Abstract
In Caenorhabditis elegans, an X chromosome-counting mechanism specifies sexual fate. Specific genes termed X-signal elements, which are present on the X chromosome, act in a concerted dose-dependent fashion to regulate levels of the developmental switch gene xol-1. In turn, xol-1 levels determine sexual fate and the activation state of the dosage compensation mechanism. The crystal structure of the XOL-1 protein at 1.55 Å resolution unexpectedly reveals that xol-1 encodes a GHMP kinase family member, despite sequence identity of 10% or less. Because GHMP kinases, thus far, have only been characterized as small molecule kinases involved in metabolic pathways, for example, amino acid and cholesterol synthesis, XOL-1 is the first member that controls nonmetabolic processes. Biochemical investigations demonstrated that XOL-1 does not bind ATP under standard conditions, suggesting that XOL-1 acts by a mechanism distinct from that of other GHMP kinases. In addition, we have cloned a XOL-1 ortholog from Caenorhabditis briggsae, a related nematode that diverged from C. elegans ∼50–100 million years ago. These findings demonstrate an unanticipated role for GHMP kinase family members as mediators of sexual differentiation and dosage compensation and, possibly, other aspects of differentiation and development.
Keywords: C. elegans, XOL-1, sexual differentiation, GHMP kinase, crystal structure
Sex determination is the critical and universal developmental pathway underlying sexual reproduction. Its manifestations are pervasive and often conspicuous. Whereas the presence or absence of the Y chromosome dictates male or female development in mammals, sexual fate in the fruit fly Drosophila melanogaster and the free-living nematode Caenorhabditis elegans is determined genetically by the number of X chromosomes relative to the number of sets of autosomes. In mammals, the primary sex determining gene is SRY, which is present only on the Y chromosome and encodes an HMG domain-containing transcription factor. In the fruit fly, the primary sex determination gene Sex-lethal (Sxl; Maine et al. 1985) is a female-specific trans-acting gene regulator that binds tra transcripts and directs alternative splicing (Inoue et al. 1990). The SRY (Werner et al. 1995) and SXL (Handa et al. 1999) interactions with polynucleotides have been characterized structurally. In C. elegans, sexual differentiation is regulated by the expression levels of the developmental switch gene xol-1. High and low levels of xol-1 result in male (XO) and hermaphrodite (XX) development (Fig. 1), respectively. XOL-1 activity is absolutely required for proper sexual differentiation and male viability (Rhind et al. 1995), but its mechanism of action is unknown.
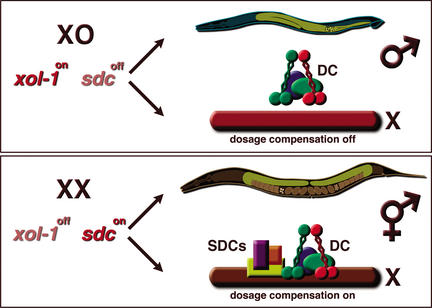
Figure 1.
Genetic control of sex determination and dosage compensation in C. elegans. xol-1 is the primary sex-determination switch gene and the direct molecular target of the X-chromosome counting mechanism. (Top) Male (XO). High xol-1 activity specifies male fate by repressing the activity of sdc-1, sdc-2, and sdc-3 genes. Overt male sexual characteristics include a one-armed gonad (yellow) and a highly specialized tail used for mating. The dosage compensation machinery (colored ovals) does not bind to the X chromosome. Thus, X chromosome gene expression is unregulated (red chromosome). DC, dosage compensation complex (i.e., DPY-26, DPY-27, DPY-28, and MIX-1). (Bottom) Hermaphrodite (XX). Low xol-1 activity specifies hermaphrodite fate by allowing the activation of sdc-1, sdc-2, and sdc-3 genes. Overt hermaphrodite sexual characteristics include a two-armed gonad (yellow, with brown oocytes and embryos) and a tapered tail. The sdc-1,sdc-2, and sds-3 gene products (colored rectangles) recruit a large complex of dosage compensation proteins (DC, colored ovals) to both X chromosomes, repressing X gene expression by half (brown chromosome).
The cooperative activity of at least four X-linked genes, termed X-signal elements, represses expression of xol-1 (for review, see Meyer 2000a). By doubling the number of X-signal elements, an XX embryo reduces xol-1 expression by ∼10-fold (Rhind et al. 1995), facilitating hermaphrodite development. Two C. elegans X-signal elements have been characterized molecularly as follows: FOX-1 (Hodgkin et al. 1994; Nicoll et al. 1997; Skipper et al. 1999), an RNA-binding protein that may regulate alternate splicing of xol-1 RNA, and SEX-1 (Carmi et al. 1998), a nuclear receptor and likely a transcription factor. Although hermaphrodites and males differ in X chromosome number, the expression of most X-linked genes must be equal to ensure viability. This is accomplished through dosage compensation, which reduces expression of X-linked genes in hermaphrodites to male levels (for reviews, see Wood et al. 1997; Hansen and Pilgrim 1999; Meyer 2000b; Boag et al. 2001). High levels of XOL-1 in males correlate with low SDC-2 expression, preventing dosage compensation (Miller et al. 1988; Rhind et al. 1995). Conversely, low levels of XOL-1 in hermaphrodites correlate with high SDC-2 expression and the assembly on the X chromosome of the dosage compensation complex, which is composed of sdc, dpy, and mix-1 gene products (Nonet and Meyer 1991; Chuang et al. 1996; Lieb et al. 1996, 1998; Davis and Meyer 1997; Dawes et al. 1999; Chu et al. 2002). Other genes downstream of xol-1, such as her, tra, and fem, whose activities are inversely related in hermaphrodites and males, coordinate sexual differentiation (Goodwin and Ellis 2002). Null mutants of xol-1 are XO-lethal, inappropriately activating dosage compensation where only one X chromosome is present, whereas XOL-1 overexpression is XX-lethal, deactivating the dosage compensation pathway and elevating the expression of X chromosome genes to lethal levels in hermaphrodites (Rhind et al. 1995).
XOL-1 is an acidic 51-kD nuclear protein (pKa 4.6), whose transcript is expressed at high levels only in pre-comma stage XO embryos (Rhind et al. 1995). XOL-1 transcripts are present in low levels throughout other larval stages in XO animals, but are nearly undetectable in XX larvae and adults of both sexes (Rhind et al. 1995). Currently, XOL-1 is annotated as a subtilisin-like protease on the basis of primary sequence (http://www.wormbase.org). BLAST searches of Genbank failed to identify any homologs that may have provided additional clues as to the function of XOL-1. Thus, we used x-ray crystallography to investigate the function of XOL-1, hypothesizing that the three-dimensional structure of the protein would yield insights into its nature, largely uncharacterized biochemically. The resulting crystal structure of XOL-1 (Fig. 2A) unambiguously and unexpectedly defines XOL-1 as a member of the GHMP kinase family, a family of proteins known to be involved in small molecule metabolism, but not known to participate directly in sexual differentiation or dosage compensation.
