Abstract
We have identified Xenopus homologs of the budding yeast Sld5 and its three interacting proteins. These form a novel complex essential for the initiation of DNA replication in Xenopus egg extracts. The complex binds to chromatin in a manner dependent on replication licensing and S-phase CDK. The chromatin binding of the complex and that of Cdc45 are mutually dependent and both bindings require Xenopus Cut5, the yeast homolog of which interacts with Sld5. On replicating chromatin the complex interacts with Cdc45 and MCM, putative components of replication machinery. Electron microscopy further reveals that the complex has a ring-like structure. These results suggest that the complex plays an essential role in the elongation stage of DNA replication as well as the initiation stage.
Keywords: DNA replication, Initiation Complex, MCM, Cdc45, S-phase CDK
Molecular mechanisms for the initiation of eukaryotic DNA replication have been investigated using various model systems. In particular, a cell-free replication system from Xenopus eggs and the single-cell systems of budding and fission yeasts have provided us with a unified view of the control of initiation (Bell and Dutta 2002; Nishitani and Lygerou 2002). A key feature of the control mechanism is that chromatin must be “licensed” for replication at the end of M phase and during the G1 period (Blow and Hodgson 2002). The licensed state is established by the sequential assembly of replication proteins onto origins. First, the binding of the ORC (origin recognition complex) to chromatin marks the replication origins. Then Cdc6 and Cdt1 bind to chromatin in a manner dependent on the ORC binding. The binding of Cdc6 and Cdt1 is, in turn, required for the loading of MCM onto chromatin, which is a crucial step for the formation of prereplicative complexes (pre-RCs). The licensed state can be defined as the formation of pre-RCs onto origins (Takisawa et al. 2000), and molecules involved in the formation of pre-RCs have been well characterized so that pre-RCs have been successfully reconstituted on purified DNA with the Xenopus system (Gillespie et al. 2001).
At the onset of S phase, pre-RCs are activated by cyclin-dependent kinase (CDK) and Dbf4-dependent Cdc7 kinase (DDK), converting them into initiation complexes (ICs; Bell and Dutta 2002; Blow and Hodgson 2002; Masai and Arai 2002). This process would involve the melting of DNA at origins, the activation of a replicative DNA helicase, and the association of single-stranded DNA-binding proteins with unwound origins. Subsequently, replication machinery is assembled at the unwound origin, and the loading of the polymerase α-primase complex initiates the synthesis of an RNA primer. Processive synthesis of DNA following the primer synthesis has been extensively studied with the SV40 DNA replication system (Waga and Stillman 1994). In particular, the function of RFC, a clamp loader, and that of PCNA, a polymerase clamp, have been well documented at the switching of polymerase α to δ. However, the molecular components of the replication machinery and the functional role of each component are only partially understood, including the function of DNA polymerase ɛ, the third essential polymerase in cellular DNA replication (Morrison et al. 1990; Kesti et al. 1999; Waga et al. 2001; Ohya et al. 2002).
Accumulating evidence suggests that Cdc45 is involved in the conversion of pre-RCs to ICs. Cdc45 binds to chromatin after the formation of pre-RCs in a CDK- and DDK-dependent manner, and is required for the loading of polymerase α and ɛ onto chromatin. In addition, it has been reported that Cdc45 induces the local unwinding of plasmids in a nuclear-free system derived from Xenopus egg extracts (Walter and Newport 2000). Dpb11/Cut5 is another essential protein involved in recruiting the polymerases. DPB11 in budding yeast was isolated as a multicopy suppressor of the mutant of DPB2, which encodes the second largest subunit of polymerase ɛ (Araki et al. 1995). Dpb11 and its homologs, including fission yeast Cut5, Drosophila Mus101, and human TopBP1, participate in DNA replication and checkpoints (Saka and Yanagida 1993; Yamamoto et al. 2000; Makiniemi et al. 2001; Yamane et al. 2002). Chromatin immunoprecipitation (ChIP) assay of Dpb11 further demonstrates that Dpb11 is essential for the loading of DNA polymerase α and ɛ onto replication origins (Masumoto et al. 2000). Recently, we have identified a Xenopus homolog of a Dpb11/Cut5, which we call Xenopus Cut5. We have shown that the binding of Xenopus Cut5 to chromatin is required for the action of S-phase CDK (S-CDK) and that it is therefore required for the chromatin binding of Cdc45, which eventually leads to the loading of polymerase α and ɛ onto chromatin (Hashimoto and Takisawa 2003). Essentially the same gene has been recently identified as a Xenopus homolog of Mus101, which is required for the loading of Cdc45 onto chromatin (Van Hatten et al. 2002).
In an effort to clarify the role of DNA polymerase ɛ, a group of six genes called SLD (the synthetic lethal mutants of dpb11-1) has been isolated by genetic screening of budding yeast (Kamimura et al. 1998). Sld1 is identical to Dpb3, the third largest subunit of polymerase ɛ; Sld4 is identical to Cdc45; and Sld6 is identical to Rad53, which is required for intra-S and damage checkpoints. SLD2, SLD3, and SLD5 are novel genes. Recent studies with Sld2/Drc1 reveal that Sld2 is phosphorylated by S-CDK and that the phosphorylation of Sld2 at the onset of S phase is essential for its association with Dpb11 and for the initiation of DNA replication (Masumoto et al. 2002; Noguchi et al. 2002). Sld3 in budding and fission yeast has been shown to be required for the association of Cdc45 with replication origins (Kamimura et al. 2001; Nakajima and Masukata 2002). Although homologs of Sld2 and Sld3 have not been identified in higher eukaryotes yet, it is expected that these proteins are conserved in eukaryotes. SLD5 is the least characterized SLD gene, and studies with budding yeast described in Takayama et al. (2003) have further identified interacting genes called PSF1 (partner of SLD five), PSF2, and PSF3, all of which are essential for growth.
Here, we report the identification and characterization of Xenopus homologs of Sld5 and its associated partners, Psf1, Psf2, and Psf3. The four proteins form a tetrameric complex in Xenopus egg extracts, and this novel complex is required for the initiation of DNA replication. Its binding to chromatin is mutually dependent on Cdc45 binding to chromatin, and both bindings require pre-RC formation and S-CDK. The complex formation of Sld5–Psf1–Psf2–Psf3 with Cdc45 and MCM on replicating chromatin suggests that Sld5–Psf1–Psf2–Psf3, cooperating with Cdc45, performs an essential role in the initiation stage of DNA replication. The function of the ring-like structure of the novel complex is also discussed.
