atnack1 mutant. However, the effects of the anq1 mutation on plant growth and plant morphology seemed to be milder than those of the atnack1 mutation. For example, in contrast to atnack1 plants, whose development ceased at the vegetative stage, nine out of 24 anq1 plants generated short inflorescence stems with two or three flowers (data not shown). Whereas anq1 mutants produced small leaves with smooth surfaces, atnack1 mutants developed small leaves with rough surfaces. As shown in Figure 6F, we found large cells with incomplete cell walls and multiple nuclei in leaves and roots of anq1 mutant plants, as well as in atnack1 mutant plants (Nishihama et al. 2002).
In addition to the growth defect, we found that anq1 homozygotes produced pollen grains that were abnormal in size and shape (62.7%, n = 67; Fig. 6E). We observed unusually large and malformed pollen grains (53.7%), and pollen grains with a tetrad structure (9.0%) were observed. No similar abnormality was found in pollen grains that were produced in anq1 heterozygotes (data not shown). This might have been caused by the supply of ANQ1 protein from the maternal heterozygous plants to pollen cells.
Discussion
NPK1, NQK1, and NRK1 constitute a protein-phosphorylation cascade, the PQR pathway, that operates at late M phase
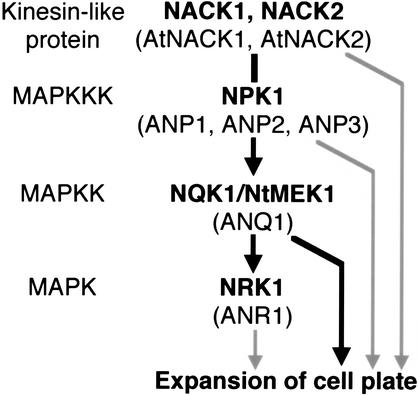
In the present study, we identified two factors that act downstream of NPK1, namely, NQK1 MAPKK and NRK1 MAPK. Our results showed that NPK1 activated NQK1 by phosphorylation and that NQK1 activated NRK1 by phosphorylation. The activities of NPK1, NQK1, and NRK1 increased coordinately during late M phase of the cell cycle in tobacco BY-2 cells (Figs. 2, 3; Nishihama et al. 2001). The timing of these increases in activity was almost identical to that of the accumulation of the NACK1 kinesin-like protein that has the ability to activate NPK1 MAPKKK (Ito et al. 2002; Nishihama et al. 2002). Thus, the accumulation of NACK1 might be the trigger for activation of these three protein kinases. In addition to the coordinated activation of NPK1, NQK1, and NRK1, our results also indicated that maintenance of the kinase activities at the late M phase might be controlled by a common regulatory mechanism that might be related to the structural state of phragmoplast MTs, because these activities decreased rapidly and simultaneously upon depolymerization of phragmoplast MTs, even though the kinase proteins themselves remained detectable (Fig. 3). Therefore, we can conclude that NPK1 (ANP1/2/3), NQK1 (ANQ1), and NRK1 (ANR1: Arabidopsis NRK1 that might correspond to AtMPK13) form a MAP kinase cascade, which may be triggered by binding of the NACK1/2 (AtNACK1/2) kinesin-like protein to NPK1 at the late M phase of the cell cycle in plant cells (Fig. 7). We have designated this cascade the NACK-PQR pathway.
Figure 7.
The MAPK pathway that controls plant cytokinesis. Components of the NACK-PQR pathway in tobacco and Arabidopsis (in parentheses) are shown. The NPK1 MAPKKK is activated by binding with the NACK1 kinesin-like protein (depicted by the vertical solid line; Nishihama et al. 2002). An active NPK1 activates NQK1 by phosphorylation, and the active NQK1, in turn, activates NRK1 by phosphorylation (depicted by solid arrows; see text). Because it is also possible that these components might independently control expansion of cell plates, such pathways are depicted by gray arrows. It remains to be shown whether the NRK1 MAPK functions downstream of NQK1 in cytokinesis (depicted by the gray arrow).