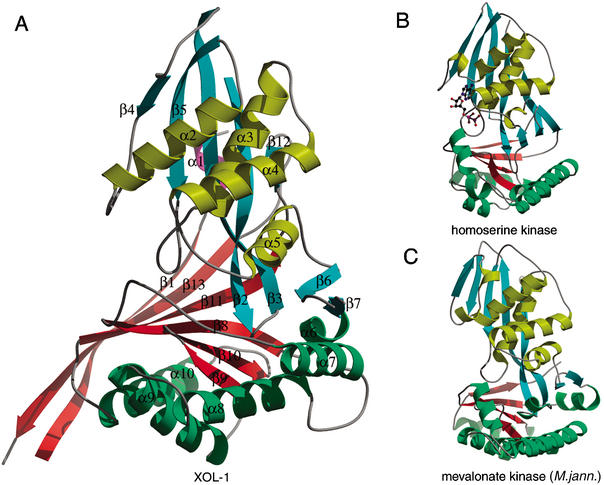
Figure 2.
Comparison of XOL-1 and GHMP kinase structures. (A) The structure of XOL-1. Ribbon diagram of XOL-1 (PDB ID: 1MG7). Domain 1 consists of β-strands 2–7 and 12 (cyan) and α-helices 1–5 (yellow). Domain 2 consists of β-strands 8–11 (red) and α-helices 6–10 (green). Additional strands β1 and β13 and helix α1 form novel elements not found in known GHMP kinase structures. (B) The structure of homoserine kinase bound to ADP (PDB ID: 1FWK). Domain 1 β-strands and α-helices are cyan and yellow, respectively. Domain 2 β-strands and α-helices are red and green, respectively. ADP is represented by a ball and stick model. (C) The structure of mevalonate kinase (M. jannaschii; PDB ID: 1KKH). Colors are as in B.
Galactokinase, homoserine kinase, mevalonate kinase, and phosphomevalonate kinase were originally identified as prototypic members of the GHMP kinase family and observed to contain a conserved Pro–Xaa3–Gly–Leu–Gly–Ser–Ser–Ala–Ala motif (Fig, 3A) that was hypothesized (Tsay and Robinson 1991; Bork et al. 1993), and later proved (Zhou et al. 2000; Krishna et al. 2001; Fu et al. 2002) to be involved in ATP binding. The nucleotide fold of GHMP kinases is distinct from those of other kinases, that is, P-loop, protein, and Hsp70-like kinases (Zhou et al. 2000; Bonanno et al. 2001; Fu et al. 2002; Romanowski et al. 2002; Yang et al. 2002), and binds ATP in both syn and anti-conformations (Zhou et al. 2000; Fu et al. 2002). GHMP kinases are found in bacteria, archaea, and eukaryotes and contain an unusual left-handed β–α–β fold similar to that observed in domain IV of elongation factor G (Zhou et al. 2000). In humans, deficiency of galactokinase, which participates in the conversion of galactose to glucose, contributes to cataract formation (Monteleone et al. 1971; Beutler 1972; Harley et al. 1972; Levy et al. 1972). Mutations in mevalonate kinase, an enzyme involved in the synthesis of sterols from acetate, are associated with mevalonic aciduria (Hoffmann et al. 1986; Schafer et al. 1992; Houten et al. 1999b) and hyperimmunoglobulinemia D/periodic fever syndrome (Drenth et al. 1999; Houten et al. 1999a, 2001; Cuisset et al. 2001; Rios et al. 2001; Simon et al. 2001). To date, no GHMP kinases have been shown to function in developmental pathways unrelated to metabolism.
Results
XOL-1 is a GHMP kinase family member
Full-length XOL-1 (417 residues), as well as various N- and C-terminal truncation mutants, were overexpressed and purified from Escherichia coli. N- and C-terminal truncations were designed on the basis of biological data (Rhind et al. 1995) and secondary structure predictions. A C-terminal truncation mutant (residues 1–374), which retained full rescue activity of xol-1 null males (Rhind et al. 1995), crystallized in the orthorhombic space group P21212 with unit cell dimensions a = 116.1 Å, b = 86.0 Å, and c = 80.2 Å, with two molecules per asymmetric unit (Vm = 2.4 Å3/Da). Initial phases and the positions of selenium atoms of a selenomethionine-derivatized XOL-1 (see Materials and Methods) were determined by MAD at 2.5 Å. The structure was further refined by phase extension using native data to 1.55 Å. The final R values for the refined model are Rcryst = 20.1 and Rfree = 21.2 (Table 1). The final model contains two XOL-1 molecules per asymmetric unit consisting of 703 amino acid residues and 628 water molecules. Superimposition of monomer 2 (chain B) onto monomer 1 (chain A) yielded an r.m.s. deviation of 0.17 Å for all equivalent backbone atoms, indicating no significant differences between the two molecules in the asymmetric unit. The following regions were omitted from the final model for lack of interpretable density: residues 1–7, 29–38, and 190–195 in monomer 1 and residues 1–7, 29–38, and 190–194 in monomer 2.
Table 1.
Crystallographic data and refinement
| Data collection
| |||||
|---|---|---|---|---|---|
| Native | Se_peak | Se_inflection | Se_remotel | Se_remote2 | |
| Space group | P21212 | P21212 | P21212 | P21212 | P21212 |
| Wavelength (Å) | 0.98 | 0.9796 | 0.9794 | 0.9611 | 0.8947 |
| Resolution (Å) | 50.00–1.55 | 50.00–2.30 | 50.00–2.00 | 50.00–2.40 | 50.00–1.81 |
| Completeness (%) | 97.3 (96.9)a | 99.2 (99.8) | 99.4 (99.6) | 83.5 (86.4) | 97.3 (99.1) |
| Unique reflections | 113,135 (12,340) | 67,634 (6,783) | 103,251 (10,355) | 51,374 (5,279) | 151,074 (15,262) |
| Rmerge (%)b | 5.4 (54.0) | 8.9 (41.1) | 7.6 (40.6) | 7.2 (35.1) | 8.1 (56.6) |
| I/σ | 25.2 (2.4) | 14.4 (2.4) | 18.8 (3.1) | 14.3 (2.4) | 21.7 (2.0) |
| Overall figure of Merit | 56.0 | ||||
| Refinement
| |
|---|---|
| Resolution (Å) | 50–1.55 |
| Rcryst(%)c | 20.3 |
| Rfree(%)d | 21.3 |
| Rmsd bonds (Å) | 0.006 |
| Rmsd angles (°) | 1.40 |
| Average B (Å2) | |
| protein | 20.4 |
| water | 30.2 |
| Ramachandran plot (%) | |
| most favored | 92.3 |
| additionally favored | 7.7 |
| generously allowed | 0.0 |
| disallowed | 0.0 |
Numbers in parenthesis refer to the highest resolution shell.
Rmerge = [Σh Σi | Ii(h) − <I(h)>|/Σh ΣiIi(h)] × 100, where <I(h)> is the mean of the I(h) observation of reflection h.
Rcryst = Σhkl | Fo − Fc | /Σhkl | Fo |
Rfree was calculated as for Rcryst, but on 5% of the data, excluded from the refinement.
