Abstract
Pre-mRNA splicing is essential for generating mature mRNA and is also important for subsequent mRNA export and quality control. The splicing history is imprinted on spliced mRNA through the deposition of a splicing-dependent multiprotein complex, the exon junction complex (EJC), at ∼20 nucleotides upstream of exon–exon junctions. The EJC is a dynamic structure containing proteins functioning in the nuclear export and nonsense-mediated decay of spliced mRNAs. Mago nashi (Mago) and Y14 are core components of the EJC, and they form a stable heterodimer that strongly associates with spliced mRNA. Here we report a 1.85 Å-resolution structure of the Drosophila Mago–Y14 complex. Surprisingly, the structure shows that the canonical RNA-binding surface of the Y14 RNA recognition motif (RRM) is involved in extensive protein–protein interactions with Mago. This unexpected finding provides important insights for understanding the molecular mechanisms of EJC assembly and RRM-mediated protein–protein interactions.
Keywords: Mago, Y14, RNA recognition motif, splicing, nuclear export, nonsense-mediated decay
Pre-mRNA splicing is coupled with subsequent cellular processes including export and nonsense-mediated decay (NMD) of spliced mRNAs (Maquat and Carmichael 2001; Dreyfuss et al. 2002; Reed and Hurt 2002; Wagner and Lykke-Andersen 2002). Splicing deposits a multiprotein complex, known as the exon junction complex (EJC), on spliced mRNAs at a position ∼20 nucleotides upstream of the exon–exon junctions. These EJC proteins have important functions in determining the fate of spliced mRNAs (Kataoka et al. 2000; Le Hir et al. 2000a,b, 2001b; Kim et al. 2001b). To date, at least eight EJC proteins have been identified. They include Y14, Mago, DEK, RNPS1, SRm160, Upf3, UAP56, and REF/Aly (Mayeda et al. 1999; Kataoka et al. 2000, 2001; Le Hir et al. 2000a, 2001a; McGarvey et al. 2000; Zhou et al. 2000; Hachet and Ephrussi 2001; Kim et al. 2001a; Luo et al. 2001; Lykke-Andersen et al. 2001). These proteins have been shown to have distinct and sometimes multiple functions in various aspects of mRNA metabolism such as splicing, nuclear export, and mRNA quality control. The EJC appears to be a dynamic complex, as the composition of EJC varies at different stages of splicing and export, and various EJC components associate with the spliced mRNA with different affinities (Reichert et al. 2002). Two proteins, a human homolog of Drosophila mago nashi, Magoh, and Y14, stably associate with spliced mRNAs in the nucleus and in the cytoplasm, and they are thought to be part of the core EJC (Kataoka et al. 2001; Le Hir et al. 2001a). Here, we focus our study on Drosophila Mago and Y14 proteins.
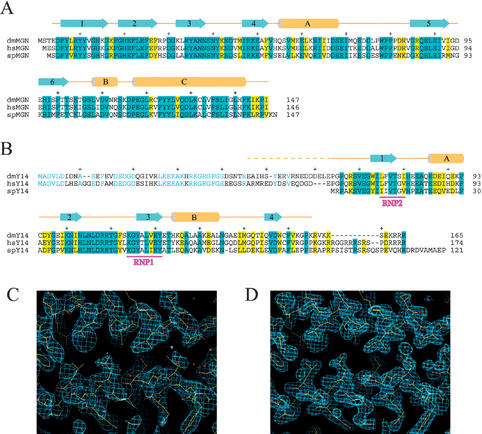
Both Mago and Y14 are highly conserved from Saccharomyces pombe to human (Fig. 1A,B). In Drosophila melanogaster, both Mago and Y14 are essential for viability and required for correct localization of oskar mRNA at the posterior pole (Micklem et al. 1997; Newmark et al. 1997; Hachet and Ephrussi 2001; Le Hir et al. 2001a), which is critical for generating cell asymmetry and specification during embryonic development. Mago interacts with Y14 stably in vitro and in vivo (Zhao et al. 2000; Hachet and Ephrussi 2001; Kataoka et al. 2001; Le Hir et al. 2001a; Mingot et al. 2001). Magoh has also been shown to interact with the mRNA export factor TAP (Kataoka et al. 2001). Y14 is a putative RNA-binding protein that shuttles between the nucleus and the cytoplasm (Kataoka et al. 2000; Mingot et al. 2001). It interacts with TAP and other EJC components REF/Aly, RNPS1, and hUpf3 (Kataoka et al. 2001; Kim et al. 2001a). The two proteins are stably associated with mRNAs during the export from the nucleus to the cytoplasm (Kataoka et al. 2001; Kim et al. 2001b; Le Hir et al. 2001a), and this association persists until the first round of translation (Dostie and Dreyfuss 2002).