Not only NACK1 and NPK1 but also NQK1 are essential for formation of the cell plate
We investigated the functions of NQK1 using two different approaches. In one case, we used an overexpression system of a dominant-negative mutant of NQK1. Because overexpression of the kinase-negative mutant form of NPK1 resulted in abortion of expansion of both the cell plate and the phragmoplast, with the subsequent generation of multinucleate cells with aberrant cell plates (Nishihama et al. 2001), we examined whether overexpression of kinase-negative NQK1 might also result in similar defects. BY-2 cells and tobacco plants that expressed this mutant protein had phenotypes that were indistinguishable from those generated by overexpression of kinase-negative NPK1 (Fig. 5; Nishihama et al. 2001). The simplest interpretation for this phenomenon is that the mutant NQK1 blocked the activation of endogenous NQK1, with resultant inhibition of the phosphorylation of a substrate protein(s) that is involved in formation of the cell plate. Thus, NQK1 seems to be a positive regulator of the formation of the cell plate. The phenotype might also have been due to an artifact caused by overexpression of the mutant NQK1 construct. However, as described below, our results support a role for NQK1 in cytokinesis.
We analyzed the phenotype associated with a loss-of-function mutation in the ANQ1 gene. Histological observations of anq1 mutant plants revealed the presence of enlarged cells with multiple nuclei and cell-wall stubs in the roots and rosette leaves, and these features are consistent with the results of overexpression of kinase-negative NQK1. The phenotype is typical of various cytokinesis-deficient mutants, such as knolle (Lukowitz et al. 1996), keule (Assaad et al. 1996), atnack1 (hinkel; Nishihama et al. 2002; Strompen et al. 2002), and anp2 anp3 (Krysan et al. 2002). The phenotype of pollen grains of anq1 mutant plants (Fig. 6E) was similar to that of pollen grains from tetraspore, stud, quartet1, and quartet2 mutants (Preuss et al. 1994; Hulskamp et al. 1997; Spielman et al. 1997). The first two mutations result in defects in male meiotic cytokinesis, whereas the latter genes are required for pollen separation. Thus, the phenotype of pollen grains from the anq1 homozygote is consistent with the hypothesis that ANQ1 might be involved in cytokinesis and/or the separation of pollens, as well as in mitotic cytokinesis.
Although the Arabidopsis mutants mentioned above have similar defects in cytokinesis, their developmental phenotypes are slightly different. The atnack1 (hinkel) mutants fail to develop inflorescence stems (Nishihama et al. 2002; Strompen et al. 2002), whereas the anq1 mutant and the anp2 anp3 double mutant do develop such stems (Krysan et al. 2002). Moreover, the anp1 anp2 anp3 triple mutant (Krysan et al. 2002) and the atnack1 atnack2 double mutant (our unpublished data) do not produce any gametes during gametogenesis, suggesting that the ANP1 (also ANP2, ANP3) and AtNACK1 (also AtNACK2) genes are essential for cytokinesis during meiosis, as well as during mitosis. Thus, the phenotype of the anq1 plants is relatively inconspicuous at various developmental stages as compared with the phenotypes associated with mutations in other genes in the NACK-PQR pathway. These observations suggest the possible existence of some other unknown factor(s) whose function overlaps with that of ANQ1. In this context, it is of particular interest that NPK1 can activate the PRKK MAPKK, which is the alfalfa ortholog of AtMKK2 and can, in turn, activate MMK3 MAPK, which is the alfalfa ortholog of NRK1/ANR1 (Cardinale et al. 2002).
For understanding of the molecular network system including NACK1, NPK1, and NQK1, it should be critical to examine functions of an NRK1 MAPK in cytokinetic control. In this context, we examined the effects of a kinase-negative NRK1 on cytokinesis of BY-2 cells, but we detected no abnormalities in cytokinesis associated with this mutant protein (data not shown). Further experiments with NRK1 and its homologs are required to clarify the roles of these MAPKs in cytokinesis.