Submission of the XOL-1 atomic coordinates to the Dali server (Holm and Sander 1993; http://www.ebi.ac.uk/dali) returned surprisingly high Z-scores of 18.5, 17.1, 15.8, 15.2, and 15.1 for five GHMP kinases; mevalonate kinase (Rattus norvegicus; PDB ID:1KVK; Fu et al. 2002), mevalonate kinase (Methanococcus jannaschii; PDB ID:1KKH; Yang et al. 2002; Fig. 2B), phosphomevalonate kinase (Streptococcus pnumoniae; PDB ID:1K47; Romanowski et al. 2002), homoserine kinase (PDB ID:1FWK; Zhou et al. 2000; Fig. 2C), and mevalonate-5-phosphate decarboxylase (PDB ID:1FI4; Bonanno et al. 2001) with r.m.s. deviations for superimposed Cα atoms of 3.8, 3.0, 3.1, 3.5, and 4.0 Å, respectively (Table 2). Despite indisputable structural similarity, XOL-1 displays remarkably little sequence similarity to other members of the family, with overall sequence identities returned from the Dali server of 9%–10%. Even within the GHMP kinase conserved motif1, motif2, and motif3 (Fig. 3A), the sequence is highly divergent. In fact, throughout the entire sequence, only one structurally equivalent residue (Gly 123 in motif2) is absolutely conserved [Figs. 3A (in red), 4B] among XOL-1 and the five GHMP kinase structures deposited in the PDB. Although, across subfamilies of GHMP kinases, sequence indentity can be surprisingly low (Burley and Bonanno 2002), sequence identity is sufficiently high within subfamilies to convincingly delineate related enzymes. Thus, XOL-1 is the first GHMP kinase family member to be identified solely from structural information. In addition, the role of XOL-1 in sexual differentiation was completely incongruous with known GHMP kinase functions.
Table 2.
Statistics from Dali Server
| GHMP Kinase
|
PDB ID
|
Z score
|
R.M.S.D.
|
% identity
|
No. equivalent residues
|
|---|---|---|---|---|---|
| Mevalonate kinase (R. norvegicus) | 1KVK | 18.5 | 3.8 | 9 | 269 |
| Mevalonate kinase (M. jannaschii) | 1KKH | 17.1 | 3.0 | 9 | 242 |
| Phosphomevalonate kinase | 1K47 | 15.8 | 3.1 | 9 | 243 |
| Homoserine kinase | 1FWK | 15.2 | 3.5 | 10 | 251 |
| Mevalonate 5-phosphate decarboxylase | 1FI4 | 15.1 | 4.0 | 9 | 257 |
Figure 3.
Sequence alignment of XOL-1. (A) Structure-based sequence alignment of XOL-1 within GHMP kinase conserved motif regions. Sequence of XOL-1 (bottom) is aligned with structurally equivalent residues from other GHMP kinases. The three GHMP conserved motifs are highlighted in yellow. In XOL-1, these residues are as follows: motif1, residues P45-S53; motif2, residues D120-P128; and motif3, V298-G302. Tandem serines of motif2 are highlighted by boxes. The sequence of XOL-1 is highly divergent, even within conserved motifs, from other GHMP kinases with only one structurally equivalent residue found to be identical (Gly 123 in motif2) throughout the entire sequence. Residues conserved identically in all structures are in red. Highly conserved residues are in blue. Secondary structure is assigned above. PMK, phosphomevalonate kinase; MVK_mj, mevalonate kinase (M. jannaschii); MVK_rn, mevalonate kinase (R. norvegicus); MVPD, mevalonate-5-phosphate decarboxylase; HSK, homoserine kinase; XOL_c, XOL-1 (C. briggsae); XOL_ce, XOL-1 (C. elegans). (B) Alignment of C. elegans and C. briggsae XOL-1 sequences. The C. elegans and C. briggsae XOL-1 sequences were aligned by the clustal method (GONNET series). Gaps and insertions incompatible with the structure were edited manually. Residues corresponding to GHMP conserved motifs are highlighted in yellow. A conserved acidic C-terminal motif is highlighted in green. Identically conserved residues are highlighted in red and conservatively substituted residues are in blue. Secondary structure is assigned above.
The overall secondary structure of XOL-1 is mixed, containing 10 α-helices and 13 β-strands, which fold into an elongated dumbbell-shape (Fig. 2A). As in other GHMP kinases (Zhou et al. 2000; Bonanno et al. 2001; Fu et al. 2002; Yang et al. 2002; Fig. 2B,C), the GHMP kinase core of XOL-1, which excludes β-strands 1 and 13 formed by the extended N and C termini (Fig. 2A), consists of two domains, each composed of four associated α-helices, associated β-strands, and a single, short, isolated α-helical segment. Domain 1 contains a bundle of four α-helices (α2, α3, α4, α5; Fig. 2A, in yellow), a β-sheet (β12, β2, β3, β5, β4) with two associated strands (β6, β7; Fig. 2A, in cyan), and a short, isolated α-helix (α1; Fig. 2A, in magenta). Domain 2 contains four α-helices packed side-by-side (α7, α8, α9, α10; Fig. 2A, in green) on one side of the β-sheet (β9, β10, β8, β11; Fig. 2A, in red), and a short α-helix (α6; Fig. 2A, in green), which connects the β-sheets from domains 1 and 2.
Identification of a XOL-1 ortholog in C. briggsae
C. briggsae diverged from C. elegans ∼50–100 million years ago. The recent, but not yet complete, sequencing of the C. briggsae genome (The Sanger Institute, http://www.sanger.ac.uk; The Genome Sequencing Center, http://genome.wustl.edu) allowed us to predict an apparent ortholog of xol-1. The existence of the expressed gene was confirmed by isolating and sequencing corresponding cDNA fragments from C. briggsae. Although the overall amino acid sequence identity is low (22% identical, 41% conservatively substituted; Fig. 3B), significant homology is found in key locations. Within strands β9 and β10 and the intervening loop that defines motif3, the sequences are 68% identical and the unusual QQGG sequence is completely conserved (Figs. 3B, 4C,D). The critical glycine (123) in motif2 is also conserved [Figs. 3B (in red), 4B]. Synteny reinforces the view that the xol-1 genes in C. briggsae and C. elegans are orthologs. In C. elegans, the xol-1 gene is 10 kb upstream of dim-1. Similarly, in C. briggsae, xol-l and dim-1 are in close proximity (32 kb) on the same chromosome (see Materials and Methods). Although the biological role of C. briggsae XOL-1 is unknown, the identification of a xol-1 ortholog indicates that the C. elegans XOL-1 protein is not simply an isolated case, and that the GHMP kinase fold has likely been adapted by a much more divergent array of protein sequences than had been assumed previously. Studies to analyze the function of C. briggsae xol-1 are underway.
Figure 4.
The putative active-site of XOL-1. (A) The cavity that represents the putative active-site of XOL-1 and, in other GHMP kinases, binds ATP is represented as an electrostatic surface nestled between domains 1 and 2. Red is electronegative and blue is electropositive (calculated in GRASP; Nicholls et al. 1991; −15 to +15 kT/e). Secondary structure elements are colored as in Figure 2A. Side chains of charged and polar residues lining the cavity are represented by ball and stick. (B) Same as A, but with view rotated by ∼180°. The location of the Cα carbon of Gly 123 is represented by a red dot. (C) The cavity that represents the putative active site of XOL-1 is represented as a surface colored by conservation. Side chains of residues identical between C. elegans and C. briggsae XOL-1 are in dark blue, whereas those conservatively substituted are in cyan with the corresponding colors mapped onto the surface. The strongly conserved region encompassing motif3 (β9–β10) is colored dark blue. (D) Same as C, but with view rotated by ∼180°.