Figure 1.
Sequence conservation and structure determination of Drosophila Mago nashi and Y14. (A) Sequence alignment of Drosophila melanogaster (dmMGN), human (hsMGN), and S. pombe (spMGN) Mago proteins. Identical residues among all three species are highlighted in cyan, and similar residues are highlighted in yellow. Every 10 residues are indicated with a “+” above the sequence. Secondary structural elements, determined from the crystallographic analysis, and their nomenclatures are shown above the sequence. (B) Sequence alignment of Y14 proteins of the same three species. The same criteria and color code as above are used to indicate sequence homology. RNP1 and RNP2 sequence motifs are underlined and labeled in magenta. Identical residues between Drosophila and human Y14 proteins that are not included in the crystallized fragment are shown in cyan letters. (C) A section of the 2.8 Å MAD phased electron density map. The map is calculated using the RESOLVE program and displayed using O at 1.2σ contour level. The refined 1.85 Å structure is superimposed as a bond model. (D) The 1.85 Å 2FO-FC map of the same region contoured at 1.5σ level.
A great deal about the function of Mago and Y14 in EJC assembly, mRNA export and decay has been learned from recent studies. However, the molecular mechanisms of the Mago–Y14 interaction and their interactions with mRNA and other EJC components are still poorly understood. To understand the structural basis of Mago(h)–Y14 interaction and the mode of protein–RNA interaction, we have solved a 1.85 Å resolution structure of a Drosophila Mago–Y14 complex. The structure reveals a novel mode of Mago–Y14 interaction through the RNA-binding surface of the Y14 RRM (RNA recognition motif), which has important implications for understanding protein–RNA and protein–protein interactions of the EJC.
Results and Discussion
Recombinant Mago–Y14 complex was produced in Escherichia coli by coexpression. Purified Mago–Y14 complex exists as a 1:1 heterodimer in solution as judged by dynamic laser scattering (data not shown). Purified Mago–Y14 complex was subjected to endoproteinase Glu-C treatment to produce a core complex containing the full-length Mago and a truncated Y14, Y14Δ, lacking 46 amino acids at the N terminus and 10 amino acids at the C terminus as revealed by mass spectrometric analyses (data not shown). The crystal structure of the Mago–Y14Δ complex was solved by seleno-methionyl (SeMet) multiwavelength anomalous diffraction (MAD; Fig. 1C,D). The refined structure contains all but three N-terminal residues of Mago and amino acids 61–155 of Y14, encompassing the entire RRM domain. Detailed statistics of the crystallographic analysis are shown in Table 1.
Table 1.
Summary of crystallographic analysis
| Data sets wavelength
|
Native (0.95 Å)
|
SeMet MAD
|
||
|---|---|---|---|---|
| (0.9786 Å)
|
(0.9780 Å)
|
(0.9500 Å)
|
||
| Diffraction dataa | ||||
| Resolution (Å) | 1.85 | 2.3 | 2.2 | 2.5 |
| Measured reflections | 155,205 | 64,822 | 72,167 | 57,058 |
| Unique reflections | 23,222 | 11,928 | 13,787 | 10,302 |
| Completeness (%) | 97.5 (81.4) | 98.2 (99.9) | 98.7 (98.4) | 99.3 (100) |
| Average I/σ | 18.0 | 22.9 | 20.2 | 20.5 |
| Rmergeb | 0.051 (0.346) | 0.048 (0.143) | 0.054 (0.227) | 0.054 (0.149) |
| SOLVE Phasing | ||||
| Resolution | 2.8 Å | |||
| Overall figure of merit | 0.65 | |||
| Overall Z-score | 37.1 | |||
| Refinement | ||||
| Resolution range (Å) | 50–1.85 | |||
| R-factorc/Rfree | 0.215/0.261 (0.29/0.36) | |||
| Number of protein atoms | 1,937 | |||
| Number of Sr2+ ions | 1 | |||
| Number of BME molecules | 2 | |||
| Number of water molecules | 216 | |||
| R.M.S. deviations | ||||
| bond lengths | 0.014 Å | |||
| bond angles | 1.77° | |||
| dihedrals | 23.6° | |||
| improper | 1.08° | |||
Numbers in parentheses are statistics of the highest resolution shell.