NQK1 and NRK1 are not localized predominantly in the phragmoplast at M phase
We showed previously that NPK1 is localized at the equatorial zone of the expanding phragmoplast but not at the cell plate. Calderini et al. (1998) and Bögre et al. (1999) reported that antibodies against NTF6 and its alfalfa ortholog, MMK3, respectively, stained developing cell plates. However, we failed to detect the accumulation of NQK1 and NRK1 in phragmoplasts or cell plates in spite of a careful examination by immunostaining, and expression of these proteins as fusions with green fluorescent protein. Subcellular fractionation of phragmoplast-associated proteins revealed that NQK1 and NRK1 were predominantly recovered from the cytoplasmic fraction (our unpublished data), although this result does not exclude the possibility that a limited amount of NQK1 and NRK1 might be recruited to the phragmoplast, where it might be activated and function in cytokinesis. It is also possible that most of the activated NQK1 that has been phosphorylated at the phragmoplast might be released from NPK1 but might function in the formation of the cell plate around the phragmoplast.
The NACK-PQR pathway is curtailed by depolymerization of phragmoplast MTs
Expansion of the cell plate is led by expansion of the phragmoplast, which involves the disassembly and reassembly of MTs at the inner and the outer edges of the phragmoplast, respectively (Yasuhara et al. 1993). It has been proposed that depolymerization of MTs on the inner side of the phragmoplast is one of the processes that is regulated by the NACK1/NPK1 complex during cytokinesis (Nishihama and Machida 2001; Nishihama et al. 2001, 2002). Arabidopsis embryos with mutations in the AtNACK1 (HINKEL) gene have aberrant phragmoplasts in which the central MTs fail to disassemble (Strompen et al. 2002). In addition, the observation that overexpression of motor domain-truncated NACK1 prevents accumulation of NPK1 at the phragmoplast and causes the generation of incomplete cell plates (Nishihama et al. 2002) indicates the importance of colocalization of NPK1 and phragmoplast MTs. In this study, we showed that the activities of all the components of the NACK-PQR pathway disappeared rapidly after the depolymerization of phragmoplast MTs even though the components themselves remained detectable (Fig. 3). This finding suggests not only that activation of the NACK-PQR pathway requires phragmoplast MTs but also that there exists a mechanism for the rapid interruption of the pathway in response to the depolymerization of MTs. A regulatory circuit might operate between activation of the NACK-PQR pathway by NACK1 and various structural states of phragmoplast MTs. If the NACK-PQR pathway were to stimulate depolymerization of phragmoplast MTs, it might be reasonable to postulate that, once MTs have been disassembled, the pathway would be rapidly suppressed to prevent further depolymerization of MTs, as would be necessary for the local control of MT dynamics within the phragmoplast. Such suppression of the pathway by the depolymerization of MTs would also operate after the complete expansion of the phragmoplast at the end of cytokinesis.
Materials and methods
Plant materials and transformation of cells and plants
Tobacco BY-2 cells were maintained in suspension culture, and proliferation of cells was synchronized as described previously (Banno et al. 1993; Nishihama et al. 2001). Transgenic BY-2 cells and tobacco plants were generated by Agrobacterium-mediated transformation (Nishihama et al. 2001). Arabidopsis thaliana plants are described elsewhere (Nishihama et al. 2001; Tanaka et al. 2001).
