Abstract
The DNA-damage response genes in bacteria are up-regulated when LexA repressor undergoes autocatalytic cleavage stimulated by activated RecA protein. Intact LexA is stable to intracellular degradation but its auto-cleavage fragments are degraded rapidly. Here, both fragments of LexA are shown to be substrates for the ClpXP protease. ClpXP recognizes these fragments using sequence motifs that flank the auto-cleavage site but are dormant in intact LexA. Furthermore, ClpXP degradation of the LexA-DNA-binding fragment is important to cell survival after DNA damage. These results demonstrate how one protein-processing event can activate latent protease recognition signals, triggering a cascade of protein turnover in response to environmental stress.
Keywords: SOS response, proteolysis, AAA+, peptide recognition, ClpP, UV response
Protein degradation plays a critical role in allowing cells to adjust to changing conditions. Intracellular proteolysis is an essential component of many response pathways that permit bacteria to survive or recover from DNA damage, heat shock, or attack by reactive oxygen species (Mizusawa and Gottesman 1983; Gerth et al. 1998; Roberston et al. 2002). In addition to stress-related regulatory functions, proteases serve to degrade damaged proteins as well as undamaged proteins that the cell no longer needs under a new set of environmental conditions. In each of these examples, one can ask the same question: what mechanisms allow intracellular proteases to degrade the appropriate substrates at the right time?
In bacteria, many intracellular proteases (e.g., ClpXP, ClpAP, and HslUV) divide the tasks of substrate recognition and proteolysis between a hexameric AAA+ ATPase and a multisubunit peptidase with an internal degradation chamber (for review, see Gottesman 1996). The ATPase (ClpX, ClpA, or HslU) recognizes specific substrates, unfolds these proteins, and translocates the denatured polypeptide into the peptidase (ClpP or HslV) chamber for degradation (Kim et al. 2000). In many instances, the ATPase components recognize substrates via specific peptide motifs, often at their N or C termini (Levchenko et al. 1995; Gonciarz-Swiatek et al. 1999; Flynn et al. 2003). For example, the ssrA degradation tag, an 11-residue C-terminal peptide sequence, targets proteins for degradation by ClpXP and ClpAP (Gottesman et al. 1998). For some substrates, such as the stationary-phase sigma factor (σs), accessory proteins are needed for efficient degradation, allowing regulation of proteolysis through the synthesis or modification of these factors (Becker at al. 1999; Zhou et al. 2001). Here we provide evidence for an alternative mechanism of regulated degradation: We find that complete degradation of the LexA repressor requires an initiating cleavage event that is regulated in response to DNA damage.
The SOS regulatory system in Escherichia coli controls the cellular response to DNA damage. Under normal conditions, LexA repressor dimers negatively regulate the expression of genes involved in DNA repair, replication, and cell division (for review, see Freidberg et al. 1995). After DNA damage, single-stranded DNA is exposed and is bound by the RecA protein. This event activates RecA, which then stimulates the auto-cleavage of LexA at a site between its N-terminal DNA-binding domain and its C-terminal dimerization domain (Philzicky and Roberts 1981; Little 1984). The resulting N- and C-terminal fragments are then degraded rapidly (Little 1983a). The C-terminal fragment is stabilized ∼10-fold in Lon-defective cells (Little 1983b), but the protease that degrades the N-terminal fragment has not previously been identified.
In recent proteomic experiments, we identified tryptic fragments from E. coli proteins that were trapped in vivo within a proteolytically inactive ClpP variant (ClpPtrap) and found that some peptides originated from LexA (Flynn et al. 2003). Western blots of the undigested trapped proteins indicated that the two auto-cleavage fragments of LexA represented the dominant captured forms. This result suggested that these auto-cleavage fragments might be ClpXP substrates. Here, we show that ClpXP degrades the auto-cleavage fragments of LexA, but not full-length LexA, both in vivo and in vitro. Recognition of these fragments for destruction occurs via peptide signals created or exposed by the initial auto-cleavage event. The use of these latent recognition signals allows specific recognition and degradation of the LexA fragments by ClpXP at the biologically appropriate time—after LexA has undergone RecA-stimulated self-cleavage in response to DNA damage.