The putative catalytic site of XOL-1 and implications for activity
The putative active site of XOL-1 is a deep cavity nestled in the interface of domains 1 and 2 (Fig. 4), a position analogous to that occupied by the active sites of other GHMP kinases (Fig. 2B). The surface of the cavity is largely electronegative with interspersed electropositive regions defined by Lys 124 and Arg 264 (Fig. 4A,B). The negatively charged residues lining the cavity are Asp 52, Asp 120, Asp 220, Glu 276, and Glu 297 (Fig. 4A,B), only one of which (Asp 120) is apparently conserved in the C. briggsae ortholog (Asp 116; Fig. 4C,D). The phosphate-binding loop (residues 120–129), which connects β5 and α3, straddles the back of the cavity (Fig. 4B). Although the XOL-1 motif2 contains two tandem serines (121, 122; Fig. 3A, in boxes), as in other GHMP kinases, the positions of these residues are not analogous. Whereas in other GHMP kinases the tandem serines are incorporated into the first turn of the ensuing α-helix, these positions are occupied by the hydrophobic residues Ala 132 and Val 133 in XOL-1, and the tandem serines are located within the loop several residues before the start of the corresponding α-helix, α3 (Fig. 4A,B). The only structurally equivalent phosphate-binding loop residue conserved between XOL-1 and other GHMP kinases appears to be Gly 123 [Figs. 3A (red within motif2), 4B). A functional requirement for a strong positive charge at position Lys 124 is suggested by conservative substitution by Arg 120 in the C. briggsae ortholog (Fig. 3B, motif2). A constellation of residues (Gly 295 to Lys 309) at the bottom of the cavity formed by the putative active site of XOL-1 is also conserved in the C. briggsae ortholog (Figs. 3B, 4C,D; β9 and β10, in dark blue), a characteristic that may correlate with functional requirements.
In homoserine kinase and mevalonate kinase (R. norvegicus), Glu 130 and Glu 193 carboxylates, respectively, coordinate the nucleotide phosphate liganded Mg2+. Glu 149 in mevalonate kinase (M. jannaschii) and Ser 153 carbonyl oxygen and Oγ in mevalonate-5-phosphate decarboxylase occupy the analogous position. In phosphomevalonate kinase, nearby Asp 144 and Asp 150 provide possible Mg2+-chelating moieties. XOL-1 presents a marked structural incongruity with regard to this important Mg2+-binding region. This location in XOL-1 is entirely hydrophobic, bounded by Val 127, Ile 128, Ile 159, Leu 164, Ile 166, and Ile 190 side chains and devoid of any charged or polar groups, even from the peptide backbone, to coordinate Mg2+.
The surprising structure of XOL-1 implies that, in this case, the GHMP kinase fold acts as a developmental gene regulator or that sexual differentiation in C. elegans is regulated in a manner only remotely considered previously, that is, by a small secondary effector molecule. Thus, the structure suggests new avenues of experimentation that otherwise would likely have been overlooked.
The XOL-1 used in the structure determination (residues 1–374) lacks the highly acidic feminizing C terminus (residues 375–417), yet retains complete ability to rescue xol-1 null males (Rhind et al. 1995). Therefore, the structure does not provide information as to the function of this short C-terminal domain, which, thus far, has not been observed in other GHMP kinases. However, it can be deduced from the structure that the GHMP kinase core of XOL-1 does not incorporate the acidic C terminus. A conserved acidic motif at the extreme C termini is apparent in the aligned C. elegans and C. briggsae XOL-1 orthologs (residues 408–413 in C. elegans XOL-1; QDDTYD, residues 416–421 in C. briggsae XOL-1; NDDTFD), the functional significance of which is unknown (Fig. 3B, highlighted in green).
XOL-1 binding to ATP is not detectable
The characterization of XOL-1 as a member of the GHMP kinase family of proteins led us to test whether XOL-1 could bind ATP. UV cross-linking failed to detect binding of XOL-1 to isotopically labeled ATP (data not shown). Likewise, residual XOL-1-catalyzed ATP hydrolysis was not detectable in an enzyme-coupled colorimetric assay (Fig. 5A). These results were confirmed by fluorescence spectroscopy. The fluorescent probe TNP-ATP, which had been used in studies of human mevalonate kinase (Cho et al. 2001), did not display any significant enhancement of fluorescence in the presence of XOL-1 (Fig. 5B). In addition, crystals were both soaked and grown in the presence of a 10-fold molar excess of either ATP or the nonhydrolyzable analog AMP-PNP. However, no additional density was observed in the putative catalytic site in electron density maps calculated from x-ray diffraction data collected from these crystals (data not shown). Therefore, XOL-1 may not be a small molecule kinase, having evolved and adapted a GHMP kinase scaffold to execute a completely novel function. The lack of sequence similarity present between XOL-1 and other GHMP kinases, even within previously identified conserved motifs (Fig. 3A), supports this conclusion. Of note, the left-handed β–α–β fold in the N-terminal domain of XOL-1 and other GHMP kinases, termed the ribosomal protein S5 domain 2-like fold, is found in numerous RNA/DNA-binding proteins, raising the intriguing possibility that XOL-1 may interact with polynucleotide substrates, although, for this to be the case, significant conformational change in the active site would seem to be required. Structural similarity has also been noted between an RNA-binding protein domain, for example, domain V of elongation factor G, and the C-terminal domain of homoserine kinase (Zhou et al. 2000). Interestingly, RNA-binding domain 2 of the Sex-lethal protein has a fold similar to that of elongation factor G. The possibility remains that XOL-1-ATP binding may require an additional uncharacterized interaction.
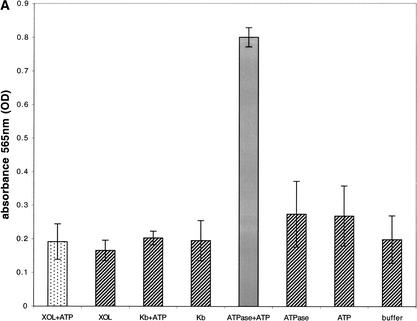
Figure 5.
XOL-1 ATP hydrolysis and binding. (A) In the presence of XOL-1 (10 μg; stippled bar), absorbance at 565 nm (Y-axis) is not significantly increased over the absorbance observed in any negative controls (hatched bars) in an enzyme-coupled colorimetric ATPase assay (see Materials and Methods). Negative controls (hatched bars) XOL-1, H-2Kb + 1 mM ATP, H-2Kb, Na+/K−-ATPase (5 milliunits), 1 mM ATP, buffer. As a positive control (gray bar), N+/K−-ATPase (5 milliunits) was incubated with 1 mM ATP. (B) The fluorescence emission of 20 mM ATP-TNP (○) is not significantly increased in the presence of 20 μM XOL-1 (▴). As a positive control, 20 μM hexokinase was incubated with 20 μM TNP-ATP (▵). Fluorescence emission intensity (Y-axis) is represented as function of wavelength (505–600 nm, X-axis). Excitation wavelength = 410 nm.