Construction of plasmids and site-directed mutagenesis
Mutant derivatives of NQK1 and NRK1 were produced by site-directed mutagenesis and the polymerase chain reaction (PCR). The replacement of amino acids and the sites of such replacement are described in Results. Details of the manipulations and primers used for mutagenesis are available from the authors upon request.
cDNAs for NQK1, NRK1, and their mutant derivatives were cloned between the BamHI and NotI sites of pET28a (Novagen, Darmstadt, Germany). A 1.1-kb BamHI–XhoI-digested fragment of pET28-NQK1 and of pET28-NQK1:KW was inserted into pGEX-KG to produce pGEX-NQK1 and pGEX-NQK1:KW, respectively. For genetic analysis using yeast cells, SalI–-NotI-digested PCR-amplified DNA fragments that included the cDNA for NQK1:KW or the cDNA for NQK1:SATA were cloned into a yeast expression vector, pNV7. For expression of His-T7-tagged NPK1Δ374 in yeast cells, a 1.3-kb XbaI–NotI-digested fragment of pET28a that included cDNA for NPK1Δ374 (Banno et al. 1993) was cloned into pKT12. For yeast two-hybrid screening, we constructed pBTM116-NQK1:KW by inserting a 1.1-kb SalI–NotI-digested fragment of pET28-NQK1:KW into a modified form of pBTM116, namely, pBTM116E1. A SalI–SpeI-digested PCR-amplified fragment that contained GST∷NQK1, GST∷NQK1:KW, or GST was inserted into the binary vector, pTA7001 (Aoyama and Chua 1997).
Functional screening using yeast cells
We used mRNA that had been prepared from BY-2 cells at the mid-logarithmic phase of growth for preparation of a cDNA library, which was constructed in pNV7. S. cerevisiae TM344 (a gift from K. Matsumoto, Nagoya University, Nagoya, Aichi, Japan; MATa, ura3, leu2, trp1, his3, lys2, pbs2Δ∷HIS3), harboring YEpGAP-NPK1 (Banno et al. 1993) and pKT11-051 (Nishihama et al. 2002), was transformed with the cDNA library and plated on synthetic medium that lacked tryptophan, uracil, and histidine (SC-WUH). Colonies of 2.3 × 105 independent transformants were replica-plated on YPGalA plates (1% yeast extract, 2% peptone, 2% galactose, 0.2% sucrose, 50 μg/mL adenine sulfate, 2% agar) that contained 0.9 M sorbitol and cultured at 30°C for 4 d. Clones growing on the plates were established on SC-WUH plates and then streaked on YPGalA and YPDA (1% yeast extract, 2% peptone, 2% glucose, 50 μg/mL adenine sulfate, 2% agar) plates supplemented with 0.9 M sorbitol to confirm that growth was galactose-dependent.
Yeast two-hybrid screening
The two-hybrid screening was performed as described by Kitakura et al. (2002). S. cerevisiae L40 [MATα his3-200 trp1-901 leu2-3,112 ade2 LYS2∷(lexAop)4-HIS3 URA3∷(lexAop)8-lacZ] was transformed with pBTM116-NQK1:KW as the “bait” plasmid. A cDNA library was prepared from tobacco BY-2 cells and plated on synthetic medium without histidine.
Production and purification of recombinant proteins
We produced His-T7-tagged recombinant proteins and proteins fused to GST in Escherichia coli BL21(DE3) and DH10B, respectively, and we purified recombinant proteins basically as described by Kitakura et al. (2002) using HiTrap Chelating HP (Amersham Pharmacia Biotech) and glutathione-Sepharose (Amersham Pharmacia Biotech).
Kinase assays in vitro and in gels
Recombinant NRK1 (250 ng) and recombinant NQK1 (50 ng) were incubated in 20 μL of kinase buffer (Nishihama et al. 2001) containing 50 μM ATP and 10 μCi of [γ-32P]ATP (Amersham Pharmacia Biotech) at 25°C for 30 min. Reactions were terminated by addition of 20 μL of 2× sample buffer for SDS-PAGE, and reaction mixtures were fractioned by SDS-PAGE. Radioactivity was visualized with an imaging analyzer (BAS-1800; Fuji Film). For kinase assays in gels, the kinase reaction was performed as described above but without [γ-32P]ATP. The reaction products were then separated by SDS-PAGE on polyacrylamide gels prepared with and without 1 mg/mL MBP (Sigma-Aldrich). Renaturation of proteins and kinase reactions in the gel were then performed as described by Usami et al. (1995).