Results and Discussion
The LexA auto-cleavage fragments are ClpXP substrates
Capture experiments using ClpPtrap were performed in E. coli exposed to the DNA-damaging agent nalidixic acid. The ClpPtrap is an inactive (S97A) affinity-tagged variant of ClpP that, when expressed in cells, allows the capture and purification of physiological ClpXP substrates (Flynn et al. 2003). A Western blot of the trapped material revealed two bands that cross-reacted with anti-LexA antibodies and had the same electrophoretic mobilities as the LexA1–84 and LexA85–202 fragments generated by auto-cleavage of purified LexA (Fig. 1A). No full-length LexA was detected in the trapped material, although we observed ∼20 ng of each cleavage fragment and could detect as little as 1 ng of LexA protein. Furthermore, when purified LexA was added to a sample of the trapped material and prepared for Western blotting, this protein was efficiently detected, indicating that full-length LexA could have survived this procedure. Trapping required ClpX, as the LexA fragments were not detected in material isolated from clpX− cells (Flynn et al. 2003). We conclude that ClpX recognizes the two auto-cleavage fragments but not full-length LexA.
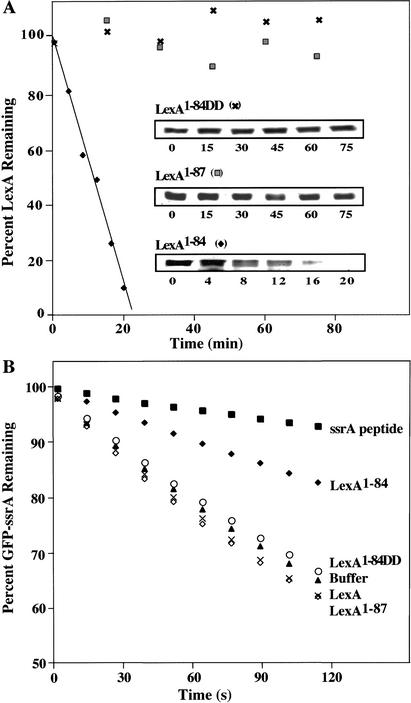
Figure 1.
LexA auto-cleavage fragments are captured and degraded by ClpXP. (A) Western blot of the LexA species captured by the ClpPtrap and generated by auto-cleavage in vitro. (B) The proteolytic stability of LexA, LexA1–84, and LexA85–202 in vivo was determined in clpX+ (MC4100) and clpX− (SG22101) strains bearing plasmid pJL42. LexA, LexA1–84, and LexA85–202 were identified by comparison with auto-cleaved LexA (lane S). It is difficult to see the continued disappearance of LexA, as expected from auto-processing, because the Western blots are mildly overexposed to allow observation of the low levels of LexA85–202. (C) ClpXP degradation of purified LexA, LexA1–84, and LexA85–202 in vitro. The band marked ATP-RS is creatine kinase.
To determine the stability of the various forms of LexA in vivo, we induced the DNA-damage response with nalidixic acid, blocked protein synthesis, and measured the half-lives of intact LexA and the two cleavage fragments. The N-terminal LexA1–84 fragment was rapidly degraded in a wild-type strain (half-life 2–5 min) but was not degraded to any detectable extent in an otherwise isogenic clpX− strain (Fig. 1B). The C-terminal LexA85–202 fragment was also unstable in wild-type cells (half-life ∼1 min) but was only stabilized modestly in the clpX− cells (half-life ∼2 min), presumably because Lon protease also contributes to its degradation (Little 1983b). Full-length LexA appeared to be reasonably stable to degradation in the wild-type and clpX− strains, although it was susceptible to continued auto-cleavage (Fig. 1B).
To determine if the LexA fragments were indeed ClpXP substrates, we assayed for degradation in vitro. Purified LexA, LexA1–84, and LexA85–202 were incubated with ClpXP and an ATP-regenerating system, and degradation was assayed by SDS-PAGE (Fig. 1C). ClpXP degraded both fragments but not LexA in ATP-dependent reactions. Hence, both auto-cleavage fragments of LexA are ClpXP substrates, whereas the full-length protein is not.