Rmerge = Σ|I − <I>|/Σ<I>, where I and <I> are the measured and averaged intensities of multiple measurements of the same reflection. The summation is over all the observed reflections.
R-factor = Σ∥FO| − |FC∥/Σ |FO|, where FO denotes the observed structure factor amplitude and FC denotes the structure factor calculated from the model.
The structure of Mago
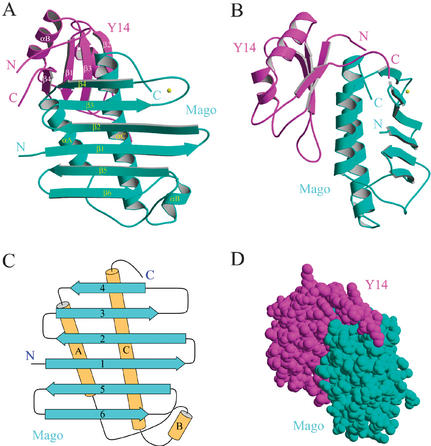
Mago is highly conserved during evolution: Drosophila and human proteins share 89% sequence identity, whereas the Drosophila and S. pombe proteins share 59% identity (Fig. 1A,B). However, Mago does not contain any recognizable sequence motifs. The crystal structure (Fig. 2A,B) reveals that Mago consists of six antiparallel β-strands (β1–β6) and three helices (αA–αC). The six β-strands form an unusually flat β-sheet, and two long antiparallel helices (αA and αC) are packed on the same side of the β-sheet (Fig. 2A–C). A metal-binding site is found in the loop between β3 and β4. Crystallization of the Mago–Y14 complex requires divalent ions such as Mg2+, Co2+, or Sr2+. In the structure, a Sr2+ ion is identified based on bonding geometry and electron density intensity. The metal ion appears to play a role in stabilizing the local conformation of the loop connecting β3 and β4, as it interacts with four residues on this loop. The Sr2+ ion is enclosed in an oxygen “cage” formed by side chain oxygen atoms of Asn 38 and Asn 43, carbonyl oxygen atoms of Asn 40, Asn 43, and Thr 45, and three water molecules. The interatomic distances between the Sr2+ ion and these oxygen atoms range from 2.2 to 2.6 Å. In a physiological environment, more abundant divalent ions such as Mg2+ or water molecules may play a similar role.
Figure 2.
Overall structure of Mago and Y14. (A) A ribbon representation of the Mago–Y14Δ complex. Mago is shown in cyan, and Y14 is shown in magenta. N and C termini are indicated in cyan and magenta letters for Mago and Y14, respectively. Secondary structures for both proteins are labeled. (B) The same ribbon model as shown in A, rotated ∼90° around a vertical axis. (C) A topological diagram of Mago. Helices are shown as brown cylinders, and strands are shown as cyan arrows. (D) A CPK model showing extensive interaction between Mago and Y14 (Mago, cyan; Y14, magenta). The model is viewed from the same direction as in B.
The structure of Y14
Y14 contains a central RRM flanked by highly charged N- and C-terminal regions. As expected, our crystal structure shows that Y14 contains a canonical RRM (amino acids. 72–149), which consists of a four-stranded antiparallel β-sheet (β1–β4) and two helices (αA, αB) in a β1–αA–β2–β3–αB–β4 arrangement (Fig. 2A,B). The Y14 RRM structure can be superimposed with the structure of the N-terminal RRM of U1A (Oubridge et al. 1994) with a root-mean-squared deviation of 0.62 Å using the Cα positions of 23 residues located on β1–β4 for alignment. Most notable differences lie within the loop connecting β2 and β3, and regions outside of the RRM fold. These differences are not unusual, though, as they are the most variable regions among different RRMs (Varani and Nagai 1998). The loop connecting β2 and β3 interacts with RNA in some RRMs, and conformational differences of this loop between native and RNA-bound forms have been documented. The N- and C-terminal extensions to many RRMs are normally disordered in the absence of RNA but become ordered upon nucleic acid binding. In the Mago–Y14Δ structure, both regions are stabilized by hydrogen bonding between residues located at the two termini. For example, the two termini interact via mainchain hydrogen bonding of Pro 65–Lys 152 and Glu 67–Phe 150. In addition, the packing of Trp 73, Pro 66, His 124, and Trp 148 provides major stabilization of the terminal regions of Y14.