Production of antibodies and immunodetection of proteins
NQK1-specific antibodies (pQC-L antibodies) and NRK1-specific antibodies (pRN-R antibodies) were raised against the oligopeptides IDFGILVSSLEPPVNFPR (pQC) and MENETNEKLEIKGVPTHE (pRN), respectively, which were synthesized by Sawady Technology. Rabbits were immunized and antibodies were purified as described previously (Nishihama et al. 2001).
Immunoblotting was performed as described by Nishihama et al. (2001). For detection of NACK1, NPK1, NQK1, and NRK1, we used specific affinity-purified antibodies (0.5 μg/mL) as described by Nishihama et al. (2001, 2002). Monoclonal T7-specific antibodies (10,000-fold dilution) were purchased from Novagen.
Preparation for lysates of yeast cells and protein extracts of BY-2 cells
Lysates of yeast cells and protein extracts of BY-2 cells were prepared with TG150 buffer (25 mM Tris-HCl at pH 7.5, 10 mM EDTA, 10 mM EGTA, 150 mM NaCl, 10% glycerol, 1 mM DTT, 20 mM β-glycerophosphate, 1 mM sodium-o-vanadate, 1 mM NaF, and 1 mM PMSF, plus 5 μg/mL each leupeptin, cymostatin, pepstatin A, and antipain), as described previously (Nishihama et al. 2001). Extracts of BY-2 cells for immunoprecipitation of NPK1 and NRK1 were prepared with TG150 buffer that had been supplemented with 1% Triton-X100 and 0.5% NP-40 and with PG150 buffer (TG150 buffer that contained 20 mM PIPES at pH 6.2 instead of Tris-HCl), respectively. Proteins in lysates of yeast cells and extracts of BY-2 cells were quantified with a protein assay kit (Bio-Rad) with BSA as the standard.
Immunocomplex kinase assay
Lysates of yeast cells and extracts of BY-2 cells (0.2 mg of total proteins) were incubated with T7-specific (1 μg), NPK1-specific (1 μg), NQK1-specific (1 μg), or GST-specific antibodies (1 μL; Sigma-Aldrich) at 4°C for 2 h. For immunoprecipitation of NRK1, extracts were incubated with NRK1-specific antibodies (1 μg) at 30°C for 2 h. Then mixtures were incubated with 20 μL of rProteinA-Sepharose beads (50% slurry; Amersham Pharmacia Biotech) at 4°C for 1 h. For precipitation of plant CDKs, extracts of BY-2 cells were incubated with 10 μL of p13SUC1-agarose beads (50% slurry; Upstate Biotechnology, Lake Placid, NY, USA) for 1 h at 4°C for 1 h. Beads were washed four times with 1 mL of the buffer used for extraction and twice with 1 mL of kinase buffer, and then they were incubated in 10 μL of kinase buffer that contained 50 μM ATP, 10 μCi of [γ-32P]ATP, and substrate proteins for 30 min at 25°C for 30 min. His-T7-NQK1:KW (1 μg), His-T7-NRK1:KW (1 μg), MBP (1 μg), and histone (2 μg; Sigma-Aldrich) were used as substrates for NPK1, NQK1, NRK1, and CDKs, respectively. Reactions were terminated by addition of 30 μL of 2× sample buffer for SDS-PAGE. Radioactivity was visualized with an imaging analyzer.