The new C-terminal sequence of LexA1–84 targets it to ClpXP
RecA-stimulated auto-cleavage of LexA creates a Val 82–Ala 83–Ala 84–COOH sequence at the end of the N-terminal fragment, which is very similar to the Leu–Ala–Ala–COOH sequence at the end of the ssrA degradation tag (Tu et al. 1995; Keiler et al. 1996). These residues of the ssrA tag are the principal determinants of recognition by ClpX (Flynn et al. 2001). To test the importance of the C-terminal residues of LexA1–84 for ClpXP degradation, we constructed a variant (LexA1–84DD) with aspartic acids replacing both alanines. In ClpXP degradation assays in vitro (Fig. 2A), LexA1–84DD was not degraded to any detectable extent, whereas LexA1–84 was degraded with a half-life of ∼10 min. Thus, these alanines play a major role in targeting the N-terminal LexA fragment for degradation by ClpXP.
Figure 2.
The new C terminus of LexA1–84 directs its degradation. (A) A time course of ClpXP degradation of LexA1–84, LexA1–87, and LexA1–84DD was quantified after SDS-PAGE. (B) ClpXP degradation of GFP-ssrA was measured by loss of fluorescence at 511 nm in the presence of various forms of LexA at 20 μM. An effective inhibitor of GFP-ssrA degradation, the ssrA peptide (20 μM), is included for comparison.
To test whether these alanines need to be at the C terminus for recognition, we constructed and purified a variant, LexA1–87, extended by the next three residues (Gly 85–Glu 86–Pro 87) of intact LexA. ClpXP did not degrade this variant (Fig. 2A), revealing that the Val 82–Ala 83–Ala 84 sequence must be at the C-terminal end for efficient degradation. LexA1–84 also inhibited degradation of another ClpXP substrate, GFP bearing an ssrA degradation tag, suggesting that LexA1–84 and ssrA-tagged proteins compete for ClpX recognition or other processes involved in degradation (Fig. 2B). In contrast, LexA1–84DD, LexA1–87, and full-length LexA failed to inhibit GFP-ssrA degradation. Together, these results demonstrate that LexA1–84 carries a functional ClpXrecognition signal, whereas LexA1–84DD, LexA1–87, and the full-length protein lack an accessible or functional signal.
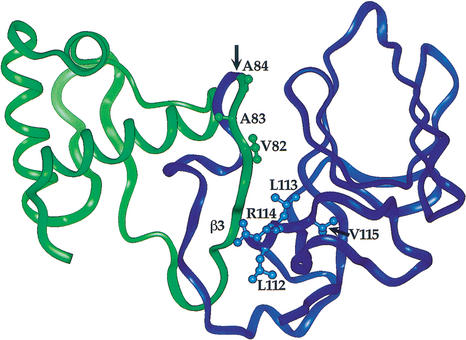
The Val 82–Ala 83–Ala 84 sequence is within a folded region in the LexA crystal structure, and the side chain of Val 82 is buried and packs against residues in the C-terminal domain (Luo et al. 2001; Fig. 3). As a consequence, it seems likely that auto-cleavage results in exposure and increased flexibility of the Val 82–Ala 83–Ala 84 sequence, allowing unimpeded interactions with ClpX. However, as the LexA1–87 fragment is not a ClpXP substrate, improved accessibility of ClpX to the Val 82–Ala 83–Ala 84 sequence is not sufficient to explain how auto-cleavage activates proteolysis of this domain. Based on peptide studies, the α-COOH group of the ssrA degradation tag has been shown to be an important determinant for ClpX recognition (Kim et al. 2000). The resistance of LexA1–87 to degradation strongly suggests that the α-COOH group of Ala 84 is similarly important for recognition of LexA1–84. Thus, our results indicate that auto-cleavage of the LexA Ala 84–Gly 85 peptide bond directly creates an essential portion of the degradation signal for the resulting N-terminal fragment.
Figure 3.
Location of latent ClpX-recognition signals in the structure of full-length LexA. The N-terminal domain is green and the C-terminal domain is blue. The proposed ClpX-recognition site in the C-terminal domain is labeled. The cleavage site is indicated by an arrow. (Structure from Luo et al. 2001.)