The most conserved regions in the RRM are the RNP2 and RNP1 sequence motifs located on β1 and β3, respectively. In all of the RRM–RNA complex structures known to date, the β-sheet forms an RNA-binding platform, and the aromatic residues in the RNP2 and RNP1 motifs stack with RNA bases (Perez-Canadillas and Varani 2001). Y14 has a perfectly conserved RNP2 motif, but a highly conserved aromatic residue in RNP1 (most frequently a phenylalanine) is absent. Instead, a leucine (Leu 118) occupies the position (Fig. 1B). This deviation alone does not appear to render the RRM unable to bind RNA, as exemplified by RNA binding of the third and fourth RRMs of polypyrimidine tract-binding protein (PTB; Conte et al. 2000), in which both of the RRMs lack the highly conserved aromatic residue. Because of the presence of a canonical RRM, it is widely believed that Y14 binds mRNA directly.
Mago–Y14 interactions
Mago and Y14 form a stable heterodimer both in vitro and in vivo, and their association is not affected by RNase A treatment (Kataoka et al. 2001). The two proteins remain associated with each other even at 1M NaCl concentration in our hands. Because Y14 contains an RRM, it is generally believed that the complex binds RNA directly. However, no direct RNA binding of the Mago–Y14 complex or each individual protein has been reported. We also cannot detect any direct RNA binding of the purified Mago–Y14 complex using a gel mobility shift assay, although purified Y14 alone exhibits some RNA-binding activities (data not shown). The structure shows that Mago and Y14Δ share an extensive interface (Fig. 2D). The interaction between Mago and Y14Δ buries a pairwise accessible surface area of 2376 Å2. Mago interacts with Y14 mainly via its two long helices, αA and αC. Surprisingly, the major interaction area on Y14 is the β-sheet surface normally involved in RNA binding in other RRMs (Fig. 3A,B). This mode of RRM-mediated protein–protein interaction differs from all known examples involving RRMs. For example, the RRM of the spliceosomal protein U2B" interacts with the leucine-rich-repeat region of U2A‘ via the surface opposite to the RNA-binding β-sheet surface (Price et al. 1998), and the atypical RRM of U2AF35 interacts with a U2AF65 peptide, also via α-helices located on the back of the presumed RNA-binding β-sheet surface (Kielkopf et al. 2001). Thus, the structure of the Mago–Y14 complex reveals a unique protein–protein interaction mode involving the conserved RNP motifs.
Figure 3.
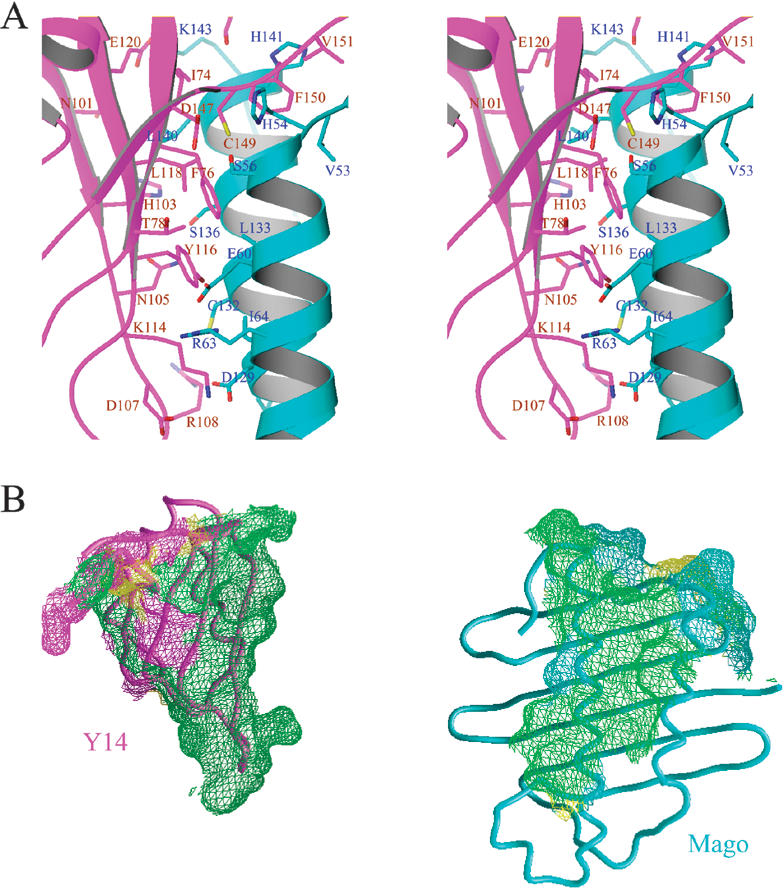
Mago–Y14 interactions. (A) Stereo view of a section of the Mago–Y14 interface surrounding the RNP motifs of Y14. The side chains of key residues are shown as stick models (nitrogen, blue; oxygen, red; sulfur, yellow; carbon atoms of Mago, cyan; carbon of Y14, magenta) superimposed with a ribbon drawing of the structure as in Figure 2B (Mago, cyan; Y14, magenta). Mago and Y14 residues are labeled in blue and brown, respectively. (B) Mago–Y14 interface involves highly conserved residues. Mago and Y14 surface regions within 4.0 Å from each other are shown in meshed surfaces. The colors on the surface indicate the conservation status: surface regions corresponding to identical residues in Figure 1A and B are shown in green, similar residues in yellow, and nonconserved residues are shown in magenta for Y14 and cyan for Mago. For viewing clarity, the two proteins are separated from each other and the proteins are rotated to allow the interaction regions to face the viewer.