Implications of crystal packing for oligimeric structure
Although the biological oligomerization state of XOL-1 is unknown, crystal packing suggests that XOL-l might be a dimer or even a tetramer. One possible mode of dimerization evident from the crystal packing of XOL-1 involves the reciprocal interactions of domains 1 and 2 from neighboring monomers (Fig. 6A) with a buried surface area of 2146 Å2 per dimer, slightly less than that calculated for R. norvegicus mevalonate kinase (2450 Å2; Fu et al. 2002). The additional structural feature formed by the unique N- and C-terminal β-strands (β1 and β13) of XOL-1 (Fig. 3A) presents a second possible interface for dimerization. XOL-1 molecules pack in the crystal such that β1 strands link β-sheets from adjacent monomers to form an extended 12-stranded antiparallel β-sheet comprised of 6 strands from each monomer (β9, β10, β8, β11, β13, β1; Fig. 6B). As a caveat, extended β-sheets that do not correlate with functional dimerization are sometimes observed in crystal structures (Schiffer et al. 1985). In addition, XOL-1 elutes as a monomer during size exclusion chromatography (data not shown).
Figure 6.
Crystal packing interactions of XOL-1 monomers. (A) Interface 1 consists of reciprocal interactions of regions 1 and 2 from neighboring monomers with a buried surface area of 2146 Å2 per dimer. (B) Interface 2 consists of anti-parallel hydrogen bonding between β1-strands of neighboring monomers, creating a 12-stranded anti-parallel β-sheet (red) comprised of 6 strands from each monomer (β9, β10, β8, β11, β13, β1). (C) The central cavity of the tetramer in the crystal has a 45 Å diameter at its core. Secondary structure elements are colored as in Figure 3.
Crystal structures of homoserine kinase (Zhou et al. 2000) and mevalonate kinase (R. norvegicus; Fu et al. 2002) reveal a possible mode of dimerization mediated through interactions between C-terminal domains. However, the crystal structure of XOL-1 does not reflect a similar mode of dimerization, as the molecules are packed in a head-to-toe array with corresponding C-terminal dimerization interfaces distal to each other, an arrangement more similar to that seen in the mevalonate decarboxylase dimer (Bonanno et al. 2001). The two potential modes of XOL-1 dimerization combine to form a tetrameric entity with a large central cavity having a diameter of ∼45 Å at its core (Fig. 6C).
Discussion
The identification of XOL-1 as a GHMP kinase family member through the elucidation of its crystal structure is difficult to reconcile with its role as a developmental regulator. However, gene regulation by GHMP kinase family members has precedent in yeast. Saccharomyces cerevisiae Gal3p, whose primary structure is highly similar to that of galactokinase, is a galactose- and ATP-dependent transcriptional inducer of GAL genes (Platt and Reece 1998; Platt et al. 2000). Moreover, GAL1 of Kluyveromyces lactis is a bifunctional protein that acts as an inducer of GAL genes and a galactokinase. (Meyer et al. 1991). As our understanding of how S. cerevisiae Gal3p and K. lactis GAL1 induce gene expression is incomplete, their relationship to XOL-1 does not enable us to predict a detailed mechanism; however, circumstantial evidence suggests a mechanism of gene regulation that is conserved across phyla.
The principles of sexual dimorphism and dosage compensation are conserved in multicellular eukaryotes, but the mechanisms are not. For instance, in female mammals, a single X chromosome is inactivated randomly to reduce the overall X-linked expression by half, thereby reducing X chromosome gene expression to male levels. In the fruit fly, the single male X chromosome is hypertranscribed, such that X chromosome gene expression is doubled to female levels. In C. elegans, transcription from both hermaphrodite X chromosomes is reduced by half. With only one XOL-1 ortholog identified, the extent to which XOL-like GHMP kinases might regulate sexual fate and dosage compensation in other eukaryotes cannot be assessed. Because dosage compensation in different eukaryotic lineages is thought to have evolved independently, regulation of sexual fate and dosage compensation by XOL-like GHMP kinases may be limited to C. elegans and related nematodes. However, XOL-like GHMP kinases may also participate considerably more extensively, not only in the regulation of sexual fate and dosage compensation, but in other modes of development and differentiation. The accelerated rate at which genes involved in sexual differentiation and dosage compensation have evolved may obscure related gene sequences. Presumably, XOL-1 is a case of rapidly divergent evolution as such a remarkable degree of structural similarity would less likely be manifested by a case of convergence. Ambiguities will likely only be resolved when both sequence and structure space are more densely populated. To this end, structural genomic (Bonanno et al. 2001; Burley and Bonanno 2002) and whole-genome sequencing projects will undoubtedly provide indispensable insights.
Nematodes, which may be free-living or parasitic, are among the most numerous animals on earth and populate terrestrial, aquatic, and marine environments in abundance. C. elegans, whose entire genome has been sequenced (The C. elegans Sequencing Consortium 1998), is the most thoroughly studied nematode species and a useful organism for the study of parasitic nematodes (Burglin et al. 1998; Favre et al. 1998; Ashton et al. 1999; Blaxter and Ivens 1999). Efforts to characterize the genomes of parasitic nematodes (The Filarial Genome Project: http://nema.cap.ed.ac.uk/fgp.html) have already resulted in the identification of the apparent homolog of C. elegans male sex determination gene her-1 in Brugia malayi (Streit et al. 1999). A her-1 homolog has also been identified in C. briggsae (Streit et al. 1999), and homologs of C. elegans hermahrodite sex determination gene tra-2 have been identified in C. briggsae (Kuwabara and Shah 1994) and Caenorhabditis ramanei (Haag and Kimble 2000).
These findings imply that, despite differing strategies of sexual reproduction between nematode species, some elements of the process are conserved. We infer from these observations that XOL-like GHMP kinases may play an important role in the development and differentiation of various nematodes. Furthermore, XOL-like GHMP kinases may regulate differentiation in related organisms such as other helminths, for example, schistosomes and, perhaps even more broadly, in other metazoans. It is likely that the precise mechanism of XOL-1 activity will eventually emerge from studies based on the discoveries described herein. In summary, the structure of XOL-1 demonstrates that the GHMP kinase family of proteins is much more diverse in sequence and function than had been presumed, and that critical pathways in development and differentiation may be controlled by a subfamily of enzymes preliminarily termed XOL-like GHMP kinases.
Materials and methods
Protein expression and purification
Full-length XOL-1 and a series of C- and N-terminal truncation mutants were generated by PCR and expressed with N-terminal polyhistidine tags in pRSET-A (Invitrogen) and PET28a (Novagen) vectors. Protein expression in BL21 DE3 strain E. coli was induced with 1 mM IPTG at a cell density of ∼0.6 OD at 600 nm. Induction took place overnight at room temperature or 16°C. Protein was purified by Ni+-affinity and anion exchange chromatography. N-terminal polyhistidine tags were removed by thrombin or enterokinase cleavage followed by an additional application of anion exchange chromatography. Selenomethionine-labeled protein was generated in the methionine-auxotrophic strain B834 DE3 (Novagen) in a similar manner, but bacteria were grown in methionine-deficient minimal medium supplemented with 0.3 mM L-selenomethionine (Sigma). Incorporation of selenomethionine into the recombinant protein was confirmed by MALDI mass spectrometry.
Crystallization and data collection
The C-terminal truncation mutant consisting of residues 1–374 (80 mg/mL) crystallized at 4°C from 0.1 M HEPES (pH 7.8), 0.2 M MgCl2, 17.5% PEG400. MPD (>5%) was used as an additive to improve crystal size and quality. Crystals were harvested from drops with nylon cryoloops (Hampton Research) and directly cryocooled to −180°C without additional cryoprotectant. Native and far remote data were collected to 1.55 Å resolution at SSRL beamline 11-1 and MAD data (peak, inflection, and remote wavelengths) were collected at ALS beamline 5.0.2 (Table 1). All data were processed and scaled in HKL2000 (Otwinowski and Minor 1997). The space group is orthorhombic P21212 with unit cell dimensions a = 116.1 Å, b = 86.0 Å, and c = 80.2 Å, and two protein molecules per asymmetric unit (Vm = 2.4 Å3/Da).
