Abstract
The signalosome is implicated in regulating cullin-dependent ubiquitin ligases. We find that two signalosome subunits, Csn1 and Csn2, are required to regulate ribonucleotide reductase (RNR) through the degradation of a small protein, Spd1, that acts to anchor the small RNR subunit in the nucleus. Spd1 destruction correlates with the nuclear export of the small RNR subunit, which, in turn, correlates with a requirement for RNR in replication and repair. Spd1 degradation is promoted by two separate CSN-dependent mechanisms. During unperturbed S phase, Spd1 degradation is independent of checkpoint proteins. In irradiated G2 cells, Spd1 degradation requires the DNA damage checkpoint. The signalosome copurifies with Pcu4 (cullin 4). Pcu4, Csn1, and Csn2 promote the degradation of Spd1, identifying a new function for the signalosome as a regulator of Pcu4-containing E3 ubiquitin ligase.
Keywords: Rad3, Chk1, replication, protein degradation, cullin, ubiquitin
The COP9 signalosome (CSN) complex was originally identified as a negative regulator of photomorphogenesis in plants (for review, see Schwechheimer and Deng 2001). Subsequently, it was purified from human cell extracts during attempts to isolate the 19S regulatory lid complex of the proteosome (Seeger et al. 1998). The human signalosome consists of eight core subunits, each sharing significant homology with a corresponding subunit in the regulatory 19S lid complex of the proteosome (Deng et al. 2000). The purified CSN complex can cleave the ubiquitin-like Nedd8 protein from cullins (Lyapina et al. 2001; Wee et al. 2002). Csn5 contains a putative metalloprotease motif that is presumed to mediate deneddylation activity (Cope et al. 2002). Cullins are subunits of E3 ubiquitin ligases (Feldman et al. 1997; Skowyra et al. 1997), and deneddylation of cullins decreases SCF E3 ubiquitin ligase activity (Osaka et al. 2000). SCF E3 complexes typically consist of a cullin, the Rbx1 RING domain protein that binds an E2 enzyme (Kamura et al. 1999; Skowyra et al. 1999), and an adapter protein, Skp1, that binds an F-box protein that determines the substrate specificity (Skowyra et al. 1997).
In Arabidopsis, the signalosome is involved in the degradation of the two bZIP transcription factors (Hy5, HyH) that lie at the top of a transcriptional cascade required to induce ∼30% of Arabidopsis genes during photomorphogenesis (Holm et al. 2002). An E2-like protein, Cop10, and an E3 RING protein, Cop1, are also required to degrade Hy5 and HyH, which occurs when seedlings are germinated in the dark (Osterlund et al. 2000; Holm et al. 2002; Suzuki et al. 2002). The biochemical role of the signalosome is unknown, although a correlation with Cop1 nuclear localization (von Arnim et al. 1997) and the associations between the signalosome and E3 ubiquitin ligases (Lyapina et al. 2001; Schwechheimer et al. 2002) suggest a regulatory role in ubiquitination that may be linked to subcellular localization (Chamovitz et al. 1996; Hellmann and Estelle 2002).
A highly conserved signalosome complex was identified in the fission yeast Schizosaccharomyces pombe (Mundt et al. 1999) and subsequently shown to be required to remove the Nedd8 ubiquitin-like protein from the cullins Pcu1 and Pcu3. Despite the fact that null mutants in csn1, csn2, csn3, csn4, and csn5 each cause constitutive Nedd8 modification of Pcu1 and Pcu3, only csn1-d and csn2-d mutants demonstrate obvious phenotypes (Zhou et al. 2001; Mundt et al. 2002). These include slow S-phase progression and a modest sensitivity to DNA-damaging agents. This suggested that some of the functions associated with the CSN complex could be independent of its role in removing Nedd8 from cullins. We have used both the slow S-phase phenotype and the DNA-damage sensitivity specific to csn1-d and csn2-d strains to investigate this novel biological role for the signalosome. Our results lead us to propose a regulatory function for the CSN complex in Pcu4-dependent ubiquitin ligase activity. We propose that the cullin Pcu4 is indirectly required for activation of ribonucleotide reductase through degradation of the protein Spd1, thereby supplying sufficient deoxyribonucleotides for DNA replication and repair.