A detailed examination of the structure shows that all of the exposed residues in the RNP1 and RNP2 motifs of Y14 are engaged in interaction with Mago (Fig. 3A,B). Among RNP2 residues, Ile 74Y interacts with Leu 140M and His 141M of Mago located at the C-terminal end of αC (Mago and Y14 residues will respectively be labeled with suffixes M and Y hereafter); Phe 76Y is surrounded by Ser 56M, Val 57M, and Glu 60M located in αA, and Leu 133M and Leu 137M located in αC; Thr 78Y is at the edge of the protein–protein interface, and it interacts with Ser 56M and Glu 60M via water-mediated and van der Waals interactions. Among RNP1 residues, Lys 114Y interacts with Asp 129M via charge interaction; Tyr 116Y interacts with Leu 133M, and Leu 118Y contacts Ser 136M and Leu 137M via hydrophobic and van der Waals interactions; Glu 120Y interacts with Lys 143M via charge interaction. In summary, all RNP residues previously known to directly interact with RNA in other RRMs are involved in interactions between Mago and Y14.
In addition to the RNP1 and RNP2 residues on β1 and β3, Asn 101Y, His 103Y, Asn 105Y, Arg 108Y, and Arg 109Y, located on β2 or a region immediately C-terminal to β2 of Y14, are involved in extensive interactions with residues located on helix αC of Mago. In contrast, only one β4 residue, Asp 147Y, is within the van der Waals radii of two Mago residues (Fig. 3A). Nevertheless, the region C-terminal to β4 (amino acids 148–155) of Y14 interacts with Mago extensively (Fig. 3A,B). The Y14 C-terminal loop wraps around the N-terminal end of αA and extends to an acidic region spanned between αA and the β-sheet (Fig. 4A,B) of Mago. The interactions include mainchain hydrogen bonding of Cys 149Y–His 54M, Phe 150Y–Phe 52M, and Lys 155Y–Asp 30M, stacking between Pro 154Y and the aromatic ring of Phe 52M. The side chain of Phe 150Y situates in a hydrophobic pocket surrounded by Ala 51M, Val 53M, His 141M, and Leu 137M. The C-terminal region of Y14 also plays important roles in stabilizing the conformation of the N-terminal loop of Y14, which is crucial for Ser 69Y, Val 70Y and Glu 71Y in this loop to interact with the C-terminal end of helix αC of Mago.
Figure 4.
The Mago–Y14 complex and models of potential protein–RNA and protein–protein interactions. (A) A surface representation of the Mago–Y14 structure. Red and blue indicate negatively and positively charged surface potentials, respectively. Neutral regions of Mago and Y14 are shown in white and light green, respectively. (B) The same surface model as shown in A, rotated ∼90° around a vertical axis. An arrow points to the location of a positively charged cleft in Mago. (C) A ribbon model showing potential Y14–RNA interactions. Y14 is rotated ∼90° around an axis perpendicular to the plane (with respect to that shown in Fig. 2B) to free the RRM surface for potential RNA binding. The direction of rotation is indicated by a curved arrow. A green curve indicates the location of a potential RNA-binding site. (D) TAP and p15 interact via their extensive β-sheet surfaces of the NTF2-like domains. (E) Interaction of two Mago molecules in the crystal lattice suggests a model of interaction with TAP.