Results
Isolation of Xenopus Sld5, Psf1, Psf2, and Psf3 homologs
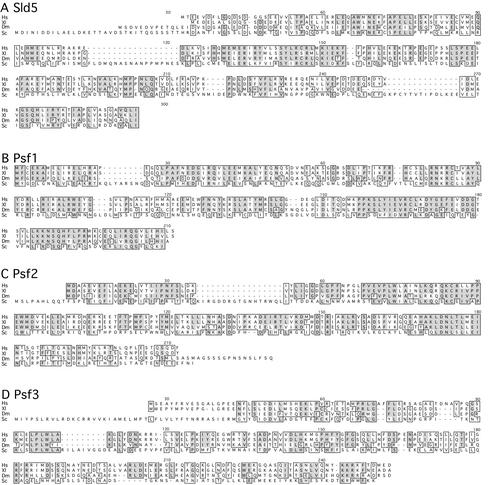
We isolated Xenopus homologs of the yeast proteins based on their sequence similarity (details are described in Materials and Methods). The open reading frames of the Xenopus Sld5, Psf1, Psf2, and Psf3 cDNAs thus obtained encode proteins consisting of 223, 196, 185, and 216 amino acids, respectively. The predicted amino acid sequences of Xenopus Sld5, Psf1, Psf2, and Psf3 show 46%, 53%, 51%, and 42% amino acid similarities to the yeast proteins. BLAST searches further showed that homologous proteins are widely distributed in eukaryotes including human, mouse, fly, worm, plants, and fission yeast, suggesting that they are evolutionarily conserved proteins (cf. Fig. 1). However, we could not find any known motifs in these proteins by sequence motif searching. Because all the yeast genes are essential for cell growth, these results suggest that Sld5, Psf1, Psf2, and Psf3 are novel proteins required for the proliferation of eukaryotic cells.
Figure 1.
Alignment of predicted amino acids sequences of Sld5 (A), Psf1 (B), Psf2 (C), and Psf3 (D) from human (Hs), Xenopus (Xl), Drosophila (Dm), and Saccharomyces cerevisiae (Sc). Identical amino acids are shaded, and highly conserved regions are boxed.
Xenopus Sld5, Psf1, Psf2, and Psf3 form a tetrameric complex in Xenopus egg extracts
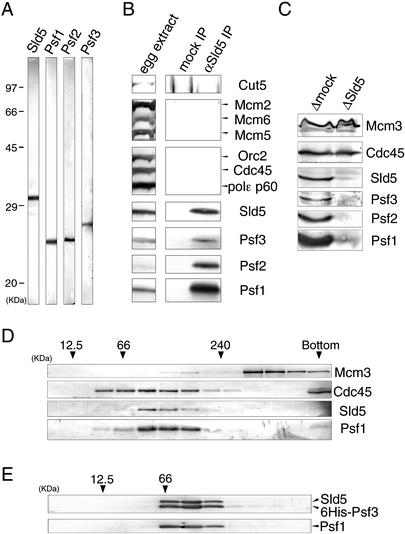
We next examined the role of Xenopus Sld5, Psf1, Psf2, and Psf3 in DNA replication using cell-free extracts of Xenopus eggs. We expressed the recombinant proteins in baculovirus-infected Sf9 cells and produced antibodies against the recombinant Xenopus proteins. The antibodies against Sld5, Psf1, Psf2, and Psf3 specifically recognized proteins in the egg extracts with apparent molecular masses of 32 kD, 24 kD, 25 kD, and 28 kD, respectively (Fig. 2A). Using the antibodies, we first investigated the physical interactions among these proteins, because genetic analysis of the yeast mutants and comprehensive analysis of the protein–protein interactions in yeast indicate that the interaction of these proteins does occur. Immunoprecipitation from egg extracts with anti-Sld5 antibody showed that Psf1, Psf2, and Psf3 coimmunoprecipitate with Sld5 (Fig. 2B). However, we could not detect the coimmunoprecipitation of Cut5, DNA polymerase ɛ, Mcm2, Mcm5, Mcm6, Orc2, and Cdc45. We also found that Psf1, Psf2, and Psf3 were concomitantly depleted from the extracts upon the depletion of Sld5 (Fig. 2C). These results indicate that Xenopus Psf1, Psf2, and Psf3 form a complex with Sld5 in egg extracts.
Figure 2.
Xenopus Sld5, Psf1, Psf2, and Psf3 form a complex in Xenopus egg extracts. (A) Specificity of antibodies. S-phase egg extract was resolved with SDS-PAGE and immunoblotted with the antibodies indicated in the figure. (B,C) Immunoprecipitation and immunodepletion of Sld5 from egg extracts. S-phase egg extracts were treated with preimmune (mock) or anti-Sld5 antibodies conjugated to protein A beads. The immunoprecipitates (B) and the depleted extracts (C) were resolved by SDS-PAGE, blotted, and probed with antibodies against the various proteins indicated in the figure. (D,E) Separation of proteins by glycerol gradient centrifugation. Soluble fractions of egg extracts (D) or purified complex of recombinant Sld5–Psf1–Psf2–Psf3 (E) were analyzed on glycerol gradients. Fractions were resolved by SDS-PAGE, blotted, and probed with antibodies against the proteins indicated in the figure. Marker proteins were centrifuged under identical conditions; peak positions of the marker proteins are indicated by arrowheads with their molecular weights.
To examine the physical properties of complexes containing Sld5, Psf1, Psf2, and Psf3, egg extracts were subjected to glycerol gradient centrifugation. The fractions containing Sld5 or Psf1 sedimented on the glycerol gradient in a peak corresponding to an apparent molecular mass of 100 kD (Fig. 2D), and similar peak fractions were observed with Psf2 and Psf3 (data not shown). The recombinant complex of Sld5, Psf1, Psf2, and Psf3 (Fig. 3B) sedimented in a similar peak to that observed with endogenous proteins (Fig. 2E). Cdc45, however, sedimented on the gradient in broad peaks ranging from 60 kD to 200 kD, and MCM sedimented in a peak of >300 kD. These results show that Xenopus Sld5, Psf1, Psf2, and Psf3 form a stable complex of ∼100 kD in the egg extracts. Hereafter, we refer to this complex as GINS (Go-Ichi-Ni-San, 5-1-2-3 in Japanese).
Figure 3.
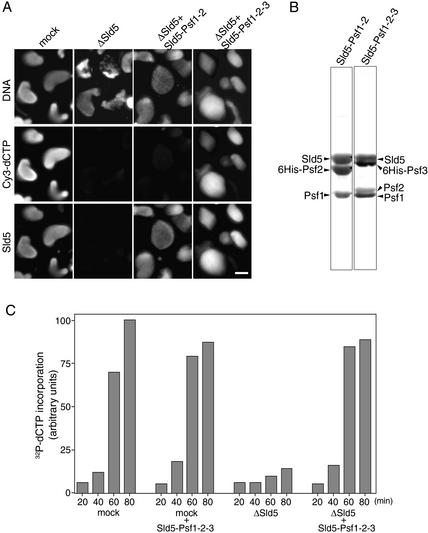
Requirement of GINS for DNA replication in Xenopus egg extracts. (A) Immunofluorescent detection of nuclear Sld5. Xenopus sperm chromatin was incubated in the presence of Cy3-dCTP in mock- or Sld5-depleted extracts supplemented with or without recombinant Sld5–Psf1–Psf2 or Sld5–Psf1–Psf2–Psf3 (GINS). After incubation at 23°C for 45 min, samples were fixed and centrifuged through 30% sucrose onto coverslips. Nuclear localization of Sld5 was visualized with rabbit anti-Xenopus-Sld5 antibody followed by Alexa488-labeled anti-rabbit IgG. DNA replication was monitored as the incorporation of Cy3-dCTP into DNA, and DNA was visualized with Hoechst 33258 dye. Bar, 10 μm. (B) Protein compositions of recombinant Xenopus Sld5–Psf1–Psf2 and Sld5–Psf1–Psf2–Psf3 complexes. Xenopus Sld5, Psf1, and Psf2 were coexpressed with and without Psf3 (either Psf2 or Psf3 was tagged with His6) in Sf9 cells using a baculovirus expression system. The protein complexes, purified with Ni-NTA resin, were resolved by SDS-PAGE and stained with Ponceau S. (C) Replication activities of Sld5-depleted extracts. Xenopus sperm chromatin was incubated in mock- or Sld5-depleted extracts in the presence of 32P-labeled dCTP, supplemented with or without recombinant GINS. At the indicated times, DNA was isolated and subjected to alkali agarose gel electrophoresis followed by autoradiography. The amount of 32P incorporated into DNA was quantified and expressed in arbitrary units.