Results
A link between the signalosome, ribonucleotide reductase, and Spd1
csn1-d- and csn2-d-null mutants are slow growing and spend an extended period of time in S phase (Mundt et al. 1999). Consistent with a defect in DNA replication, csn1-d and csn2-d are synthetically lethal with the loss of rad3 function. Rad3 is a PI3-like protein kinase homologous to Saccharomyces cerevisiae Mec1 and human ATR (Bentley et al. 1996). In Schizosaccharomyces pombe, Rad3 is required for all DNA-structure-dependent checkpoints. In response to S-phase perturbations, Rad3 activates the DNA replication checkpoint kinase Cds1, whereas upon DNA damage in late S/G2, Rad3 acts through the Chk1 kinase (Lindsay et al. 1998; Martinho et al. 1998). Cells harboring the rad3-ts allele are checkpoint-proficient at 27°C and checkpoint-deficient at 35.5°C. We previously reported that rad3-ts csn1-d double-mutant cells rapidly lose viability and accumulate evidence of mitotic catastrophe upon a temperature shift from 27°C to 35.5°C (Mundt et al. 1999).
To identify the cause of the slow S phase in signalosome mutants, we screened for multicopy suppressors of rad3-ts csn1-d lethality at 35.5°C. This analysis identified multiple independent clones of suc22 that efficiently rescued the growth defect of rad3-ts csn1-d cells. suc22 encodes the small subunit of S. pombe ribonucleotide reductase (RNR). Active RNR is a tetrameric enzyme composed of two small subunits (Suc22) and two large subunits (Cdc22) that generates deoxyribonucleotides. To confirm the suppression, we integrated a second suc22 gene at the ura4 locus, with its transcription driven by the constitutively active ADH promoter. Overexpression of suc22 (∼10-fold increase in Suc22 protein) allowed csn1-d cells to progress normally through S phase (Fig. 1A) without activating Cds1 kinase, an indication of checkpoint activation (Fig. 1B). suc22 overexpression also complemented the UV- and IR-sensitivity of csn1-d cells (data not shown; cf. Fig. 4D, below). These data suggest a role for the signalosome in the regulation of RNR activity that is important for S-phase progression and DNA repair.
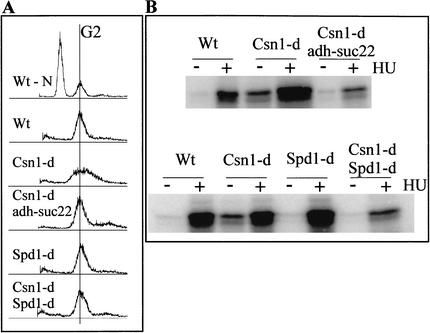
Figure 1.
Suppression of Csn1 S-phase delay. (A) FACS analysis of csn1-d cells overexpressing suc22 from an integrated ADH promoter or deleted for spd1, an inhibitor of S phase (Woollard et al. 1996). Control nitrogen-starved cells with a G1 and G2 peak are shown at the top (Wt-N). Logarithmically growing wild-type (Wt) cells show a G2 peak. csn1-d cells show a broad peak indicative of a delay in S phase (Mundt et al. 1999). Overexpression of suc22 or deletion of spd1 restores a normal FACS profile in csn1-d cells. Deletion of spd1 alone does not affect the FACS profile. (B) Cds1 kinase activity is suppressed by either suc22 overexpression or spd1 deletion. The same strains as used in A were tested for in vitro kinase activity using equal amounts of immunoprecipitated Cds1 against MBP as substrate. Cds1 derived from csn1-d cells is constitutively active because of the delay in S phase (Mundt et al. 1999). Either suc22 overexpression or spd1 deletion suppresses the constitutive activity as well as reducing the extent of the activation caused by treatment with 10 mM HU.