Previous deletion and pull-down experiments showed that the N-terminal region of human Y14 (amino acids 1–73) interacts with Magoh, while no interactions involving the middle (amino acids 74–139) and C-terminal regions (amino acids 140–174) of Y14 were detected (Kataoka et al. 2001). The crystal structure shows that the minimal RRM core structure, from the first residue of β1 to the last residue of β4, encompasses amino acids 73–148. Thus, the absence of interactions between the middle and C-terminal regions of Y14 with Magoh in the GST-pull down experiment is most likely due to disruption of the RRM fold by deletions. The N-terminal fragment of Y14 (amino acids. 1–73) may contain an additional Magoh interacting region, as this fragment interacts with Magoh in yeast two-hybrid and GST-pull down experiments (Kataoka et al. 2001). However, the N-terminal 63 residues of Y14 is not present in the structure, and thus we cannot determine whether these residues form additional Magoh interaction regions from a structural standpoint.
Structural implications
Mago and Y14 are present in the fully assembled spliceosome during pre-mRNA splicing and remain stably bound to spliced mRNA (Kataoka et al. 2000, 2001; Reichert et al. 2002). Because Y14 contains an RRM, it is thought that the heterodimer binds RNA via the RRM domain of Y14. However, the crystal structure presented here shows that the presumed RNA-binding surface of Y14 is masked by the protein–protein interactions between Mago and Y14. Therefore, new models of protein–RNA interaction involving the Mago–Y14 complex must be considered. We discuss several possible scenarios of mRNA binding by the Mago–Y14 complex below.
In one scenario, the Mago–Y14 complex may bind to spliced mRNA indirectly. Although Mago and Y14 have been shown to associate with mRNA in numerous studies, no evidence of direct interaction has been documented. In fact, Y14 failed to chemically cross-link to mRNA (Reichert et al. 2002). However, no suitable protein candidates that can mediate the interaction between the Mago–Y14 complex and mRNA have been identified. Most known EJC components cannot serve this role, as they dissociate from spliced mRNA during or soon after mRNA export, whereas Mago and Y14 remain associated with mRNA. RNPS1 and Upf3 appear to associate with mRNA until a late stage, and they both interact with Y14. However, Upf3 is unlikely to mediate the association of Mago and Y14 with mRNA, as it joins the EJC after Mago and Y14 (Reichert et al. 2002). This leaves RNPS1 as the only known candidate that can potentially mediate the interaction between Mago–Y14 and mRNA. It remains to be tested whether RNPS1 or other as yet unidentified EJC proteins can mediate a stable association of Mago and Y14 with mRNA.
In another scenario, the Mago–Y14 complex may directly bind RNA via the RRM of Y14, but a major conformational change of the heterodimer is required to expose the RNA-binding surface of the Y14 RRM. Because the RRM surface interacts strongly with Mago, a great deal of energy is required to open up the heterodimer. The process of pre-mRNA splicing may provide means and energy to open up the heterodimer for binding RNA, which is consistent with the observation that splicing is required for the association of Mago–Y14 with RNA (Kataoka et al. 2001; Le Hir et al. 2001a). In this scenario, it is possible that the heterodimer can remain associated via the N and C termini of Y14 whereas the RRM β-sheet surface and the helical side of Mago can open like a book to allow access of RNA (Fig. 4C). A related scenario would be that Y14 first binds to mRNA as a monomer, and then Mago joins the complex. However, no evidence for the existence of monomeric Mago or Y14 has been reported. Mago and Y14 appear to be imported into the nucleus as a heterodimer (Mingot et al. 2001), and recombinant Magoh injected into the cytoplasm of Xenopus oocytes failed to enter the nucleus (Kataoka et al. 2001). In all studies carried out to date, Mago(h) and Y14 appear to associate with each other and function together (Hachet and Ephrussi 2001; Kataoka et al. 2001; Le Hir et al. 2001a). It would be extremely interesting to know whether separate populations of Y14 and/or Mago exist.
A third scenario would be that the Mago–Y14 dimer can bind RNA in the present form but the Y14 RRM only plays a peripheral role in RNA binding. In support of this, the structure shows a positively charged cleft on the helical side of Mago (Fig. 4B). The cleft extends from the C-terminal end of Mago and β2 of Y14 to αB of Mago. It is possible that this cleft can accommodate the binding of RNA. It is interesting to note that mutations in the mago nashi gene affect the proper localization of oskar mRNA in Drosophila (Micklem et al. 1997; Newmark et al. 1997). However, Mago is not known to be an RNA-binding protein, and no RNA binding by the Mago–Y14 heterodimer has been demonstrated. This is perhaps not surprising, because splicing is required for RNA binding. However, this scenario of RNA binding cannot be excluded by all existing data, because splicing may be needed to load the RNA onto the Mago–Y14 heterodimer. Splicing may generate a specific RNA structure near the exon–exon boundary required for the binding of the Mago–Y14 heterodimer.