Sequences that target the C-terminal LexA fragment to ClpXP
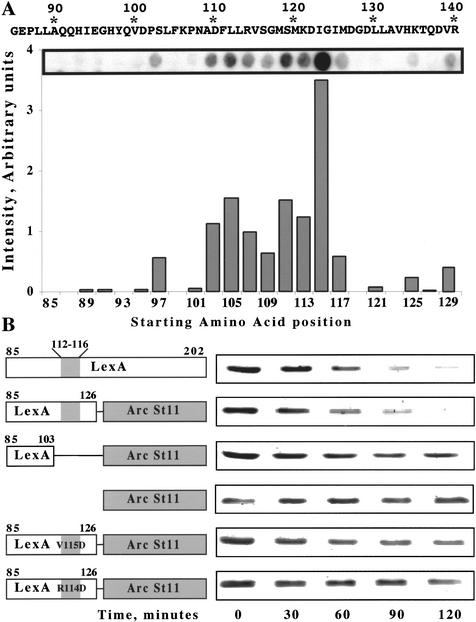
Although most LexA85–202 molecules seem to be degraded by Lon protease in vivo (Little 1983b), our experiments show that LexA85–202 is also degraded by ClpXP (Fig. 1B,C). To search for peptide sequences within LexA85–202 that might mediate ClpX recognition, we tested which regions of this fragment bound to ClpX (Fig. 4A). We prepared a covalent array of synthetic 12-residue peptides of the LexA85–202 fragment, each sharing a 10-residue overlap with its sequence neighbors. The peptide filter was incubated with ClpX and washed, and bound ClpX was detected using an anti-ClpX antibody. Peptides corresponding to the first 26 amino acids of LexA85–202 bound ClpX poorly. In contrast, a cluster of adjacent peptides containing sequences from 103 to 126 bound ClpX reasonably well. Hence, this region of the LexA sequence may contain a ClpX-recognition signal.
Figure 4.
Identification of residues in LexA85–202 important for ClpXP recognition. (A) An array of LexA peptides was incubated with purified ClpX protein, and bound ClpX was detected using anti-ClpX antibody. The chart shows the relative intensity of ClpX binding to individual sequences. The starting amino acid positions of the 12-residue peptides are listed below the chart. The signal intensity of the peptide beginning at residue 115 may be artificially high. In other experiments, we have observed high signals from different peptides with the same three initial residues (VSG). (B) Degradation of LexA85–202 and LexA-Arc-st11 fusion proteins by ClpXP in vitro.
To investigate the function of this sequence, we constructed two fusion proteins consisting of LexA residues 85–103 or 85–126 attached to the Arc-st11 protein, a variant of the Arc repressor with a stabilizing C-terminal sequence (Milla et al. 1993). ClpXP degraded the LexA85–126 fusion protein with a half-life (50 ± 9 min) similar to the half-life for LexA85–202 (60 ± 3 min; Fig. 4B). This result shows that sequence information between residues 85 and 126 of LexA is sufficient to target a protein to ClpXP for degradation. In contrast, the LexA85–103 fusion protein was degraded substantially more slowly (t1/2 = 184 ± 24 min), indicating that the most important determinants for ClpXP degradation are located between residues 104 and 126, and not between residues 85 and 103.
The 104–126 region contains a sequence (Leu 112–Leu 113–Arg 114–Val 115–Ser 116) with some similarity to a peptide motif proposed to function as a ClpX-recognition sequence (Flynn et al. 2003). To test the importance of the LexA sequence motif, we constructed LexA85–126 fusion proteins containing the Arg 114 → Asp or Val 115 → Asp mutations and found that both slowed ClpXP degradation (t1/2 = 118 ± 22 and 122 ± 30 min, respectively; Fig. 4B). Taken together, these data support the conclusion that ClpX recognizes a region of the polypeptide chain about 30 residues distal from the new N terminus of LexA85–202. Other sequences, however, may also contribute to ClpX recognition of LexA85–202.
Unlike LexA1–84, the ClpX-targeting signal on the C-terminal domain is not directly adjacent to the site of auto-cleavage. To account for the fact that intact LexA, which contains the same peptide sequences as LexA85–202, does not interact with ClpX, we propose that auto-cleavage disrupts the structure of the C-terminal domain in some fashion, helping to expose the 112–116 peptide signal. Although we do not know the structure of the isolated C-terminal domain, it is reasonable to propose that the 112–116 region will be more flexible and exposed after cleavage. In the structure of the full-length protein, a β-strand (β3) from the N-terminal domain forms an integral part of the C-terminal domain and contacts the proposed recognition signal. The loss of these contacts following cleavage may result in exposure of this signal to ClpXP (Fig. 3).