Xenopus GINS is required for DNA replication in egg extracts
The role of GINS in DNA replication was examined by performing in vitro DNA replication of Xenopus sperm chromatin with mock- or Sld5-depleted egg extracts. In both extracts, nuclear envelopes were formed around decondensed sperm chromatin within 30 min of incubation. In the mock-depleted extracts, the chromatin showed highly decondensed morphology, accompanied by the incorporation of Cy3-dCTP upon further incubation, and Sld5 became colocalized with chromatin after the formation of the nuclear envelope (Fig. 3A). In Sld5-depleted extracts, neither Sld5 signals in the nuclei nor the incorporation of Cy3-dCTP into DNA was detected (Fig. 3A, middle panels), showing that Sld5 and possibly its associated proteins Psf1, Psf2, and Psf3 are required for DNA replication.
To identify the proteins required for replication in the Sld5-depleted extracts, we produced recombinant Xenopus Sld5, Sld5–Psf1–Psf2 and Sld5–Psf1–Psf2–Psf3 (GINS) complexes using baculovirus-infected insect cells and purified the proteins to near homogeneity (see details in Materials and Methods). Figure 3B shows the protein composition of the recombinant Sld5–Psf1–Psf2 and GINS complexes, consisting of nearly equal amounts of each protein. When these recombinant proteins were added back to the Sld5-depleted extracts, the GINS complex but neither Sld5 nor the Sld5–Psf1–Psf2 complex could rescue the replication activity of the depleted extracts (Fig. 3A; data not shown). These results show that GINS is necessary and sufficient for the recovery of DNA replication in the Sld5-depleted extracts.
Similar results were obtained by examining the time course of incorporation of 32P-labeled dCTP into DNA (Fig. 3C). With Sld5-depleted extracts, the incorporation rate was markedly reduced, and the amount of incorporation after 80 min of incubation was <20% of that in the mock-depleted extracts. The addition of recombinant GINS to the depleted extracts caused efficient recovery of replication activity to a similar level to the mock-depleted extracts, whereas the same addition to mock-depleted extracts did not affect their incorporation of dCTP (Fig. 3C). Analysis of replication products by agarose gel electrophoresis further showed that no small replication products were generated with Sld5-depleted extracts (data not shown). These results show that the GINS complex is required for DNA replication in egg extracts.
Pre-RC- and S-CDK-dependent binding of GINS to chromatin
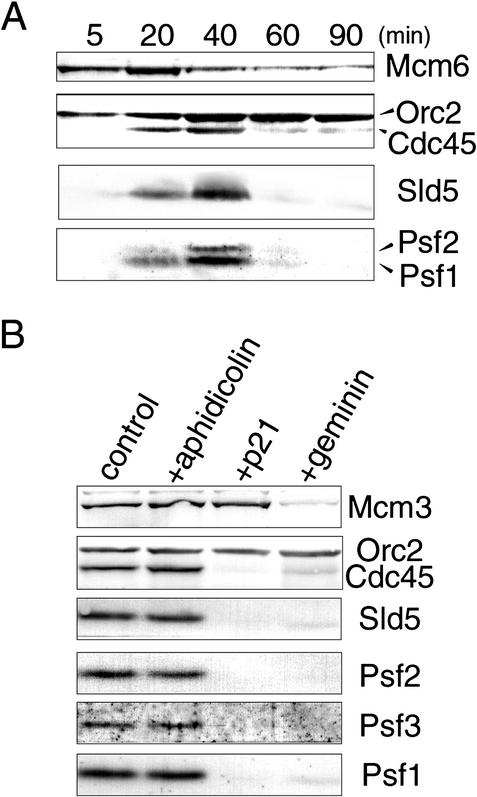
Because Sld5 appears to be a nuclear protein (Fig. 3A), we next examined the chromatin binding of the GINS complex during DNA replication in egg extracts (Fig. 4A). When sperm chromatin was incubated in the extracts, ORC and MCM complexes were assembled onto decondensed chromatin to form pre-RCs within 5 min, at a time before the formation of nuclear envelopes. After the formation of pre-RCs, Cdc45 was loaded onto the chromatin at ∼20 min, at the same time as when replication is initiated (data not shown). Xenopus Sld5, Psf1, and Psf2 became associated with chromatin after 20 min and subsequently dissociated from chromatin during the progression of DNA replication, with a very similar time course to that of Cdc45. In contrast, Orc2 remained on chromatin throughout the incubation. These results suggest that the GINS complex is involved in DNA replication after the formation of pre-RCs. Some Mcm6 still remained on chromatin after Cdc45 had come off. These remaining Mcm molecules likely represent protein bound to unreplicated chromatin, because the first cycle of replication is sometimes incomplete in this system (Blow and Laskey 1986; Hutchison et al. 1988).
Figure 4.
Requirement of replication licensing and S-CDK activity for the chromatin binding of Xenopus GINS. (A) The time course of chromatin binding of replication-related proteins. Sperm chromatin was incubated in egg extracts, and samples were collected at the indicated times. Chromatin fractions isolated by centrifugation were resolved by SDS-PAGE and immunoblotted with antibodies as indicated in the figure. (B) Effect of inhibitors of DNA replication on the chromatin binding of Sld5, Psf1, Psf2, and Psf3. Sperm chromatin was incubated in egg extracts in the absence of inhibitors (control) and the presence of aphidicolin (20 μg/mL), p21 (50 μg/mL), or geminin (15 μg/mL). After incubation for 45 min, chromatin fractions were isolated, resolved by SDS-PAGE, and immunoblotted with the indicated antibodies.
To determine the precise requirement for the chromatin binding of GINS, we used three types of inhibitors of DNA replication: geminin, p21, and aphidicolin. Geminin inhibits the formation of pre-RCs by binding to Cdt1 (Wohlschlegel et al. 2000; Tada et al. 2001). The CDK inhibitor p21 is known to prevent the loading of Cdc45 onto chromatin (Mimura and Takisawa 1998). Aphidicolin is a potent inhibitor of DNA polymerases, namely, DNA polymerase α, δ, and ɛ, thus preventing the elongation of DNA replication. Both geminin and p21 markedly blocked the binding of the Xenopus Sld5, Psf1, Psf2, and Psf3 onto chromatin, but aphidicolin did not affect the chromatin binding of these proteins (Fig. 4B). These results show that pre-RC formation, monitored by the binding of Mcm3 to chromatin, and also S-CDK activity are required for the loading of the GINS complex onto chromatin.