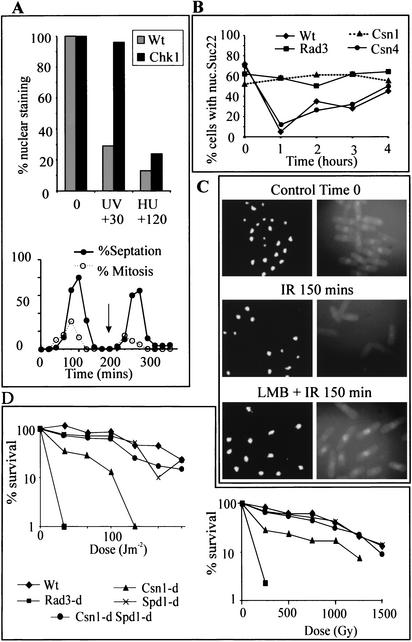
Figure 4.
Nuclear localization of Suc22 following checkpoint activation. (A) Wild-type and chk1-d cells were synchronized in G2 by elutriation, and during the second G2 (see arrow) cells were treated with either 100 J/m2 of UV and harvested after 30 min, or with 10 mM HU and harvested after 2 h. Suc22 nuclear localization was determined by indirect immunofluorescence. The bottom panel shows the same culture of chk1-d cells scored for septation and mitotic index as an example of the synchrony. (B) Asynchronous wild-type and the deletion mutant cells indicated were treated with 100 J/m2 of UV and scored as above for Suc22 localization at hourly intervals. (C) Samples of synchronized cells late in the second G2 were either irradiated (IR, 500 Gy) or irradiated immediately after the addition of Leptomycin B (LMB + IR). Samples were collected at 30-min intervals for indirect immunofluorescence using anti-Suc22 antibodies (right) and parallel DAPI staining (left). The 150-min time point is shown. (D) DNA damage sensitivity (left, UV; right, IR) of csn1-d mutants is rescued by the spd1 deletion. Single spd1-d mutants are not sensitive to DNA damage. An rad3-d null mutant is included as a control.
In S. cerevisiae, the essential function of MEC1 can be alleviated by the overexpression of a large RNR subunit or by the deletion of SML1, which encodes a 104-amino-acid inhibitor of RNR (Desany et al. 1998; Zhao et al. 1998; Chabes et al. 1999). rad3 is not an essential gene in S. pombe, but becomes essential in csn1-d cells. This essential function can be suppressed by overexpression of an RNR subunit (Fig. 1; data not shown). This similarity between S. pombe csn1-d mutants and S. cerevisiae led us to explore the possibility that an Sml1-like inhibitor exists in S. pombe. We predicted that loss of such an inhibitor might rescue rad3-ts csn1-d cells at 35.5°C and its overexpression should delay cell cycle progression in S phase and be toxic to checkpoint-defective cells. Such a gene, spd1 (S-phase delayed), encoding a small protein (124 amino acids), has previously been characterized as a negative regulator of S phase in fission yeast (Woollard et al. 1996), and its overexpression is toxic to checkpoint-defective cells (Borgne and Nurse 2000). Although there is no significant sequence homology between Sml1 and Spd1, both proteins are small (104 and 124 amino acids, respectively) and predicted to contain similar α-helical regions. We therefore combined the deletion of spd1 with either csn1-d (Fig. 1) or csn2-d (data not shown). In both cases, loss of spd1 phenocopied suc22 overexpression in csn1-d cells: a normal cell cycle profile was restored (Fig. 1A), endogenous activation of Cds1 was suppressed (Fig. 1B), and the lethality of csn1-d in an rad3-defective mutant background was alleviated (data not shown).