Structure determination and refinement
Positions of Se atoms (20 of 22 possible) were determined and refined in SOLVE and RESOLVE (Terwilliger and Berendzen 1996). Phase extension and automated chain tracing using the native data were performed in ARP/WARP (Lamzin and Wilson 1993). Further refinement was performed in CNS (Brünger et al. 1998) with intervening rounds of manual model building using the program O (Jones et al. 1991). Water molecules were added automatically using CNS (Brünger et al. 1998) and verified by manual inspection in O (Jones et al. 1991). Additional analysis of the final model was performed with PROCHECK (Laskowski et al. 1993). The final model consists of 703 amino acid residues and 628 water molecules (Table 1).
Domain analysis
The XOL-1 structure has a complex topology. Automated domain analysis programs TOPS (Westhead et al. 1999) and DOMID [Guogusng, Lund University (http://bioinfo1.mbfys.lu.se/Domid)] classified it as a single domain protein. However, visual inspection of the structure hinted at the presence of two domains. Therefore, we used a more time-consuming algorithm (Godzik et al. 1993) on the basis of the analysis of a protein distance map to analyze the domain organization of the structure. A protein domain, by definition, is a compact substructure that would form a stable structure when excised from the whole protein. Upon distance mapping, such domains can be identified as boxes along the diagonal. Visual inspection of the XOL-1 structure distance map identified two domains, located approximately between residues 45 and 195 (domain 1) and 202 and 360 (domain 2). Structures of both domains were submitted separately to DALI (Holm and Sander 1993; http://www.ebi.ac.uk/dali) and CE (Shindyalov and Bourne 1998; http://www.cl.sdsc.edu/ce.html) structure comparison servers. In both cases, strong structural similarities to the domains of GHMP kinases were detected.
Cloning of C. briggsae xol-1
The C. elegans XOL-1 amino acid sequence was compared with the unfinished genomic sequence of C. briggsae (The Sanger Institute, http://www.sanger.ac.uk; The Genome Sequencing Center, Washington University in St Louis, http://genome.wustl.edu) by TBLASTN with standard parameters. A short stretch of potential similarity was identified in C. briggsae cosmid CB030H15. Using the coding sequence predicted from the C. briggsae genome, gene-specific primers to the most highly conserved regions were used in conjunction with RACE and conventional PCR to isolate cDNA fragments corresponding to the transcribed xol-1 gene. When aligned with the C. elegans XOL-1 amino acid sequence by the clustal method (GONNET series), the ORF from the C. briggsae ortholog displays low sequence identity (22%). The C. briggsae dim-1 gene is found on cosmid CB027N19. Cosmids CB030H15 and CB027N19 are both found on contig cb25.fpc4044. In C. briggsae, xol-1 is ∼32 kb upstream of dim-1. In C. elegans, xol-1 is ∼10 kb upstream of dim-1.
ATPase assay
ATPase activity was measured using a maltose phosphorylase-glucose oxidase-horseradish peroxidase enzyme-coupled colorimetric assay (Piper Phosphatase Assay, Molecular Probes) according to the manufacturer's protocol. Briefly, XOL-1 (10 μg), MHC Class I H-2Kb (10 μg; negative control) and Na+/K−-ATPase (5 milliunits; positive control) were incubated in the presence and absence of 1 mM ATP for various times up to 2 h at 37°C in 1× reaction buffer containing 100 μM Amplex red, 0.6 unit maltose phosphorylase, 0.3 unit glucose oxidase, and 0.6 unit horseradish peroxidase in a volume of 150 μL. Assays were performed and repeated in triplicate. Absorbance at 565 nm was measured in 96-well plates using a Molecular Devices SpectraMax 250 plate reader. Additional negative controls were ATP (1 mM) alone and buffer only.
Steady-state fluorescence spectroscopy
Data were acquired using a SLM-AMINCO 8100 spectrofluorimeter. The concentration of protein and ligand (TNP-ATP, Molecular Probes) were 20 μM in 20 mM tris-HCl (pH 8.0), 5 mM MgCl2 at 25°C (volume = 150 μL). Emission was collected at 90° to the direction of excitation. To correct for fluctuation in the output from the 450 W xenon lamp, the experimental signal was divided by that of a quantum counter (3 g/L rhodamine G in ethylene glycol). To eliminate polarization artifacts, the excitation polarizer was set to the vertical position, and the emission polarizer was oriented 54.7° to the vertical (magic angle conditions). An excitation wavelength of 410 nm (8 nm = bandpass) and an emission maximum of 544 nm (8 nm = bandpass) for the bound ligand were determined experimentally. The sensitivity of the emission channel was set to 30% of maximum, resulting in a PMT voltage of 1050 V and a signal gain of 10 (voltage = 560 V and signal gain = 1 for reference channel). Emission spectra were collected from 490 to 600 nm with an integration time of 1 sec. Experiments were performed in triplicate, and spectra were corrected by subtracting the background fluorescence of the protein in the absence of ligand.
Coordinates
Structure factors and atomic coordinates (accession no. 1MG7) have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb).
Acknowledgments
We thank Robyn Stanfield, Dennis Wolan, Xiaoping Dai, and Markus G. Rudolph for assistance with data collection; Markus G. Rudolph and Andreas Heine for help with phase derivation from MAD data; and Michael Bailey of the Millar Laboratory for assistance with fluorescence spectroscopy. We also thank the staff of The Stanford Synchrotron Radiation Laboratory beamline 11-1 and The Advanced Light Source beamline 5.0.2 for their support. We thank Raymond Stevens and staff of Syrrx, Inc. for their assistance with robotic crystal screening. Figures were prepared using GRASP (Nicholls et al. 1991), MOLSCRIPT (Kraulis 1991), RASTER3D (Merritt and Murphy 1994), and BOBSCRIPT (Esnouf 1997). This work was supported by The National Institutes of Health grants GM30702 (B.J.M.) and GM38273 (I.A.W.), The Skaggs Institute (I.A.W.), a National Institutes of Health training fellowship AI-07244 (J.G.L.), a Skaggs Institute post-doctoral fellowship (J.G.L.), and a Helen Hay Whitney Fellowship (C.A.H.). B.J.M. is an investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Corresponding authors.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1082303.