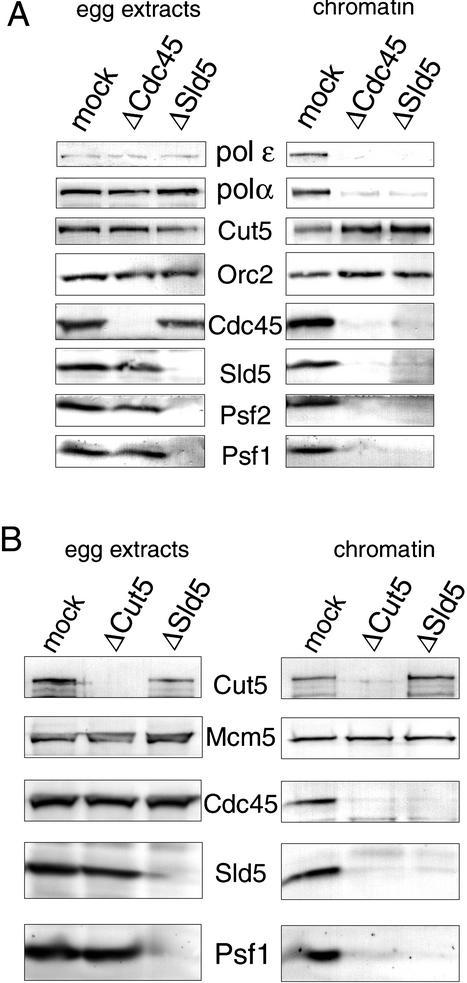
Interdependent binding of Xenopus GINS and Cdc45 to chromatin
In eukaryotic DNA replication, several replication proteins should be loaded onto origins in a precise order at specific stages of the cell cycle to initiate replication. Xenopus GINS bound to chromatin after pre-RC formation in an S-CDK-dependent manner. Xenopus Cdc45 binds to chromatin under the same conditions and with very similar timing to Sld5, Psf1, and Psf2. To clarify the order of chromatin binding of the GINS and Cdc45, we prepared the extracts immunodepleted of Sld5 or Cdc45 and examined the chromatin binding of these proteins in the depleted extracts (Fig. 5A). The depletion of either Sld5 or Cdc45 did not affect the level of the other protein in the extracts (Fig. 5A, left panels) and we could not detect the coimmunoprecipitation of Cdc45 with Sld5 (Fig. 2B). These results indicate that Sld5 and Cdc45 do not stably associate with each other in the extracts.
Figure 5.
(A) Interdependent binding of Cdc45 and GINS to chromatin. Cdc45 or Sld5 was immunodepleted from the extracts as described in the legend for Figure 2C and chromatin was assembled with mock-, Cdc45-, and Sld5-depleted extracts at 23°C for 45 min. The depleted extracts (egg extracts) and the chromatin fractions (chromatin) were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (B) Cut5-dependent binding of Cdc45 and Sld5 to chromatin. Xenopus Cut5 or Sld5 was immunodepleted from the extracts, and chromatin was assembled in the depleted extracts at 23°C for 45 min. The depleted extracts (egg extracts) and the chromatin fractions (chromatin) were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
When sperm chromatin was incubated in Sld5-depleted extracts, neither Sld5 nor Cdc45 bound to chromatin, suggesting that the binding of the GINS complex to chromatin is required for Cdc45 binding. Furthermore, the GINS complex failed to bind to chromatin in Cdc45-depleted extracts (Fig. 5A, right panels). These results show that the chromatin binding of the GINS complex and that of Cdc45 are mutually dependent processes, although they do not form a stable complex in egg extracts. In accord with the mutual behavior of GINS and Cdc45, both DNA polymerase α and ɛ failed to bind to chromatin in Sld5- or Cdc45-depleted extracts, whereas similar amounts of the polymerases were detected in mock-, Sld5-, and Cdc45-depleted extracts (Fig. 5A). Previously, we have shown that Cdc45 is required for loading of DNA polymerase α and ɛ onto chromatin (Mimura et al. 2000). The present finding therefore demonstrates that the mutual binding of Cdc45 and GINS to chromatin is required for the loading of the polymerases onto chromatin.
Recent studies have shown Xenopus Cut5/Mus101, a homolog of yeast Dpb11, is required for CDK-dependent binding of Cdc45 to chromatin (Van Hatten et al. 2002; Hashimoto and Takisawa 2003). We therefore examined whether Cut5 is required for the association of the GINS complex with chromatin. As shown in Figure 5B, not only Cdc45 but also Sld5 and Psf1 failed to bind to chromatin in Cut5-depleted extracts, although the depleted extracts contained similar amounts of Sld5, Psf1, and Cdc45 as mock-depleted extracts. On the other hand, the binding of Cut5 onto chromatin was slightly increased in Sld5-depeleted extracts. In both the depleted extracts, pre-RC formation was not affected, confirming that these proteins are not required for pre-RC formation.
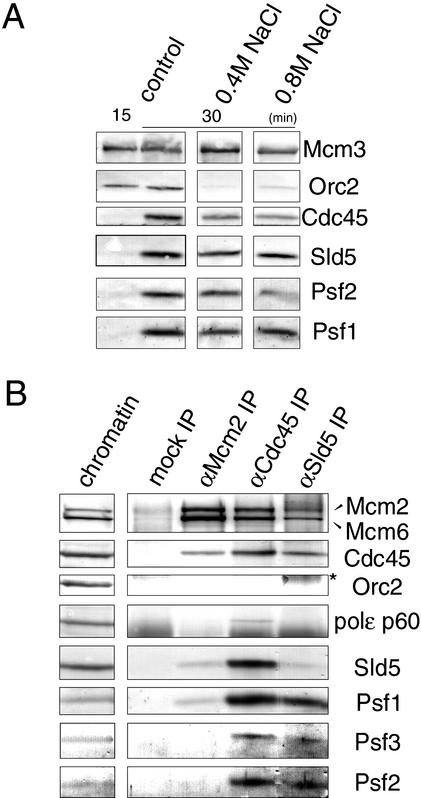
To establish the characteristics of chromatin binding of GINS, we compared the resistance of chromatin binding of various proteins, including Sld5, Psf1, and Psf2, to treatment with high salt (Fig. 6A). Under standard conditions for preparing the chromatin fractions, the components of pre-RCs, such as Orc2 and Mcm3, bound to chromatin before the initiation of DNA replication (15 min), at which point only small amounts of Sld5, Psf1, Psf2, and Cdc45 were found in the chromatin fractions. After the initiation of DNA replication (30 min), appreciable amounts of Cdc45, Sld5, Psf1, and Psf2 were found to be associated with chromatin in addition to pre-RC components. When the chromatin fractions were isolated in the presence of 0.4 M NaCl, almost all Orc2 was removed from the chromatin, whereas Mcm3 remained in the fractions, as is consistent with previous reports (Rowles et al. 1999). Cdc45, Sld5, Psf1, and Psf2 also remained in the fractions. Further increase in the concentration of NaCl to 0.8 M gave similar results, although the amounts of the proteins bound to chromatin were slightly reduced. These results show that the GINS complex is tightly associated with replicating chromatin.
Figure 6.