Dynamic changes in RNR localization during S phase are Csn1- and Spd1-dependent
Our data suggest that Spd1 inhibits RNR activity and that the CSN complex is required to release RNR from this inhibition during DNA replication or repair. To explore the mechanism by which Spd1 regulates RNR, we studied Suc22 by Western blot and indirect immunofluorescence. The suc22 gene expresses two transcripts (1.5 and 1.9 kb), both of which encode the same size protein. The 1.5-kb transcript is constitutively expressed, whereas the 1.9-kb transcript is only expressed at low levels during S phase and is undetectable in other cell cycle stages in unperturbed cells. However, in response to DNA damage or hydroxyurea (HU) treatment (which blocks cells in early S phase), the 1.9-kb transcript is induced (Harris et al. 1996). Surprisingly, no significant changes in Suc22 protein level were detected during S phase in synchronized cells or in cells blocked in S phase by HU treatment (Fig. 2A). However, Suc22 is predominantly nuclear throughout the cell cycle, but the nuclear localization of Suc22 decreases during unperturbed S phase (Fig. 2B,C) and is lost in cells exposed to HU (Fig. 3B). We next asked if loss of Suc22 nuclear staining during unperturbed S phase was dependent on Rad3. Consistent with the lack of an essential function for Rad3 in S. pombe, rad3-null mutants showed a similar Suc22 localization profile as wild-type cells. This indicates that Rad3 is not required to regulate Suc22 during the normal unperturbed cell cycle (Fig. 2D).
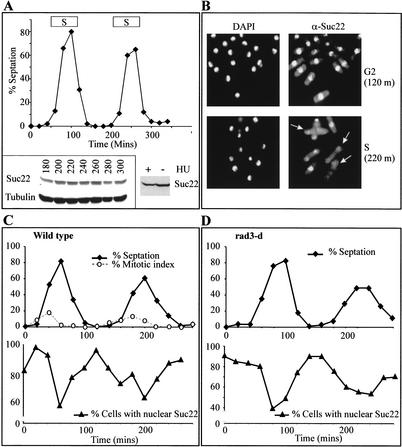
Figure 2.
Subcellular localization of Suc22. (A) Total Suc22 levels do not change significantly through the cell cycle. Cells were synchronized in G2 by elutriation and sampled for septation index at 20-min intervals (top). As cells proceeded from the second G2 period (180 min), through mitosis and S phase (the estimated time of S is indicated above the peaks of septation), extract was prepared from cell aliquots at 20-min intervals, and 50 μg of protein was separated by SDS-PAGE and probed by Western blot for Suc22 and, as a loading control, tubulin (bottom left). (Bottom right) Asynchronous cells were treated with 10 mM hydroxyurea (HU) for 2 h, and 50 μg of extracts was similarly probed for Suc22. (B) Indirect immunofluorescence of a sample from time points 120 min (early G2) and 220 min (S phase). (Right panels) Visualization of anti-Suc22 antibody by FITC-conjugated secondary antibody. (Left panel) Stained for DNA content by DAPI. S-phase (binucleate) cells show a reduced level of nuclear staining for Suc22 (for examples, see arrows). (C,D) Quantification of nuclear staining in synchronized cultures of wild-type (C) and rad3-d (D) cells. The top panel shows the progression through the cell cycle, the bottom panel shows the percentage of cells with nuclear Suc22 staining. The percentage of cells staining positive drops coincidentally with the peak of septation, which has previously been established as a reliable marker for S phase (levels do not reach zero because synchrony is not perfect and S phase is rapid in fission yeast; Christensen et al. 2000).
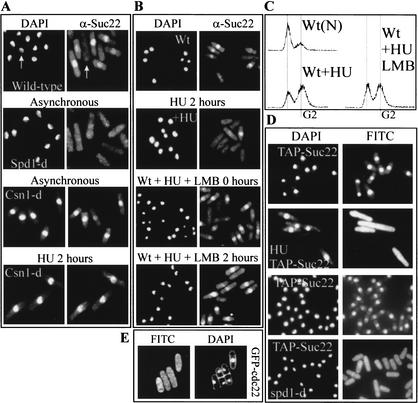
Figure 3.