References
- Ashton FT, Li J, Schad GA. Chemo- and thermosensory neurons: Structure and function in animal parasitic nematodes. Vet Parasitol. 1999;84:297–316. doi: 10.1016/s0304-4017(99)00037-0. [DOI] [PubMed] [Google Scholar]
- Beutler E. Cataracts and galactokinase deficiency. N Engl J Med. 1972;287:202–203. doi: 10.1056/NEJM197207272870427. [DOI] [PubMed] [Google Scholar]
- Blaxter M, Ivens A. Reports from the cutting edge of parasitic genome analysis. Parasitol Today. 1999;15:430–431. doi: 10.1016/s0169-4758(99)01541-0. [DOI] [PubMed] [Google Scholar]
- Boag PR, Newton SE, Gasser RB. Molecular aspects of sexual development and reproduction in nematodes and schistosomes. Adv Parasitol. 2001;50:153–198. doi: 10.1016/s0065-308x(01)50031-7. [DOI] [PubMed] [Google Scholar]
- Bonanno JB, Edo C, Eswar N, Pieper U, Romanowski MJ, Ilyin V, Gerchman SE, Kycia H, Strudier FW, Sali A, et al. Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis. Proc Natl Acad Sci. 2001;98:12896–12901. doi: 10.1073/pnas.181466998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A. Convergent evolution of similar enzymatic function on different protein folds: The hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993;2:31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu R, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burglin TR, Lobos E, Blaxter ML. Caenorhabditis elegans as a model for parasitic nematodes. Int J Parasitol. 1998;28:395–411. doi: 10.1016/s0020-7519(97)00208-7. [DOI] [PubMed] [Google Scholar]
- Burley SK, Bonanno JB. Structural genomics of proteins from conserved biochemical pathways and processes. Curr Opin Struct Biol. 2002;12:383–391. doi: 10.1016/s0959-440x(02)00330-5. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Carmi I, Kopczynski JB, Meyer BJ. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature. 1998;396:168–173. doi: 10.1038/24164. [DOI] [PubMed] [Google Scholar]
- Cho YK, Rios SE, Kim JJ, Miziorko HM. Investigation of invariant serine/threonine residues in mevalonate kinase. J Biol Chem. 2001;276:12573–12578. doi: 10.1074/jbc.M011478200. [DOI] [PubMed] [Google Scholar]
- Chu DS, Dawes HE, Lieb JD, Chan RC, Kuo AF, Meyer BJ. A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes & Dev. 2002;16:796–805. doi: 10.1101/gad.972702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang PT, Lieb JD, Meyer BJ. Sex-specific assembly of a dosage compensation complex on the nematode X chromosome. Science. 1996;274:1736–1739. doi: 10.1126/science.274.5293.1736. [DOI] [PubMed] [Google Scholar]
- Cuisset L, Drenth JP, Simon A, Vincent MF, van der Velde Visser S, van der Meer JW, Grateau G, Delpech M. Molecular analysis of MVK mutations and enzymatic activity in hyper-IgD and periodic fever syndrome. Eur J Hum Genet. 2001;9:260–266. doi: 10.1038/sj.ejhg.5200614. [DOI] [PubMed] [Google Scholar]
- Davis TL, Meyer BJ. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development. 1997;124:1019–1031. doi: 10.1242/dev.124.5.1019. [DOI] [PubMed] [Google Scholar]
- Dawes HE, Berlin DS, Lapidus DM, Nusbaum C, Davis TL, Meyer BJ. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science. 1999;284:1800–1804. doi: 10.1126/science.284.5421.1800. [DOI] [PubMed] [Google Scholar]
- Drenth JP, Cuisset L, Grateau G, Vasseur C, van de Velde-Visser SD, de Jong JG, Beckmann JS, van der Meer JW, Delpech M. Mutations in the gene encoding mevalonate kinase cause hyper-IgD and periodic fever syndrome. International Hyper-IgD Study Group. Nat Genet. 1999;22:178–181. doi: 10.1038/9696. [DOI] [PubMed] [Google Scholar]
- Esnouf RM. An extensively modified version of Molscript that includes greatly enhanced coloring capabilites. J Mol Graph Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- Favre R, Cermola M, Nunes CP, Hermann R, Muller M, Bazzicalupo P. Immuno-cross-reactivity of CUT-1 and cuticlin epitopes between ascaris lumbricoides, Caenorhabditis elegans, and Heterorhabditis. J Struct Biol. 1998;123:1–7. doi: 10.1006/jsbi.1998.4012. [DOI] [PubMed] [Google Scholar]
- Fu Z, Wang M, Potter D, Miziorko HM, Kim JJ. The structure of a binary complex between a mammalian mevalonate kinase and ATP. Insights into the reaction mechanism and human inherited disease. J Biol Chem. 2002;277:18134–18142. doi: 10.1074/jbc.M200912200. [DOI] [PubMed] [Google Scholar]
- Godzik A, Skolnick J, Kolinski A. Regularities in interaction patterns of globular proteins. Protein Eng. 1993;6:801–810. doi: 10.1093/protein/6.8.801. [DOI] [PubMed] [Google Scholar]
- Goodwin EB, Ellis RE. Turning clustering loops: Sex determination in Caenorhabditis elegans. Curr Biol. 2002;12:R111–R120. doi: 10.1016/s0960-9822(02)00675-9. [DOI] [PubMed] [Google Scholar]
- Haag ES, Kimble J. Regulatory elements required for development of Caenorhabditis elegans hermaphrodites are conserved in the tra-2 homologue of C. remanei, a male/female sister species. Genetics. 2000;155:105–116. doi: 10.1093/genetics/155.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- Hansen D, Pilgrim D. Sex and the single worm: Sex determination in the nematode C. elegans. Mech Dev. 1999;83:3–15. doi: 10.1016/s0925-4773(99)00024-6. [DOI] [PubMed] [Google Scholar]
- Harley JD, Irvine S, Gupta JD, Hertzberg R, Rowe PB. Galactokinase deficiency and cataract formation. Med J Aust. 1972;1:1326–1327. doi: 10.5694/j.1326-5377.1972.tb116507.x. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Zellan JD, Albertson DG. Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans. Development. 1994;120:3681–3689. doi: 10.1242/dev.120.12.3681. [DOI] [PubMed] [Google Scholar]
- Hoffmann G, Gibson KM, Brandt IK, Bader PI, Wappner RS, Sweetman L. Mevalonic aciduria–an inborn error of cholesterol and nonsterol isoprene biosynthesis. N Engl J Med. 1986;314:1610–1614. doi: 10.1056/NEJM198606193142504. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Houten SM, Kuis W, Duran M, de Koning TJ, van Royen-Kerkhof A, Romeijn GJ, Frenkel J, Dorland L, de Barse MM, Huijbers WA, et al. Mutations in MVK, encoding mevalonate kinase, cause hyperimmunoglobulinaemia D and periodic fever syndrome. Nature Genet. 1999a;22:175–177. doi: 10.1038/9691. [DOI] [PubMed] [Google Scholar]
- Houten SM, Romeijn GJ, Koster J, Gray RG, Darbyshire P, Smit GP, de Klerk JB, Duran M, Gibson KM, Wanders RJ, et al. Identification and characterization of three novel missense mutations in mevalonate kinase cDNA causing mevalonic aciduria, a disorder of isoprene biosynthesis. Hum Mol Genet. 1999b;8:1523–1528. doi: 10.1093/hmg/8.8.1523. [DOI] [PubMed] [Google Scholar]
- Houten SM, Koster J, Romeijn GJ, Frenkel J, Di Rocco M, Caruso U, Landrieu P, Kelley RI, Kuis W, Poll-The BT, et al. Organization of the mevalonate kinase (MVK) gene and identification of novel mutations causing mevalonic aciduria and hyperimmunoglobulinaemia D and periodic fever syndrome. Eur J Hum Genet. 2001;9:253–259. doi: 10.1038/sj.ejhg.5200595. [DOI] [PubMed] [Google Scholar]
- Inoue K, Hoshijima K, Sakamoto H, Shimura Y. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 1990;344:461–463. doi: 10.1038/344461a0. [DOI] [PubMed] [Google Scholar]