(A) Tight association of GINS with chromatin. Sperm chromatin was incubated in egg extracts at 23°C for 15 or 30 min, and the extracts were then diluted with the chromatin isolation buffer containing 0 M, 0.4 M, and 0.8 M NaCl. The isolated chromatin fractions were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (B) Coimmunoprecipitation of GINS with other replication proteins from replicating chromatin. Sperm chromatin was incubated in egg extracts at 23°C for 45 min, and the isolated chromatin fractions were digested with micrococcal nuclease. The solublilized chromatin fractions were then immunoprecipitated with control, anti-Sld5, anti-Cdc45, and anti-Mcm2 antibodies. Immunoprecipitates were resolved by SDS-PAGE and immunoblotted. (*) A smeared band of cross-linked products of heavy and light chains of IgG eluted from the antibody-conjugated beads. In lane 5, only a small amount of Sld5 was extracted from anti-Sld5 antibody conjugated beads, and most Sld5 remained associated with the beads under the present extraction conditions.
The chromatin binding of GINS and Cdc45 is a mutually dependent process, for which the pre-RC formation, Cut5, and CDK activity are all required. In addition, both of their chromatin bindings are resistant to high-salt treatment. These results suggest physical interaction of these proteins on chromatin. To explore this possibility, we prepared the chromatin fraction in the absence of aphidicolin and isolated it just after the initiation of DNA replication (45 min of incubation). The chromatin fractions, fragmented by nuclease treatment, were then immunoprecipitated with antibodies against Sld5 or Cdc45 (Fig. 6B). We found that GINS and Cdc45 were coimmunoprecipitated with each other, but Orc2 was not coprecipitated with Sld5 or Cdc45. We also found the coprecipitation of Mcm2 and Mcm6 with Sld5. Therefore, the interaction between MCM and GINS was further examined by chromatin immunoprecipitation with anti-Mcm2 antibody. Figure 6B shows that small amounts of Sld5, Psf1, and Cdc45 were coprecipitated with the MCM complex. Similar results were obtained when the chromatin was prepared after 60 min of incubation in the presence of aphidicolin, which allowed the maximal binding of these proteins to chromatin (data not shown). In contrast, we detected only a small amount of the 60-kD subunit of DNA polymerase ɛ coprecipitated with Cdc45. These results demonstrate the close interaction among GINS, Cdc45, and MCM on replicating chromatin.
Electron microscopic observation of the GINS complex
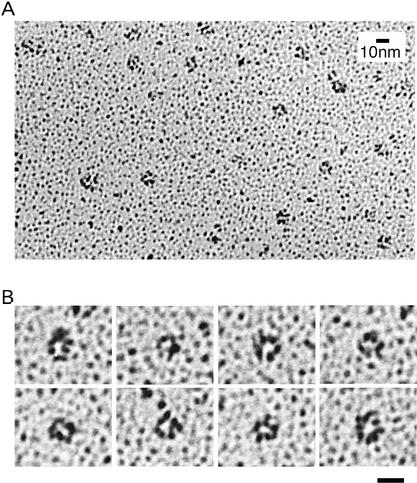
The apparent molecular masses of endogenous and recombinant GINS suggest that it is a tetrameric complex. To obtain further insight into the function of the complex, we tried to visualize the shape of the recombinant GINS complex by electron microscopy (Fig. 7). Electron microscopic images of rotary-shadowed GINS complex suggest that it has a globular structure with an average diameter of ∼10 nm (Fig. 7A). We also found much smaller distorted or larger aggregated structures, which comprised ∼40% of the total images (data not shown). Close examination of the globular structure further revealed that more than one-third of the globular images appeared to be ring-like structures with a pore in the center, or C-shaped structures (Fig. 7B). By analyzing the collected images of the ring or C-shaped structures, we obtained average diameters of 9.5 nm for the ring-like structure, and 4 nm for the pore (calculated from 108 images). It should be noted that the measured size is larger than the true molecular size because of the thickness (∼1 nm) of shadowed platinum. When we examined the rotary-shadowed images of the trimer complex (Sld5–Psf1–Psf2) under the electron microscope, we could identify only amorphous structures (data not shown). These data therefore suggest that the ring-like images of the GINS complex represent the structure of the tetrameric complex.
Figure 7.
Electron micrographs of the GINS complex. The recombinant complex of GINS was expressed in insect cells and purified on Ni-NTA resin. The purified recombinant GINS complex was rotary-shadowed and observed using transmission electron microscopy. (A) A low-magnification field of GINS complexes. (B) A gallery of high-magnification images of the complex. The images showing a ring-like structure were selected. Bar, 10 nm.
Discussion
In eukaryotic DNA replication, various proteins are involved in the initiation process. These proteins should assemble on the replication origins in a precise order to initiate DNA replication, and the process is strictly controlled by two kinds of S-phase-promoting kinases, CDK and DDK. Above all, Dpb11/Cut5 and Cdc45 play critical roles in the conversion from pre-RCs to ICs. However, the conversion process has not been fully understood yet. Here, we report that a novel complex of Xenopus proteins, conserved from yeast to human, is involved in the conversion process. Unique biochemical and structural features of the complex suggest that it plays a crucial role in the formation of the initiation complex, cooperating with Cdc45.
Identification of Xenopus Sld5, Psf1, Psf2, and Psf3 and their complex formation
Xenopus egg extracts provide us with a powerful tool for examining the function of essential gene products such as replication proteins. We have recently identified a Xenopus homolog of Dpb11, which we call Xenopus Cut5, as a novel protein involved in the initiation of DNA replication in Xenopus egg extracts (Hashimoto and Takisawa 2003). The genetic screening of the yeast dpb11-1 mutant further reveals a new class of genes called SLD (synthetic lethality of dpb11-1; Kamimura et al. 1998). SLD5 is the least characterized gene, and we began by identifying a Xenopus homolog of Sld5 based on sequence similarity to yeast Sld5. During the course of the cloning of Xenopus Sld5, we found additional yeast proteins interacting with Sld5 by searching the Comprehensive Yeast Genome Database. The yeast genes thus obtained are called PSF1 and PSF2, both of which were also identified by the genetic screening of the sld5-12 mutant (Takayama et al. 2003). In addition, the genetic screening of psf1-1 revealed the third partner of Sld5, called Psf3. By searching the EST database with the primary sequences of the yeast proteins, we readily obtained the partial sequences of potential human or Xenopus homologs, suggesting that they are evolutionarily conserved proteins. Comparisons with the predicted sequences of Xenopus homologs of Sld5, Psf1, Psf2, and Psf3 show that highly homologous proteins exist in mammals, insects, worm, plants, and fission yeast. Some of these sequences are aligned with the Xenopus sequences (Fig. 1). The fact that the budding yeast genes are all essential for growth strengthens the view that these proteins must perform an essential role in the proliferation of eukaryotic cells.