CSN is required for S-phase-dependent nuclear export of Suc22. (A) Suc22 localization in wild-type, spd1-d, and csn1-d cells. The left panels show DNA content by DAPI staining, and the right panels show Suc22 as detected by indirect immunofluorescence. In asynchronous wild-type cells, Suc22 is nuclear except in S-phase cells (the white arrow indicates an S-phase cell). In asynchronous spd1-d cells no nuclear localization of Suc22 is observed even though ∼70% of cells are in G2. In csn1-d cells, Suc22 is always nuclear, even in cells blocked in S phase by treatment with 10 mM HU for 2 h (bottom). (B,C) Loss of Suc22 staining during S-phase arrest requires active Crm1-dependent nuclear export. (B) DAPI staining (left) and Suc22 localization (right) in wild-type cells before and after treatment with 10 mM HU for 2 h (top four panels) and of wild-type cells that were pretreated with Leptomycin B (+LMB) 30 min before they were either not further treated or incubated in presence of 10 mM HU for 2 h (bottom four panels). (C) The FACS profile of these S-phase-arrested cells. [Wt(N)] Nitrogen-starved cells to show 1N and 2N peaks. (Wt + HU) Cells after 2 h in 10 mM HU. (Wt + HU + LMB) Cells similarly treated but with the addition of Leptomycin B together with HU. (D) An N-terminal TAP-tagged Suc22 localizes to the nucleus in G2 cells and to the whole cell, including the cytoplasm, after 2.5 h of HU exposure (left, DAPI; right, FITC). In spd1-d cells, Suc22 is constitutively spread over the whole cell, and this does not alter during the cell cycle. (E) Cells in which the endogenous cdc22 loci was modified to encode an N-terminal GFP tag under the control of the native cdc22 promoter were used to visualize Cdc22 localization by indirect immunofluorescence using an anti-GFP antibody. G2- and S-phase cells are shown stained for DAPI (left) and FITC (right).
In contrast to rad3-d, deletion of csn1 (or csn2; data not shown) resulted in constitutive nuclear localization of Suc22 with no decrease during S phase (Fig. 3A), even when cells were arrested with HU for 2 h (Fig. 3B). In spd1-d cells we did not observe any specific nuclear localization of Suc22 at any stage in the cell cycle (Fig. 3A). A similar profile is seen in spd1-d csn1-d double-mutant cells (data not shown). These data suggest an antagonistic role for Spd1 and the signalosome in RNR regulation: Spd1 maintains Suc22 in the nucleus, whereas the signalosome appears to assist in its nuclear export.
Suc22 relocalization requires active nuclear export
If crm1-dependent nuclear export is required for Suc22 loss from the nucleus, cells treated with Leptomycin B (LMB), an inhibitor of crm1-dependent nuclear export, should retain Suc22 nuclear staining after HU treatment. Indeed, this is the case (Fig. 3B,C). However, although we see crm1-dependent loss of nuclear Suc22 in HU-treated cells, we do not observe an accompanying increase in the cytoplasmic signal. Our anti-Suc22 antibody was raised to a C-terminal peptide. This region of the small RNR subunit is tightly associated with the large subunit in active mammalian RNR complexes (Lycksell et al. 1994). Thus, the lack of cytoplasmic staining likely reflects epitope masking in the cytoplasm. To test this possibility, we examined Suc22 localization after HU treatment using an N-terminal TAP-tagged Suc22 (Werler et al. 2003). Loss of TAP-Suc22 from the nucleus was accompanied by the simultaneous appearance of TAP-Suc22 in the cytoplasm (Fig. 3D). If the Suc22 localized in the cytoplasm is not detectable with our α-Suc22 antibody because of epitope masking, this suggests that the large subunit of RNR (Cdc22) is resident in the cytoplasm. We have been unable to obtain a polyclonal antibody to Cdc22, but using a strain expressing GFP-Cdc22 (Fig. 3E) from the cdc22 locus and under the control of the native cdc22 promoter, we, indeed, observed that Cdc22 is constitutively localized throughout the cell, without obvious exclusion from, or concentration in, the nucleus.