ClpX helps cells survive DNA damage
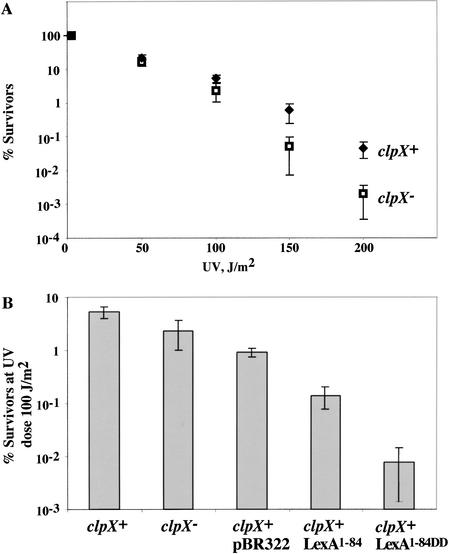
To test the importance of ClpX in the overall cellular response to DNA damage, we assayed cell survival after exposure to increasing doses of UV irradiation (Fig. 5A). These experiments showed that clpX− cells were more sensitive to UV irradiation than their wild-type counterparts, with the effect being most pronounced at the highest UV doses. When exposed to a UV dose of 200 J/m2, clpX− cells had a roughly 10-fold lower survival frequency than wild-type cells. Because the N-terminal DNA-binding domain of LexA can function as a repressor on its own (Schmidt-Dorr et al. 1991), one interpretation of this result is that ClpXP degradation of this fragment may be required to allow maximal expression of one or more DNA-damage inducible gene products, thereby improving survival following near-lethal UV doses. Alternatively, the loss of ClpX could perturb the levels of proteins other than the LexA1–84, thus affecting survival.
Figure 5.
Cells accumulating LexA1–84 are UV sensitive. (A) Survival after UV irradiation of clpX+ (MC4100) and clpX− (SG22101) cells. The percentage of surviving cells is plotted against UV dose. Each point is the average (±S.D.) of three trials. (B) Survival of cells containing the empty parent vector or plasmids directing expression of LexA1–84 or LexA1–84DD was compared at a UV dose of 100 J/m2.
To determine if excess levels of LexA1–84 causes increased UV sensitivity, we tested the effect of expressing LexA1–84 and LexA1–84DD on the percentage of cells surviving a fixed UV dose. We assayed survival of wild-type cells containing plasmid-borne genes for either LexA1–84 or LexA1–84DD under the control of the native LexA promoter (Fig. 5B); Western analysis revealed that these fragments were modestly overexpressed (two- to fourfold, with LexA1–84DD accumulating to higher levels than LexA1–84) compared with the chromosomal-encoded LexA (data not shown). Cells expressing LexA1–84DD had ∼10-fold lower survival than those with the LexA1–84-producing plasmid. Furthermore, both strains were more sensitive to UV irradiation than cells bearing the empty plasmid. Thus, accumulation of the LexA DNA-binding domain is deleterious to cell survival after DNA damage.
These data reveal that timely destruction of LexA1–84 following damage-induced auto-cleavage is likely to play an important biological role. We suggest that failure to degrade the LexA1–84 fragment following DNA damage results in the retention of some repressor activity and a consequent failure to fully induce one or more DNA-damage genes. This residual repressor activity, in turn, may account for our finding that ClpX improves bacterial survival following near-lethal UV doses. Our observation that cells expressing even modest levels of the nondegradable LexA1–84DD are more sensitive to UV irradiation than those expressing LexA1–84 supports this theory, although we cannot currently rule out more indirect explanations. In preliminary work, we did not detect reproducible differences in expression of the LexA-controlled sulA, recA, and uvrA genes following DNA damage in wild-type versus ClpX-defective cells (data not shown), but there are more than 20 genes under LexA control and some may be more sensitive than others to repression mediated by the LexA1–84 fragment.