In egg extracts, Xenopus Sld5, Psf1, Psf2, and Psf3 formed a stable tetrameric complex. An antibody against Sld5 depleted not only Sld5 but also Psf1, Psf2, and Psf3 from the extracts, and the immunoprecipitates obtained appeared to contain all these proteins (Fig. 2B). In addition, glycerol gradient analysis of recombinant as well as endogenous complex showed that all these proteins migrated in a similar peak with an apparent molecular mass of ∼100 kD (Fig. 2D,E). It should also be noted that the endogenous complex was immunoprecipitated with anti-Sld5 antibody, whereas the recombinant complex was purified through His-tagged Psf3, thus indicating that Sld5 and Psf3 and presumably Psf1 and Psf2 comigrate together. These data therefore suggest that the complex is a heterotetramer consisting of Sld5, Psf1, Psf2, and Psf3. During the reconstitution of recombinant proteins, we also found that Psf2 could form a stable complex with Sld5 but Psf3 could not, and that Psf3 was readily dissociated from the tetrameric complex (Fig. 2B). In budding yeast, high copy of PSF1 and PSF2 but not PSF3 could suppress the Sld5 mutant, thus suggesting a strong interaction between Sld5, Psf1, and Psf2 but a weak interaction between Psf3 and Sld5 (Takayama et al. 2003). These results therefore suggest that the subunit interaction of GINS is conserved from yeast to frog.
Essential role of Xenopus GINS in DNA replication
Xenopus GINS was required for DNA replication in Xenopus egg extracts (Fig. 3). The depletion of the complex abolished the incorporation of dNTP into sperm chromatin and the recombinant GINS complex—but neither Sld5 alone nor the Sld5–Psf1–Psf2 complex—could rescue the replication activity. The examination of replicated products further showed that neither nascent DNA nor short DNA fragments were formed in the depleted extracts, suggesting that the complex is required for the initiation stage of replication. Furthermore, we found that the chromatin binding of GINS and that of Cdc45 are mutually dependent, and previous studies showed that Cdc45 is essential for the loading of DNA polymerase α and ɛ onto chromatin (Mimura et al. 2000). Indeed, we could not detect the binding of these polymerases onto chromatin in Sld5-depleted extracts (Fig. 5A). The data obtained therefore suggest that GINS is required for the formation of the initiation complex.
The association of Cdc45 and GINS on replicating chromatin suggests that they cooperate with each other on the chromatin. This is further supported by the findings that both Cdc45 and GINS bind to chromatin in a manner dependent on pre-RC formation, Cut5, and S-CDK activity. In addition, the bindings of both proteins are resistant to high-salt treatment. In accordance with the present observations with Xenopus proteins, a ChIP assay in budding yeast suggests that GINS associates with origins at the onset of S phase, dependent on Dpb11 and Sld3 (Takayama et al. 2003). Because Sld3 has been shown to be an essential component for the binding of Cdc45 to origins (Kamimura et al. 2001), we assume that Cdc45 is required for the origin association of GINS. In addition, Psf1 is required for the chromatin binding of Cdc45 in the budding yeast. These results further suggest the conserved role for GINS in the initiation of DNA replication.
An apparent discrepancy between the results obtained with Xenopus and those with budding yeast is the requirement of the GINS complex for the binding of Cut5/Dpb11. In the egg extracts, GINS is not required for the chromatin binding of Cut5/Dpb11, but it appears to be required for the origin association of Dpb11 in yeast. This may be because of the difference in the binding assay rather than an inherent difference between yeast replication and early embryonic replication of eggs. With the egg extracts, only the chromatin binding of Cut5 was analyzed, whereas with yeast the origin association but not the chromatin binding of Dpb11 has been discussed. It is therefore possible that Dpb11/Cut5 binds to chromatin in the absence of Psf1 without any association with origins. It is also possible that the origin unwinding, which may require the function of Psf1, is necessary for the stable association of Dpb11 with origins. In any event, present data obtained with Xenopus egg extracts do not directly contradict the data obtained with yeast. It is conceivable that the transient association of chromatin-bound Cut5/Dpb11 with origins is required for the formation of initiation complexes in the Xenopus system, and that Dpb11/Cut5 may associate with chromatin before the initiation reaction in the yeast system.
Functional role of GINS in the initiation of DNA replication
The following features of the GINS complex suggest that it is a component of replication machinery at replicating forks: (1) GINS is tightly associated with replicating chromatin fractions. (2) GINS interacts with Cdc45 and MCM on replicating chromatin. (3) GINS is required for the loading of Cdc45 onto chromatin. Previous studies with degron mutants of budding yeast clearly demonstrate that both Cdc45 and MCM are required for the elongation stage of DNA replication, in addition to the initiation stage (Labib et al. 2000; Tercero et al. 2000). ChIP assays also suggest that MCM and Cdc45 behave similarly to DNA polymerase ɛ, which is presumably at replication forks (Aparicio et al. 1997). In Takayama et al. (2003), a ChIP assay with anti-Psf1 antibody demonstrates that Psf1 associates first with origins and then with origin-proximal regions upon the progression of S phase, showing that Psf1 behaves quite similarly to MCM, Cdc45, and DNA polymerase ɛ. Taking the complex formation of Psf1 with Sld5, Psf2, and Psf3 into account, the results obtained with Xenopus and budding yeast strongly suggest that GINS is a component of replication machinery that forms at replication forks.
A remaining question is the function of GINS in the initiation and the elongation of DNA replication. We have previously proposed that Xenopus Cdc45 functions as a loader of DNA polymerase α onto the unwound origins through a physical interaction with the polymerase (Mimura and Takisawa 1998; Mimura et al. 2000). Sld5 was originally identified based on its genetic interaction with Dpb11, which, in turn, interacts with Dpb2, a subunit of DNA polymerase ɛ (Araki et al. 1995; Kamimura et al. 1998). Similar behavior of Psf1 and polymerase ɛ at replication forks therefore suggests that GINS functions in the loading of polymerase ɛ. Electron micrographs of Xenopus GINS further support the idea that the complex may act in the loading of the polymerase. We found that the complex appears to form a ring- or C-like structure. The observed diameter of 7–8 nm (corrected for the metal thickness) is larger than the value (4–5 nm) predicted for a spherical molecule with similar molecular mass (100 kD). If this complex is an 8-nm ring with a 4-nm hole and 2-nm depth, it would have a volume similar to a 4–5-nm spherical molecule. Therefore, it is highly possible that the observed ring-like structure represents a bona fide image of the tetrameric complex of GINS. These structural features of the complex are reminiscent of the structure of PCNA (Krishna et al. 1994; Gulbis et al. 1996), a replication clamp that tethers DNA polymerase δ at the end of lagging or leading strands in replicating DNA. In analogy, it is possible that GINS acts as a clamp of DNA polymerase ɛ. However, the absence of a tight association between the 60-kD subunit of the polymerase and Cdc45 in the present study suggests that the interaction between the polymerase and the complex containing Cdc45 is not stable.
The GINS complex may also have some role in the activation of the putative MCM helicase. We have recently found that a tight complex of MCM–Cdc45 is formed upon the initiation of DNA replication, and that complex formation is closely related to the helicase activity of chromatin immunoprecipitates of Cdc45 or MCM (Masuda et al. 2003). The chromatin immunoprecipitation in the present study shows that GINS coprecipitates with MCM and Cdc45 from replicating chromatin. Moreover, preliminary experiments with the detergent extracts of chromatin fractions showed that Sld5 and Psf1 were tightly associated with MCM and Cdc45. Such a tight complex formation suggests that the GINS complex is an auxiliary component of the putative MCM helicase. Previous studies showed that Dna43/Mcm10, Sld2, and Sld3 are also required for the initiation of DNA replication (Kamimura et al. 1998, 2001; Wang and Elledge 1999; Homesley et al. 2000; Wohlschlegel et al. 2002). The complexity of the initiation machinery is not surprising, and it convinces us that the initiation of DNA replication is one of the most elaborated mechanisms in the evolution of eukaryotes.