RNR localization in response to the G2 DNA damage checkpoint
csn1-d and csn2-d cells are DNA-damage-sensitive. To ascertain the behavior of Suc22 following DNA damage, we first analyzed Suc22 localization after treatment of wild-type cells or chk1-d mutant cells with 100 J/m2 of UV light (Fig. 4A) or with 250 Gy of IR (data not shown). Irradiation of cells synchronized in G2 resulted in a rapid chk1-dependent loss of nuclear Suc22 staining. Similar analysis of asynchronous cell populations (Fig. 4B) shows this effect is also dependent on rad3 and csn1, but not on csn4. Pretreating cells with LMB (Fig. 4C) before irradiation shows that Suc22 is exported from the nucleus in a signalosome- and checkpoint-dependent manner, presumably to achieve RNR activation in response to DNA damage.
If the relocalization of Suc22 from the nucleus to the cytoplasm reflects activation of RNR in G2 in response to DNA damage, failure to relocalize Suc22 should cause DNA damage sensitivity. Indeed, csn1-d cells (which do not relocalize Suc22 following damage) are UV- and IR-sensitive (Mundt et al. 1999). Because deletion of spd1 results in constitutive loss of Suc22 nuclear localization even in a csn1-d background, we tested the UV and IR sensitivity of csn1-d spd1-d double-mutant cells (Fig. 4D) and of csn1-d mutants overexpressing suc22 (data not shown). Loss of spd1 or overexpression of Suc22 in a csn1-d background restored UV and IR sensitivity to close to wild-type levels. These data suggest that the CSN- and checkpoint-dependent relocalization of Suc22 from the nucleus to the cytoplasm is required for an efficient DNA damage response.
Relocalization of Suc22 corresponds to degradation of Spd1
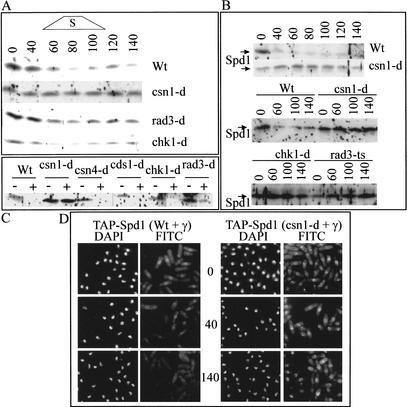
The data presented above indicate that Spd1 negatively controls nuclear export of Suc22 but Csn1 stimulates it. This predicts that Csn1 controls either a modification of Spd1 or its level in the cell. Using an antibody against Spd1 (Borgne and Nurse 2000) and epitope-tagged strains (see Materials and Methods), we characterized Spd1 levels throughout the unperturbed cell cycle, in cells treated with HU and in G2 cells following treatment with either UV or IR (Fig. 5A–C). Spd1 levels mirrored the nuclear export of Suc22: during a synchronized unperturbed cell cycle, Spd1 levels declined transiently in S phase in a manner that was dependent on csn1 but independent of rad3 and chk1 (Fig. 5A). This is consistent with previously reported data (Borgne and Nurse 2000). In response to ionizing irradiation of G2 cells, Spd1 levels decreased in a manner dependent on rad3, chk1, and csn1 (Fig. 5B). In response to HU, Spd1 levels dropped in a rad3-dependent and csn1-dependent manner as cells accumulated in S phase (Fig. 5C). Indirect immunofluorescence established that Spd1 is a nuclear protein, which disappeared in a signalosome-dependent manner upon ionizing radiation of G2 cells (Fig. 5D).