Changes in environmental conditions often result in the modification of regulatory proteins by phosphorylation or proteolytic cleavage. Similarly, protein-binding partners often change during the progression of a biochemical pathway. The results presented here show that ClpXP degrades the two LexA cleavage fragments but not the intact protein. By mapping recognition signals in LexA1–84 and LexA85–202, we show that auto-cleavage activates otherwise dormant protease recognition signals in both fragments. Our analysis of the mechanisms used to target LexA to ClpXP thus highlights how changes in protein primary or tertiary structure can be coupled to the exposure of recognition signals for a destructive protease, thereby triggering a cascade of protein turnover in response to environmental change.
LexA is a member of a protein family that includes several phage repressors that also undergo RecA-mediated auto-cleavage (Sauer et al. 1982; Little 1984; Eguchi et al. 1988). It will be interesting to see if ClpXP plays a role in degrading the auto-cleavage fragments of these proteins. The fragments of unrelated proteins that undergo proteolytic processing events may also be targeted for degradation by a similar mechanism. For example, RseA is an antisigma factor that is sequentially cleaved by two membrane proteases in response to periplasmic stress (Alba et al. 2002). The N-terminal cytoplasmic domain of RseA released by cleavage is captured by ClpXPtrap (Flynn et al. 2003) and has been proposed to contain a Val–Ala–Ala–COOH sequence identical to the signal that targets LexA1–84 for ClpXP degradation (Alba et al. 2002). Thus, we suspect that dormant degradation signals like those we have characterized in LexA will be used to couple the destruction of other proteins to changes that occur in the intracellular or extracellular environment.
Materials and methods
Strains and plasmids
Strains MC4100 and SG22101 (MC4100 clpX∷kan) were used in UV survival assays. The pBR322-based plasmid pJWL42 contains wild-type lexA under the control of its native promoter (Markham et al. 1981). Plasmid pJWL228 was a construct for overexpression of LexA and consisted of wild-type lexA under the control of the T7 promoter (Shepley and Little 1996). Plasmids pSBN15 and pSBN24 were used for overexpression of LexA1–84 and LexA1–87, respectively and were derivatives of pJWL228. Stop codons were inserted in the coding sequence after LexA residue 84 or 87, respectively, using a Quickchange kit (Stratagene). Plasmid pSBN16 was derived from pJWL228 for overexpression of the N-terminal fragment of LexA with aspartates replacing the two terminal alanines (LexA1–84DD) using Quickchange. The plasmid for the expression of LexA85–202 (pSBN17) was constructed by PCR amplification of the sequence coding for amino acids 85–202 from pJWL228. The resulting PCR product was digested with BamHI and NdeI and ligated into BamHI/NdeI-digested pET11a. Plasmid pSBN19 was created for the expression of a fusion of LexA amino acids 85–126 to Arc-st11 (LexA85–126–Arc). It was made by amplifying the region of LexA coding for amino acids 85–126, digesting with NdeI and NheI, and ligating into NdeI/NheI-digested pET11a-arc-st11 (Flynn et al. 2003). The resulting N-terminal sequence was MGEPLLAQQHIEGHYQVDPSLFKPNADFLLRV SGMSMKDIGIMASMGK (LexA in italics, Arc in bold). Plasmid pSBN20 consisted of LexA residues 85–103 fused to Arc-st11 using an oligonucleotide cassette. The N-terminal sequence of this fusion was MGEPLLAQQHIEGHYQVDPSMGK. Plasmids pSBN21 and pSBN22 were R114D and V115D variants, respectively, of the LexA portion of pSBN19 and were constructed using the Quickchange kit. Plasmids pSBN22 and pSBN23 are variants of pJL42 for the expression of LexA1–84 and LexA1–84DD, respectively, under the control of the native promoter and were constructed as per pSBN15 and pSBN16. All constructs were verified by DNA sequencing.
Proteins/peptide array
Purification of GFP-ssrA, Arc-st11, ClpX (Flynn et al. 2003 and references therein), and ClpP (Kim et al. 2000) have been described. LexA85–202, LexA, LexA1–84, and LexA1–84DD were purified from overproducing strains essentially as described for LexA (Little et al. 1994). A LexA peptide array was prepared by the MIT biopolymers facility using an Abimed instrument. Peptides interacting with ClpX were detected as described (Flynn et al. 2003).