Isolation of a T-DNA-tagged anq1 mutant
We used PCR to screen 133,440 T-DNA-tagged Arabidopsis plants that had been generated at the Arabidopsis Knockout Facility at the University of Wisconsin in a search for an anq1mutant, following the instructions at http://www.biotech.wisc.edu/Arabidopsis and using primers specific for the T-DNA left-border (JL-202), ATQS2 (5′-GGATTGAGAGAAAGCGT TAAGATCAGAG-3′), and ATQSR2 (5′-TATCTACGCGTCT TGAGATCTTGAAATG-3′).
Fluorescence microscopy and light microscopy
All techniques for microscopic analysis were described previously (Nishihama et al. 2001; Tanaka et al. 2001)
Acknowledgments
We thank Professor K. Matsumoto (Nagoya University, Nagoya, Aichi, Japan) for providing yeast strains and Dr. S. Usami (Hiroshima Unversity) for providing the BY-2 cDNA library. This work was supported in part by Grants-in-Aid for Scientific Research on Priority Areas (nos. 10182102 and 14036216) from the Japanese Ministry of Education, Science, Culture and Sports; by a grant from the Research for the Future Program of the Japan Society for the Promotion of Science; and by a grant from Program for Promotion of Basic Research Activities for Innovative Biosciences. R.N. was supported by Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL yas@bio.nagoya-u.ac.jp; FAX 81-52-789-2966.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1071103.
References
- Aoyama T, Chua N-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- Assaad FF, Mayer U, Wanner G, Jürgens G. The KEULE gene is involved in cytokinesis in Arabidopsis. Mol Gen Genet. 1996;253:267–277. doi: 10.1007/pl00008594. [DOI] [PubMed] [Google Scholar]
- Banno H, Hirano K, Nakamura T, Irie K, Nomoto S, Matumoto K, Machida Y. NPK1, a tobacco gene that encodes a protein with a domain homologous to yeast BCK1, STE11 and Byr2 protein kinases. Mol Cell Biol. 1993;13:4745–4752. doi: 10.1128/mcb.13.8.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Calderini O, Binarova P, Mattauch M, Till S, Kiegerl S, Jonak C, Pollaschek C, Barker P, Huskisson NS, et al. A MAP kinase is activated late in plant mitosis and becomes localized to the plane of cell division. Plant Cell. 1999;11:101–113. [PMC free article] [PubMed] [Google Scholar]
- Boguslawski G, Polazzi JO. Complete nucleotide sequence of a gene conferring polymyxin B resistance on yeast: Similarity of the predicted polypeptide to protein kinases. Proc Natl Acad Sci. 1987;84:5848–5852. doi: 10.1073/pnas.84.16.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- Calderini O, Bögre L, Vicente O, Binarova P, Heberle-Bors E, Wilson C. A cell cycle regulated MAP kinase with a possible role in cytokinesis in tobacco cells. J Cell Sci. 1998;111:3091–3100. doi: 10.1242/jcs.111.20.3091. [DOI] [PubMed] [Google Scholar]
- Calderini O, Glab N, Bergounioux C, Heberle-Bors E, Wilson C. A novel tobacco mitogen-activated protein (MAP) kinase kinase, NtMEK1, activates the cell cycle-regulated p43Ntf6 MAP kinase. J Biol Chem. 2001;276:18139–18145. doi: 10.1074/jbc.M010621200. [DOI] [PubMed] [Google Scholar]
- Cardinale F, Meskiene I, Ouaked F, Hirt H. Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell. 2002;14:703–711. [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Gibson TB, Robinson F, Silvestro L, Pearson G, Xu B, Wright A, Vanderbilt C, Cobb MH. MAP kinases. Chem Rev. 2001;101:2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- Graves LM, Guy HI, Kozlowski P, Huang M, Lazarowski E, Pope RM, Collins MA, Dahlstrand EN, Earp HS, III, Evans DR. Regulation of carbamoyl phosphate synthetase by MAP kinase. Nature. 