The structure also provides important insights into Mago and Y14 interactions with other EJC components. Y14 interacts with Ref/Aly, TAP, RNPS1, and hUpf3, whereas Mago interacts avidly with TAP. It is interesting to note that Ref/Aly, RNPS1, and hUpf3 are highly basic proteins. The structure shows that the exposed surface of Y14 helix αA, located on the opposite side of the β-sheet surface, has a distinct patch of negatively charged residues (Glu 86Y, Asp 87Y, Glu 88Y, and Glu 91Y; Fig. 4A,B). This negatively charged region of Y14 may interact with positively charged EJC partners. Mago also interacts with in vitro translated TAP in GST-pull down assays. This interaction is most likely direct, as RNase A treatment had no effect on this interaction (Kataoka et al. 2001). The structure of Mago, together with the crystal structure of a TAP–p15 complex (Fribourg et al. 2001), provides an interesting molecular model of Mago–TAP interaction.
The C-terminal half of TAP interacts with the nuclear pore complex and contains an NTF2-like domain. The NTF2-like domain of TAP interacts with p15. Both the TAP NTF2-like domain and p15 contain a six-strand antiparallel β-sheet (Fribourg et al. 2001), and the two proteins interact extensively via their β-sheet surfaces (Fig. 4D). It is conceivable that Mago may interact with TAP also via its β-sheet surface in a manner similar to that between TAP and p15. The β-sheet surface of Mago is capable of mediating protein–protein interactions, as shown in the packing interaction between two symmetry-related Mago molecules in the crystal lattice (Fig. 4E). Thus, replacing one Mago β-sheet in Figure 4E with one from TAP would generate an intriguing model of Mago–TAP interaction. In this model, Mago occupies the p15-binding site in TAP, and p15 will not be able to bind TAP. It is possible that Mago can substitute the function of p15 in stabilizing the NTF2-like domain of TAP necessary for interacting with nucleoporin (Braun et al. 2002). Alternative models of Mago–TAP interactions are also possible, and the crystal structure of Mago should be helpful in facilitating biochemical dissection of the molecular basis of Mago–TAP interactions.
Materials and methods
Protein expression, purification, and crystallization
The cDNA fragments encoding full-length Mago and Y14 were amplified by PCR from a Drosophila total cDNA preparation. The Mago cDNA was cloned into a modified pGEX-KG vector to produce GST-fused Mago. The Y14 cDNA was first cloned into a pET28a (Novagen) vector and then transferred into a pMR101 vector (ATCC) to produce polyhistidine-tagged Y14. The two vectors, pGEX-KG and pMR101, have compatible replicons and different antibiotic selection markers, and thus are suitable for coexpression of Mago and Y14 in E. coli. Recombinant Mago–Y14 complex was first purified by glutathione-sepharose column. The GST tag and His tag were then removed by in-column thrombin digestion, and the eluted Mago–Y14 complex was further purified by HiTrap-Q and Superdex-75 (Pharmacia) column chromatography. The purified full-length Mago–Y14 complex was then treated with endoproteinase Glu-C overnight at room temperature. Mass spectrometry shows that the Glu-C treatment removes 46 residues at the N terminus and 10 residues at the C terminus of Y14 (Y14Δ) while Mago remains intact. The Mago–Y14Δ complex was further purified by HiTrap-Q and Superdex-75 column chromatography. Purified Mago–Y14Δ complex was then concentrated to ∼25–30 mg/mL in a buffer containing 10 mM Hepes at pH 8.0, 5% glycerol, and 500 mM NaCl for crystallization. Best diffracting crystals were grown in conditions containing 100 mM sodium acetate at pH 5.8, 30% 2-methyl-2,5-pentanediol (MPD), and 10 mM SrCl2. SeMet substituted Mago–Y14Δ complex was produced using E. coli strain DL41(DE3) in a defined medium containing 30 mg/L of SeMet. Purification and crystallization of SeMet Mago–Y14Δ were similar to that of the native proteins.