Materials and methods
Cloning and sequencing of Xenopus cDNAs
To clone Xenopus Sld5, we obtained a partial sequence of potential human Sld5 (gi, 97705599) by homology search of the EST database with yeast Sld5 (YDR489w). The fragment of human Sld5 was then amplified from cDNA of HeLa cells by PCR using 5′ primer (GCCTGGATGAATGAAAAGTTTGCC) and 3′ primer (CGACACCGCAAGTAGCTGCTGGAG). The amplified fragment was used as a probe to clone a full-length Xenopus cDNA. To clone Xenopus Psf1, we performed a homology search of the Protein and DNA database with yeast Psf1 (YDR013w), and highly conserved regions of Psf1 were identified by comparing the sequences of putative Psf1 homologs in human (KIAA0186), Drosophila (CG9187), Caenorhabditis elegans (CER53–6R53.6), Arabidopsis (F28B13.27), and Schizosaccharomyces pombe (SPBP23A10.09). We designed degenerate primers for the conserved amino acid sequences of Psf1, which are ALRWEYG, DYGEFE, and KKNSQH; a fragment of Xenopus Psf1 was then amplified from Xenopus oocyte cDNA using 5′ primer (GCNYTNMGITGGARTAYGG) and 3′ primer (ACYTCRAAYTCNCCRTARTC). The DNA fragment obtained was further amplified by nested PCR using the same 5′ primer with 3′ primer (RAARTGYTGISRTTYTTYTT), and the amplified DNA fragment was used as a probe. To clone Xenopus Psf2 and Psf3, we obtained partial sequences of potential Xenopus Psf2 (BF611013) and Psf3 (BE025983) by searching the EST database for homology with yeast Psf2 (YOL146w) and Psf3 (YJL072c). The fragments of BF611013 and BE025983 were obtained by PCR-amplifying Xenopus oocyte cDNA using 5′ primers (CGGGGTACCGCGGGAGATTTAGTCATGGA and GG TACCAATGTGGGAACCCTACATGCCA, respectively) and 3′ primers (CCCAAGCTTTGTCCTTCCTCTGGGTTCTG and AAGCTTCCCAGCTCTGACATGCATTC, respectively). The amplified fragments were used as probes. With these probes, we screened a λ ZAPII-derived cDNA library of Xenopus oocyte mRNAs and obtained the full-length Xenopus cDNAs for Sld5, Psf1, Psf2, and Psf3. The clones obtained were sequenced for both strands with an automatic DNA sequencer (ABI, 377).
Protein expression and antibody production
To prepare full-length recombinant proteins, the ORF sequences of Xenopus Sld5, Psf1, Psf2, and Psf3 (accession nos. AB097167, AB097168, AB097169, and AB097170, respectively) were subcloned into pFastBac vectors. For Sld5, the fragment from cDNA amplified using 5′ primer (TTTGAATTCACCA TGGAAGATGAGCTGGCG) and 3′ primer (GCTCTAGATT AAATGAGTTTCACAGCTCC) was subcloned into the EcoRI–XbaI site of the vector. For Psf1, the cDNA was digested with EcoRI and XhoI, and the digested fragment was subcloned into the vector. For His-tagged versions of Sld5 and Psf1, 5′ primers (TTTGAATTCACCATGGAAGATGAGCTGGCG and TTTG AATTCACCATGTTCTGTGAGAAAGCCATT, respectively) and 3′ primers containing sequences for six histidines (CCCC TCGAGCTAGTTAATGGTGATGGTGATGGTGAATGAGTT TCACAGCTCCTG and TTTCTCGAGTTAGTGATGGTGATG GTGATGAGACAGTACGTGCTCTAGAAC, respectively) were used to amplify fragments. These were subcloned into the EcoRI–XhoI site of the vector. For Psf2, the cDNA was amplified by PCR using 5′ primer (CGGGATCCCGCCCCTTCCTATCC CCAATA) and 3′ primer (GGGGTACCCCTTGTGTGTTCTC AGCAGCCA) and then subcloned into pFastBac with or without six histidines in the coding region. For Psf3, the cDNA was digested with BamHI and XhoI and likewise subcloned into pFastBac with or without six histidines in the coding region.
His6-tagged versions of the proteins were expressed in baculovirus-infected Sf9 cells for production of antibodies against each protein. To obtain a complete Xenopus GINS (Sld5–Psf1–Psf2–Psf3) complex, Sld5, Psf1, Psf2, and Psf3 with 6 × His at the N terminus (His6-Psf3) were coexpressed in baculovirus-infected Sf9 cells. To obtain an Sld5–Psf1–Psf2 complex, Psf2 with 6 × His at the N terminus (His6-Psf2) was used. Cell lysates from the infected Sf9 cells were immediately applied to an Ni-NTA column (QIAGEN), and the proteins were purified with standard procedures. The purified proteins were resolved by SDS-PAGE, and each protein band was identified with the corresponding antibody. Polyclonal rabbit antiserum were raised against the recombinant proteins (Hokudo Inc.) and further affinity-purified with the recombinant proteins immobilized on Affi-Gel 10 (Bio-Rad).
Preparation of the egg extract and chromatin fraction
Xenopus egg extracts and sperm chromatin were prepared as described previously (Kubota and Takisawa 1993). To prepare the chromatin fraction, sperm chromatin (4000 sperm heads/μL extract) was incubated with the extracts for appropriate times at 23°C. Aliquots were diluted with 5–10 times volume of ice-cold EB containing 0.25% NP-40; then the chromatin was isolated by centrifugation through a 10% sucrose cushion. For high-salt washes, the indicated concentrations of NaCl were added to the dilution buffer. For immunoprecipitation from chromatin, isolated chromatin fractions were suspended in EB containing 2 mM CaCl2, protease inhibitors (1 μg/mL each of leupeptin, pepstatin, and aprotinin), and micrococcal nuclease (2 units/μL) and incubated at 30°C for 15 min. The reaction was stopped by adding 5 mM EDTA to the reaction mixture, then centrifuged at 10,000g at 4°C for 10 min. The supernatants thus obtained were used as solubilized chromatin fractions for immunoprecipitation.
Immunodepletion and immunoprecipitation
The immunodepletions of Xenopus proteins were carried out as described (Mimura and Takisawa 1998) except that rProtein A Sepharose Fast Flow (Amersham Biosciences) was used instead of Affi-Prep protein A matrix (Bio-Rad). The immunoprecipitations of egg extracts and chromatin fraction were performed as described (Mimura et al. 2000).
Glycerol gradient centrifugation
The egg extract was centrifuged at 80,000 rpm at 4°C for 30 min with a Hitachi S100AT rotor to obtain the clear soluble fraction. The soluble fraction of the extract (50 μL) was centrifuged through 15%–40% glycerol gradients containing 2.5 mM MgCl2 and 50 mM HEPES-KOH (pH 7.5). Centrifugation was performed with a Beckman SW50.1 rotor at 45,000 rpm at 4°C for 21 h. Marker proteins were centrifuged under identical conditions. Fractions of 200 μL were collected, and 20 μL of each fraction was resolved by SDS-PAGE and blotted.