2000;403:328–332. doi: 10.1038/35002111. [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Parekh NS, Grini P, Schneitz K, Zimmermann I, Lolle SJ, Pruitt RE. The STUD gene is required for male-specific cytokinesis after telophase II of meiosis in Arabidopsis thaliana. Dev Biol. 1997;187:114–124. doi: 10.1006/dbio.1997.8554. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang S, Hirt H, Wilson C, et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002;7:301–308. doi: 10.1016/s1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell. 2002;13:1891–1905. doi: 10.1105/TPC.010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Axtell MJ, Dahlbeck D, Ekwenna O, Zhang S, Staskawicz B, Baker B. NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev Cell. 2002;3:291–297. doi: 10.1016/s1534-5807(02)00205-8. [DOI] [PubMed] [Google Scholar]
- Jonak C, Ökrész L, Bögre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signaling. Curr Opin Plant Biol. 2002;5:415–424. doi: 10.1016/s1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- Kitakura S, Fujita T, Ueno Y, Terakura S, Wabiko H, Machida Y. The protein encoded by oncogene 6b from Agrobacterium tumefaciensinteracts with a nuclear protein of tobacco. Plant Cell. 2002;14:451–463. doi: 10.1105/tpc.010360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR. Identification of transferred DNA insertions within Arabidopsisgenes involved in signal transduction and ion transport. Proc Natl Acad Sci. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Jester PJ, Gottwald JR, Sussman MR. An Arabidopsismitogen-activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell. 2002;14:1109–1120. doi: 10.1105/tpc.001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLEgene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or binding of an SH3-containing osmosenser. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- Machida Y, Nakashima M, Morikiyo K, Soyano T, Nishihama R. MAPKKK-related protein kinase NPK1: Involvement in the regulation of the M phase of plant cell cycle. J Plant Res. 1998;111:243–246. [Google Scholar]
- Minshull J, Sun H, Tonks NK, Murray AW. A MAP kinase-dependent spindle assembly checkpoint in Xenopusegg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- Nakashima M, Hirano K, Nakashima S, Banno H, Nishihama R, Machida Y. The expression pattern of the gene for NPK1 protein kinase related to mitogen-activated protein kinase kinase kinase (MAPKKK) in a tobacco plant: Correlation with cell proliferation. Plant Cell Physiol. 1998;39:690–700. doi: 10.1093/oxfordjournals.pcp.a029423. [DOI] [PubMed] [Google Scholar]
- Nishihama R, Machida Y. Expansion of the phragmoplast during plant cytokinesis: A MAPK pathway may MAP it out. Curr Opin Plant Biol. 2001;4:507–512. doi: 10.1016/s1369-5266(00)00208-9. [DOI] [PubMed] [Google Scholar]
- Nishihama R, Banno H, Kawahara E, Irie K, Machida Y. Possible involvement of differential splicing in regulation of the activity of ArabidopsisANP1 that is related to mitogen-activated protein kinase kinase kinases (MAPKKKs) Plant J. 1997;12:39–48. doi: 10.1046/j.1365-313x.1997.12010039.x. [DOI] [PubMed] [Google Scholar]
- Nishihama R, Ishikawa M, Araki S, Soyano T, Asada T, Machida Y. The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes & Dev. 2001;15:352–363. doi: 10.1101/gad.863701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R, Soyano T, Ishikawa M, Araki S, Tanaka H, Asada T, Irie K, Ito M, Terada M, Banno H, et al. Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell. 2002;109:87–99. doi: 10.1016/s0092-8674(02)00691-8. [DOI] [PubMed] [Google Scholar]