Trapping/detection of LexA
Trapping was carried out essentially as described using strain JF162 (Flynn et al. 2003) except that cultures were treated with 50 μg/mL of nalidixic acid for 2 h before expression of the ClpPtrap was induced. ClpP cross-reacts with the LexA antibody and therefore was removed before Western blotting. Samples were dialyzed against 8 M urea for 6 h, then incubated with Ni-NTA beads (Quiagen) with rocking for 1 h. Supernatant was removed, dialyzed against LexA storage buffer, concentrated using a centricon YM-3 concentrator (Microcon), and used for Western blotting (see following). In a control experiment, this treatment did not affect our ability to detect full-length LexA, indicating that the protein is stable under these conditions.
Degradation in vitro and in vivo
For in vitro degradation, ClpX6 (0.3 μm), ClpP14 (0.8 μm), and an ATP-regenerating system (4 mM ATP, 50 μg/mL creatine kinase, 2.5 mM creatine phosphate) were incubated in buffer NB (50 mM Tris-HCl, 100 mM KCl, 10 mM MgCl2, 1 mM DTT) at 30°C for 2 min. Substrate (10 μm) was added, and samples were removed at specific times, added to SDS loading buffer, and frozen on liquid nitrogen. After heating at 100°C for 5 min, samples were analyzed by 15% (N-LexA, C-LexA, Arc fusions) or 12.5% (LexA) SDS-PAGE. Gels were visualized by staining with Sypro orange (Molecular Probes) and scanning on a Molecular Dynamics Fluorimager 595. Imagequant (Molecular Dynamics) was used to quantify degradation. Inhibition of GFP-ssrA degradation was measured as described using 0.5 μM GFP-ssrA and 20 μM inhibitor (Flynn et al. 2001).
For in vivo degradation, samples were prepared from MC4100 and SG22101 cells containing plasmid pJL42. Cultures at an OD600 of 0.2 were treated with 50 μg/mL nalidixic acid (Sigma) and incubated with shaking at 37°C for 30 min, then chloramphenicol (Sigma) was added to 100 μg/mL to stop protein synthesis. For each time point, cells were harvested by centrifugation, resuspended in SDS loading buffer, and rapidly frozen on liquid nitrogen. Cells were lysed by boiling and centrifuged in a microfuge at top speed for 5 min, and the resulting extract was separated by 15% SDS-PAGE. Western blots were performed with an ECF Western blotting kit in accordance with the manufacturer's guidelines (Amersham) using rabbit polyclonal LexA antibody at a 1:5000 dilution (John Little). Blots were imaged using a Molecular Dynamics Fluorimager 595. LexA fragments were prepared from purified LexA as described (Little 1984).
UV survival assays
Cells were grown to an OD600 of 0.5 in Luria broth (LB), gently pelleted, washed once, and then resuspended in an equal volume of 0.85% saline. Suspensions were irradiated at a UV dose of 1.5 J/m2/sec for set times using a 15W G15T8 germicidal lamp (GE). The UV intensity was measured using a UVX radiometer (UVP). Appropriate dilutions were plated on LB agar plates (or LB agar plus 100 μg/mL ampicillin, as appropriate) and colonies were counted after 24 h. The decreased plating efficiency of cells carrying the pBR322 plasmid, compared with the nonplasmid-containing cells, was attributed to the need to maintain selection for the plasmid, based on parallel platings on media lacking antibiotic. Each assay was done at least in triplicate.
Acknowledgments
This work was supported by NIH grant AI-16892 and Howard Hughes Medical Institute. T.A.B. is an employee of Howard Hughes Medical Institute. We thank members of the Baker and Sauer labs for helpful discussions and careful reading, Graham Walker and members of the Walker lab for advice and use of equipment, and John Little for antibodies and plasmids.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tabaker@mit.edu; FAX (617) 252-1852.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1078003.