Crystallographic analysis
All diffraction data were collected at 100°K using a CCD detector (ADSC) at beamline X26C of National Synchrotron Light Source, Brookhaven National Laboratory. The HKL software package (Otwinowski and Minor 1997) was used for data processing. The Mago–Y14Δ complex crystallizes in spacegroup C2 with cell dimensions of 105.5 Å × 52.4 Å × 52.9 Å and β = 104.9°. The structure was solved by SeMet MAD. Three MAD data sets, with resolutions ranging from 2.2 to 2.5 Å, were collected at wavelengths of 0.9786 Å, 0.9780 Å, and 0.9500 Å, corresponding to inflection point, peak, and remote wavelengths of the Se K edge. A 1.85 Å resolution native data set was collected at λ = 0.95 Å. Detailed data statistics are shown in Table 1. Three ordered Se positions were identified in anomalous and isomorphous Patterson maps, and phasing and solvent flattening were carried out using SOLVE (Terwilliger and Berendzen 1999). Iterative cycles of model building and refinement were carried out using O (Jones et al. 1991), CNS (Brünger et al. 1998), and CCP4 programs. Ten percent of the data was used for cross-validation in CNS. Refinement statistics are shown in Table 1. Figures were prepared using MOLSCRIPT (Kraulis 1991), Raster3D (Merritt and Bacon 1997), RIBBONS (Carson 1997), and GRASP (Nicholls et al. 1991) programs.
The PDB accession code is 1OO0.
Acknowledgments
We thank Frances Hannan for Drosophila cDNA, Annie Heroux and Dieter Schneider for technical help during data collection, Michael Myers for help in protein mass analyses, and Michelle Hastings, Lisa Manche, and Zuo Zhang for comments on the manuscript. The work is supported in part by the W.M. Keck Foundation and an NIH grant (GM55874).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL xur@cshl.org; FAX (516) 367-8873.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.260403.
References
- Braun IC, Herold A, Rode M, Izaurralde E. Nuclear export of mRNA by TAP/NXF1 requires two nucleoporin-binding sites but not p15. Mol Cell Biol. 2002;22:5405–5418. doi: 10.1128/MCB.22.15.5405-5418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carson M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- Conte MR, Grune T, Ghuman J, Kelly G, Ladas A, Matthews S, Curry S. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 2000;19:3132–3141. doi: 10.1093/emboj/19.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Fribourg S, Braun IC, Izaurralde E, Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr Biol. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon–exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science. 2001a;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J. 2001b;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript—A program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000a;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon–exon junctions. Genes & Dev. 2000b;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001a;2:1119–1124. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001b;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Maquat LE, Carmichael GG. Quality control of mRNA function. Cell. 2001;104:173–176. doi: 10.1016/s0092-8674(01)00202-1. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Badolato J, Kobayashi R, Zhang MQ, Gardiner EM, Krainer AR. Purification and characterization of human RNPS1: A general activator of pre-mRNA splicing. EMBO J. 1999;18:4560–4570. doi: 10.1093/emboj/18.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T, Rosonina E, McCracken S, Li Q, Arnaout R, Mientjes E, Nickerson JA, Awrey D, Greenblatt J, Grosveld G, et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon–product complexes. J Cell Biol. 2000;150:309–320. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, Gonzalez-Reyes A, St Johnston D. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol. 1997;7:468–478. doi: 10.1016/s0960-9822(06)00218-1. [DOI] [PubMed] [Google Scholar]
- Mingot JM, Kostka S, Kraft R, Hartmann E, Gorlich D. Importin 13: A novel mediator of nuclear import and export. EMBO J. 2001;20:3685–3694. doi: 10.1093/emboj/20.14.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Mohr SE, Gong L, Boswell RE. Mago nashi mediates the posterior follicle cell-to-oocyte signal to organize axis formation in Drosophila. Development. 1997;124:3197–3207. doi: 10.1242/dev.124.16.3197. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Perez-Canadillas JM, Varani G. Recent advances in RNA–protein recognition. Curr Opin Struct Biol. 2001;11:53–58. doi: 10.1016/s0959-440x(00)00164-0. [DOI] [PubMed] [Google Scholar]
- Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B"-U2A‘ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- Reichert VL, Le Hir H, Jurica MS, Moore MJ. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes & Dev. 2002;16:2778–2791. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- Wagner E, Lykke-Andersen J. mRNA surveillance: The perfect persist. J Cell Sci. 2002;115:3033–3038. doi: 10.1242/jcs.115.15.3033. [DOI] [PubMed] [Google Scholar]
- Zhao XF, Nowak NJ, Shows TB, Aplan PD. MAGOH interacts with a novel RNA-binding protein. Genomics. 2000;63:145–148. doi: 10.1006/geno.1999.6064. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]