- Jones TA, Cowan S, Zou JY, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of proteins. J Applied Crystallogr. 1991;24:946–950. [Google Scholar]
- Krishna SS, Zhou T, Daugherty M, Osterman A, Zhang H. Structural basis for the catalysis and substrate specificity of homoserine kinase. Biochemistry. 2001;40:10810–10818. doi: 10.1021/bi010851z. [DOI] [PubMed] [Google Scholar]
- Kuwabara PE, Shah S. Cloning by synteny: identifying C. briggsae homologues of C. elegans genes. Nucleic Acids Res. 1994;22:4414–4418. doi: 10.1093/nar/22.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr. 1993;D49:129–149. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Levy NS, Krill AE, Beutler E. Galactokinase deficiency and cataracts. Am J Ophthalmol. 1972;74:41–48. doi: 10.1016/0002-9394(72)91123-3. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Capowski EE, Meneely P, Meyer BJ. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science. 1996;274:1732–1736. doi: 10.1126/science.274.5293.1732. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. MIX-1: An essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- Maine EM, Salz HK, Cline TW, Schedl P. The Sex-lethal gene of Drosophila: DNA alterations associated with sex-specific lethal mutations. Cell. 1985;43:521–529. doi: 10.1016/0092-8674(85)90181-3. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Murphy MEP. Raster3D Version 2.0—A program for photorealistic graphics. Acta Crystallogr. 1994;D50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Meyer BJ. Sex in the wormcounting and compensating X-chromosome dose. Trends Genet. 2000a;16:247–253. doi: 10.1016/s0168-9525(00)02004-7. [DOI] [PubMed] [Google Scholar]
- ————— Sex and death of a worm: Assessing and repressing X chromosomes. Harvey Lect. 2000b;95:85–105. [PubMed] [Google Scholar]
- Meyer J, Walker-Jonah A, Hollenberg CP. Galactokinase encoded by GAL1 is a bifunctional protein required for induction of the GAL genes in Kluyveromyces lactis and is able to suppress the gal3 phenotype in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5454–5461. doi: 10.1128/mcb.11.11.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LM, Plenefisch JD, Casson LP, Meyer BJ. xol-1: A gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell. 1988;55:167–183. doi: 10.1016/0092-8674(88)90019-0. [DOI] [PubMed] [Google Scholar]
- Monteleone JA, Beutler E, Monteleone PL, Utz CL, Casey EC. Cataracts, galactosuria and hypergalactosemia due to galactokinase deficiency in a child. Studies of a kindred. Am J Med. 1971;50:403–407. doi: 10.1016/0002-9343(71)90230-0. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Nicoll M, Akerib CC, Meyer BJ. X-chromosome-counting mechanisms that determine nematode sex. Nature. 1997;388:200–204. doi: 10.1038/40669. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Meyer BJ. Early aspects of Caenorhabditis elegans sex determination and dosage compensation are regulated by a zinc-finger protein. Nature. 1991;351:65–68. doi: 10.1038/351065a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Platt A, Reece RJ. The yeast galactose genetic switch is mediated by the formation of a Gal4p–Gal80p–Gal3p complex. EMBO J. 1998;17:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Ross HC, Hankin S, Reece RJ. The insertion of two amino acids into a transcriptional inducer converts it into a galactokinase. Proc Natl Acad Sci. 2000;97:3154–3159. doi: 10.1073/pnas.97.7.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind NR, Miller LM, Kopczynski JB, Meyer BJ. xol-1 acts as an early switch in the C. elegans male/hermaphrodite decision. Cell. 1995;80:71–82. doi: 10.1016/0092-8674(95)90452-2. [DOI] [PubMed] [Google Scholar]
- Rios SE, Cho YK, Miziorko HM. Characterization of mevalonate kinase V377I, a mutant implicated in defective isoprenoid biosynthesis and HIDS/periodic fever syndrome. Biochim Biophys Acta. 2001;1531:165–168. doi: 10.1016/s1388-1981(01)00105-6. [DOI] [PubMed] [Google Scholar]
- Romanowski MJ, Bonanno JB, Burley SK. Crystal structure of the Streptococcus pneumoniae phosphomevalonate kinase, a member of the GHMP kinase superfamily. Proteins. 2002;47:568–571. doi: 10.1002/prot.10118. [DOI] [PubMed] [Google Scholar]
- Schafer BL, Bishop RW, Kratunis VJ, Kalinowski SS, Mosley ST, Gibson KM, Tanaka RD. Molecular cloning of human mevalonate kinase and identification of a missense mutation in the genetic disease mevalonic aciduria. J Biol Chem. 1992;267:13229–13238. [PubMed] [Google Scholar]
- Schiffer M, Chang CH, Stevens FJ. Formation of an infinite β-sheet arrangement dominates the crystallization behavior of λ-type antibody light chains. J Mol Biol. 1985;186:475–478. doi: 10.1016/0022-2836(85)90120-2. [DOI] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Simon A, Cuisset L, Vincent MF, van Der Velde-Visser SD, Delpech M, van Der Meer JW, Drenth JP. Molecular analysis of the mevalonate kinase gene in a cohort of patients with the hyper-igd and periodic fever syndrome: Its application as a diagnostic tool. Ann Intern Med. 2001;135:338–343. doi: 10.7326/0003-4819-135-5-200109040-00010. [DOI] [PubMed] [Google Scholar]
- Skipper M, Milne CA, Hodgkin J. Genetic and molecular analysis of fox-1, a numerator element involved in Caenorhabditis elegans primary sex determination. Genetics. 1999;151:617–631. doi: 10.1093/genetics/151.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A, Li W, Robertson B, Schein J, Kamal IH, Marra M, Wood WB. Homologs of the Caenorhabditis elegans masculinizing gene her-1 in C. briggsae and the filarial parasite Brugia malayi. Genetics. 1999;152:1573–1584. doi: 10.1093/genetics/152.4.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Correlated phasing of multiple isomorphous replacement data. Acta Crystallogr. 1996;D52:749–757. doi: 10.1107/S0907444996000832. [DOI] [PubMed] [Google Scholar]
- Tsay YH, Robinson GW. Cloning and characterization of ERG8, an essential gene of Saccharomyces cerevisiae that encodes phosphomevalonate kinase. Mol Cell Biol. 1991;11:620–631. doi: 10.1128/mcb.11.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY–DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- Westhead DR, Slidel TW, Flores TP, Thornton JM. Protein structural topology: Automated analysis and diagrammatic representation. Protein Sci. 1999;8:897–904. doi: 10.1110/ps.8.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB, Streit A, Li W. Dosage compensation: X-repress yourself. Curr Biol. 1997;7:R227–R230. doi: 10.1016/s0960-9822(06)00107-2. [DOI] [PubMed] [Google Scholar]
- Yang D, Shipman LW, Roessner CA, Scott AI, Sacchettini JC. Structure of the Methanococcus jannaschii mevalonate kinase, a member of the GHMP kinase superfamily. J Biol Chem. 2002;277:9462–9467. doi: 10.1074/jbc.M110787200. [DOI] [PubMed] [Google Scholar]
- Zhou T, Daugherty M, Grishin NV, Osterman AL, Zhang H. Structure and mechanism of homoserine kinase: Prototype for the GHMP kinase superfamily. Structure Fold Des. 2000;8:1247–1257. doi: 10.1016/s0969-2126(00)00533-5. [DOI] [PubMed] [Google Scholar]