References
- Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigmaE-dependent extracytoplasmic stress response. Genes & Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker G, Klauck E, Hengge-Aronis R. Regulation of RpoS proteolysis in Escherichia coli: The response regulator RssB is a recognition factor that interacts with the turnover element RpoS. Proc Natl Acad Sci. 1999;96:649–644. doi: 10.1073/pnas.96.11.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi Y, Ogawa T, Ogawa H. Stimulation of RecA-mediated cleavage of phage phi 80 cI repressor by deoxydinucleotides. J Mol Biol. 1988;204:69–77. doi: 10.1016/0022-2836(88)90600-6. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn, J.M., Neher, S.B., Kim, Y.I., Sauer, R.T., and Baker, T.A. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell (In press). [DOI] [PubMed]
- Freidberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington, DC.: ASM Press; 1995. [Google Scholar]
- Gerth U, Kruger E, Derre I, Msadek T, Hecker M. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol Microbiol. 1998;28:787–802. doi: 10.1046/j.1365-2958.1998.00840.x. [DOI] [PubMed] [Google Scholar]
- Gonciarz-Swiatek M, Wawrzynow A, Um SJ, Learn BA, McMacken R, Kelley WL, Georgopoulos C, Sliekers O, Zylicz M. Recognition, targeting, and hydrolysis of the lambda O replication protein by the ClpP/ClpX protease. J Biol Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes & Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Luo L, Baker TA. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes & Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- Little JW. The SOS regulatory system: Control of its state by the level of RecA protease. J Mol Biol. 1983a;167:791–808. doi: 10.1016/s0022-2836(83)80111-9. [DOI] [PubMed] [Google Scholar]
- ————— . Variations in the in vivo stability of LexA repressor during the SOS regulatory cycle. In: Friedberg EC, Bridges BA, editors. Cellular responses to DNA damage. New York: Alan Liss; 1983b. pp. 369–378. [Google Scholar]
- ————— Autodigestion of lexA and phage lambda repressors. Proc Natl Acad Sci. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW, Kim B, Roland KL, Smith MH, Lin LL, Slilaty SN. Cleavage of LexA repressor. Methods Enzymol. 1994;244:266–284. doi: 10.1016/0076-6879(94)44022-0. [DOI] [PubMed] [Google Scholar]
- Luo Y, Pfuetzner R, Mosimann S, Paetzel M, Frey E, Cherney M, Kim B, Little JW, Strynadka N. Crystal structure of LexA: A conformational switch for regulation of self-cleavage. Cell. 2001;106:585–594. doi: 10.1016/s0092-8674(01)00479-2. [DOI] [PubMed] [Google Scholar]
- Markham BE, Little JW, Mount DW. Nucleotide sequence of the lexA gene of Escherichia coli K-12. Nucleic Acids Res. 1981;9:4149–4161. doi: 10.1093/nar/9.16.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla ME, Brown BM, Sauer RT. P22 Arc repressor: Enhanced expression of unstable mutants by addition of polar C-terminal sequences. Protein Sci. 1993;2:2198–2205. doi: 10.1002/pro.5560021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S, Gottesman S. Protein degradation in Escherichia coli: The lon gene controls the stability of sulA protein. Proc Natl Acad Sci. 1983;80:358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philzicky EM, Roberts JW. Induction of SOS function: Regulation of proteolytic activity of E. coli RecA protein by interaction with DNA and nucleoside triphosphate. Cell. 1981;25:259–267. doi: 10.1016/0092-8674(81)90251-8. [DOI] [PubMed] [Google Scholar]
- Roberston G, Ng W, Foley J, Gilmour R, Winker M. Global transcriptional analysis of ClpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J Bacteriol. 2002;184:3508–3520. doi: 10.1128/JB.184.13.3508-3520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Ross MJ, Ptashne M. Cleavage of the lambda and P22 repressors by recA protein. J Biol Chem. 1982;257:4458–4462. [PubMed] [Google Scholar]
- Schmidt-Dorr T, Oertel-Buchheit P, Pernelle C, Bracco L, Schnarr M, Granger-Schnarr M. Construction, purification, and characterization of a hybrid protein comprising the DNA binding domain of the LexA repressor and the Jun leucine zipper: A circular dichroism and mutagenesis study. Biochemistry. 1991;30:9657–9664. doi: 10.1021/bi00104a013. [DOI] [PubMed] [Google Scholar]
- Shepley DP, Little JW. Mutant LexA proteins with specific defects in autodigestion. Proc Natl Acad Sci. 1996;93:11528–11533. doi: 10.1073/pnas.93.21.11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu GF, Reid GE, Zhang JG, Moritz RL, Simpson RJ. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J Biol Chem. 1995;270:9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. The RssB response regulator directly targets sigmaS for degradation by ClpXP. Genes & Dev. 2001;15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]