- Pages G, Lenormand P, L'Allemain G, Chambard JC, Meloche S, Pouyssegur J. Mitogen-activated protein kinases p42mapk and p44mapk are required for fibroblast proliferation. Proc Natl Acad Sci. 1993;90:8319–8323. doi: 10.1073/pnas.90.18.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: Scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- Pruess D, Rhee SY, Davis RW. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science. 1994;264:1458–1460. doi: 10.1126/science.8197459. [DOI] [PubMed] [Google Scholar]
- Samaj J, Ovecka M, Hlavacka A, Lecourieux F, Meskiene I, Lichtscheidl I, Lenart P, Salaj J, Volkmann D, Bögre L, et al. Involvement of the mitogen-activated protein kinase SIMK in regulation of root hair tip growth. EMBO J. 2002;21:3296–3306. doi: 10.1093/emboj/cdf349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro PS, Vaisberg E, Hunt AJ, Tolwinski NS, Whalen AM, McIntosh JR, Ahn NG. Activation of the MKK/ERK pathway during somatic cell mitosis: Direct interactions of active ERK with kinetochores and regulation of the mitotic 3F3/2 phosphoantigen. J Cell Biol. 1998;142:1533–1545. doi: 10.1083/jcb.142.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman M, Preuss D, Li FL, Browne WE, Scott RJ, Dickinson HG. TETRASPORE is required for male meiotic cytokinesis in Arabidopsis thaliana. Development. 1997;124:2645–2657. doi: 10.1242/dev.124.13.2645. [DOI] [PubMed] [Google Scholar]
- Strompen G, El Kasmi F, Richter S, Lukowitz W, Assaad FF, Jürgens G, Mayer U. The Arabidopsis HINKELgene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr Biol. 2002;12:153–158. doi: 10.1016/s0960-9822(01)00655-8. [DOI] [PubMed] [Google Scholar]
- Takenaka K, Gotoh Y, Nishida E. MAP kinase is required for the spindle assembly checkpoint but is dispensable for the normal M phase entry and exit in Xenopusegg cell cycle extracts. J Cell Biol. 1997;136:1091–1097. doi: 10.1083/jcb.136.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka K, Moriguchi T, Nishida E. Activation of the protein kinase p38 in the spindle assembly checkpoint and mitotic arrest. Science. 1998;280:599–602. doi: 10.1126/science.280.5363.599. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Onouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Machida C, Machida Y. A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsisembryos and juvenile plants. Development. 2001;128:4681–4689. doi: 10.1242/dev.128.23.4681. [DOI] [PubMed] [Google Scholar]
- Usami S, Banno H, Ito Y, Nishihama R, Machida Y. Cutting activates a 46-kilodalton protein kinase in plants. Proc Natl Acad Sci. 1995;92:8660–8664. doi: 10.1073/pnas.92.19.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XM, Zhai Y, Ferrell JE., Jr A role for mitogen-activated protein kinase in the spindle assembly checkpoint in XTC cells. J Cell Biol. 1997;137:433–443. doi: 10.1083/jcb.137.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- Wilson C, Anglmayer R, Vicente O, Heberle-Bors E. Molecular cloning, functional expression in Escherichia coli, and characterization of multiple mitogen-activated-protein kinases from tobacco. Eur J Biochem. 1995;233:249–257. doi: 10.1111/j.1432-1033.1995.249_1.x. [DOI] [PubMed] [Google Scholar]
- Wright JH, Munar E, Jameson DR, Andreassen PR, Margolis RL, Seger R, Krebs EG. Mitogen-activated protein kinase kinase activity is required for the G2/M transition of the cell cycle in mammalian fibroblasts. Proc Natl Acad Sci. 1999;96:11335–11340. doi: 10.1073/pnas.96.20.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara H, Sonobe S, Shibaoka H. Effects of taxol on the development of the cell plate and of the phragmoplast in tobacco BY-2 cells. Plant Cell Physiol. 1993;34:21–29. [Google Scholar]
- Zecevic M, Catling AD, Eblen ST, Renzi L, Hittle JC, Yen TJ, Gorbsky GJ, Weber MJ. Active MAP kinase in mitosis: Localization at kinetochores and association with the motor protein CENP-E. J Cell Biol. 1998;142:1547–1558. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]