Assay for DNA replication activity
The replication activities of the egg extracts were measured as incorporation of [α-32P]dCTP into sperm DNA. [α-32P]dCTP incorporation was carried out as described (Mimura and Takisawa 1998) except that the autoradiography was quantified by NIH image.
Fluorescence microscopy
Samples for fluorescence microscopy were prepared as follows. Sperm nuclei (4000/μL) were incubated in 10 μL of the egg extract containing 10 μM Cy3-dCTP (Amersham Biosciences) for appropriate times at 23°C. The samples were diluted with 90 μL of EB and fixed by adding 11 μL of 37% formaldehyde for 30 min on ice. The nuclei were then recovered by centrifugation at 1200g for 5 min through PBS containing 30% sucrose, onto poly-lysine-coated coverslips. The coverslips were washed with TTBS (0.9% NaCl, 0.1% Tween 20, and 100 mM Tris-HCl at pH 7.5) and incubated at 4°C overnight with the primary antibody in 10% skimmed milk/TTBS. They were then washed three times with TTBS and incubated at room temperature for 2 h with the secondary antibody (Alexa 488-labeled anti-rabbit IgG) in 10% skimmed milk/TTBS. After sequential washing with TTBS, PBS, and dH2O, they were mounted on glass slides in 3 μL of fixing solution (3% formaldehyde, 2 μg/mL Hoechst dye 33342, 80 mM KCl, 15 mM NaCl, 50% glycerol, 15 mM Pipes at pH 7.2) containing 0.5% 2-mercaptoethanol. Fluorescence images were captured with the OpenLab imaging program (Improvision).
Electron microscopy
Xenopus GINS and Sld5–Psf1–Psf2 complexes were examined by electron microscopy following low-angle rotary shadowing as previously described (Arata 1998). The complexes were diluted with 50% (v/v) glycerol containing 0.1 M ammonium acetate to give a final concentration of 0.05 mg/mL. The protein solution was sprayed onto a freshly cleaved mica surface. The mica was then dried under vacuum, rotary-shadowed with platinum at an angle of 8°, and supported with carbon at an angle of 90° (JEE-5B, JEOL). The shadowed platinum films were removed from the mica by soaking in water and mounted on copper grids. The replicas were examined at magnification of 50,000× with a JEOL JEM-1010 electron microscope operating at 80 kV. Images were digitized by a film scanner and further processed with Adobe Photoshop software.
Acknowledgments
We thank Shou Waga for antibody against the p60 subunit of Xenopus pol ɛ, and Taro S. Masuda and Catherine Merrick for critical reading of the manuscript. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture, Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL takisawa@bio.sci.osaka-u.ac.jp; FAX (81) 66850-5554.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1070003.
References
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase IIɛ in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arata T. Electron microscopic observation of monomeric actin attached to a myosin head. J Struct Biol. 1998;123:8–16. doi: 10.1006/jsbi.1998.4019. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Hodgson B. Replication licensing—Defining the proliferative state? Trends Cell Biol. 2002;12:72–78. doi: 10.1016/s0962-8924(01)02203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Li A, Blow JJ. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2001;2:15. doi: 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Hashimoto, Y. and Takisawa, H. 2003. Xenopus Cut5 is essential for CDK-dependent process in the initiation of DNA replication. EMBO J. (In press). [DOI] [PMC free article] [PubMed]
- Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye BK. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes & Dev. 2000;14:913–926. [PMC free article] [PubMed] [Google Scholar]
- Hutchison CJ, Cox R, Ford CC. The control of DNA replication in a cell-free extract that recapitulates a basic cell cycle in vitro. Development. 1988;103:553–566. doi: 10.1242/dev.103.3.553. [DOI] [PubMed] [Google Scholar]
- Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 2001;20:2097–2107. doi: 10.1093/emboj/20.8.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase ɛ catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Takisawa H. Determination of initiation of DNA replication before and after nuclear formation in Xenopus egg cell free extracts. J Cell Biol. 1993;123:1321–1331. doi: 10.1083/jcb.123.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Makiniemi M, Hillukkala T, Tuusa J, Reini K, Vaara M, Huang D, Pospiech H, Majuri I, Westerling T, Makela TP, et al. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J Biol Chem. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- Masai H, Arai K. Cdc7 kinase complex: A key regulator in the initiation of DNA replication. J Cell Physiol. 2002;190:287–296. doi: 10.1002/jcp.10070. [DOI] [PubMed] [Google Scholar]
- Masuda T, Mimura S, Takisawa H. CDK- and Cdc45-dependent priming of the MCM complex on chromatin during S-phase in Xenopus egg extracts: Possible activation of MCM helicase by association with Cdc45. Genes Cells. 2003;8:145–161. doi: 10.1046/j.1365-2443.2003.00621.x. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases α and ɛ and the autonomously replicating sequence region of budding yeast. Mol Cell Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Masuda T, Matsui T, Takisawa H. Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells. 2000;5:439–452. doi: 10.1046/j.1365-2443.2000.00340.x. [DOI] [PubMed] [Google Scholar]
- Morrison A, Araki H, Clark AB, Hamatake RK, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Nakajima R, Masukata H. SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in DNA replication in fission yeast. Mol Biol Cell. 2002;13:1462–1472. doi: 10.1091/mbc.02-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z. Control of DNA replication licensing in a cell cycle. Genes Cells. 2002;7:523–534. doi: 10.1046/j.1365-2443.2002.00544.x. [DOI] [PubMed] [Google Scholar]
- Noguchi E, Shanahan P, Noguchi C, Russell P. CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr Biol. 2002;12:599–605. doi: 10.1016/s0960-9822(02)00739-x. [DOI] [PubMed] [Google Scholar]
- Ohya T, Kawasaki Y, Hiraga S, Kanbara S, Nakajo K, Nakashima N, Suzuki A, Sugino A. The DNA polymerase domain of pol(ɛ) is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J Biol Chem. 2002;277:28099–28108. doi: 10.1074/jbc.M111573200. [DOI] [PubMed] [Google Scholar]
- Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, Y., Kamimura, Y., Okawa, M., Muramatsu, S., Sugino, A., and Araki, H. 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Takisawa H, Mimura S, Kubota Y. Eukaryotic DNA replication: From pre-replication complex to initiation complex. Curr Opin Cell Biol. 2000;12:690–696. doi: 10.1016/s0955-0674(00)00153-8. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Labib K, Diffley JF. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 2000;19:2082–2093. doi: 10.1093/emboj/19.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hatten RA, Tutter AV, Holway AH, Khederian AM, Walter JC, Michael WM. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J Cell Biol. 2002;159:541–547. doi: 10.1083/jcb.200207090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- Waga S, Masuda T, Takisawa H, Sugino A. DNA polymerase ɛ is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc Natl Acad Sci. 2001;98:4978–4983. doi: 10.1073/pnas.081088798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Wang H, Elledge SJ. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol Cell. 2002;9:233–240. doi: 10.1016/s1097-2765(02)00456-2. [DOI] [PubMed] [Google Scholar]
- Yamamoto RR, Axton JM, Yamamoto Y, Saunders RD, Glover DM, Henderson DS. The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C-terminus domains. Genetics. 2000;156:711–721. doi: 10.1093/genetics/156.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]