Figure 5.
Spd1 levels during S phase and after DNA damage. (A) Wild-type and null mutant cells (as indicated) were synchronized in G2 (see Fig. 2A), and Spd1 levels were monitored through the second S phase. An indication of the time of S phase is given above. Fifty micrograms of total protein extracts from each time point were separated by SDS-PAGE and detected with anti-Spd1 (Borgne and Nurse 2000) antibodies, which routinely give significant background “spots.” (B) Wild-type and the indicated mutant cells were synchronized in G2, and half the culture was irradiated (IR, 250 Gy, timing as in Fig. 4B), and extracts prepared from untreated (0 time) and treated (40–140 min) cells were analyzed by Western blot and probing with α-Spd1. rad3-ts cells were treated at the restrictive temperature of 36°C. (C) Asynchronous wild-type and indicated mutant cells were either not treated or incubated with HU for 2.5 h, and Spd1 was detected (as above) with α-Spd1. (D) TAP-tagged Spd1 (the TAP-epitope integrated at the spd1 locus and under the control of the spd1 endogenous promoter) was detected by indirect immunofluorescence in either csn+ or csn1-d cells before, 40, and 140 min following 500 Gy of ionizing radiation. In the absence of Csn1, Spd1 remains nuclear after DNA damage. Also seen in csn1-d cells are bright foci that appear to be spindle pole bodies. The significance of this, if any, is unknown.
Proteosome and polyubiquitination-dependent Spd1 degradation
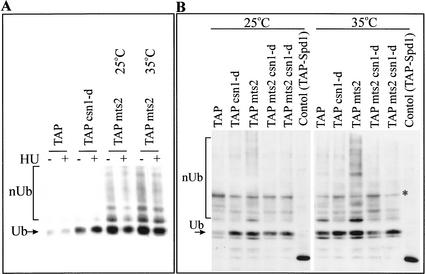
To confirm that Spd1 is degraded in a ubiquitin- and proteosome-dependent manner, we examined ubiquitin modification of an endogenous TAP-Spd1 protein (expressed from its own promoter at the spd1 locus) using an assay in which His6-Ub is expressed in cells and ubiquitinated proteins are purified by IMAC chromatography. Probing the purified fractions derived from HU-treated csn1-d, mts2-1, and mts3-1 single-mutant cells (Gordon et al. 1997) and mts2-1 csn1-d double-mutant extracts for TAP-Spd1 (Fig. 6A,B) demonstrates that, when the proteosome function is attenuated, polyubiquitinated TAP-Spd1 can be detected. Mts2 and Mts3 are subunits of the regulatory particle of the proteosome. Whereas in mts+ cells polyubiquitinated proteins are rapidly degraded and are thus not detected, mts2 or mts3 mutants accumulate polyubiquitinated TAP-Spd1 at the restrictive temperature. Consistent with a role of the CSN complex in Spd1 degradation, polyubiquitinated TAP-Spd1 is absent in two independent mts2-1 csn1-d double mutants at the restrictive temperature (Fig. 6B). Interestingly, monoubiquitination of TAP-Spd1 is not significantly decreased in the csn1-d mutant background.
Figure 6.
The dependency of Spd1 degradation on Pcu4 and copurification of Pcu4 with the signalosome. (A) Ubiquitinated TAP-Spd1 forms are evident in mts3 mutant cells at both the permissive and restrictive temperatures. Wild-type, csn1-d, and mts3-1 mutant cells were used to express His6-ubiquitin. Soluble extracts were prepared from each strain (mts3-1 cells were analyzed at both the permissive 25°C and restrictive 35°C temperatures) either with (+) or without (−) prior incubation in HU for 2.5 h. His6-ubiquitin was purified on Ni2+ beads; the beads were boiled and the eluate was resolved by 12% SDS-PAGE and probed for the TAP-tag. nUb indicates the multiple-ubiquitinated forms of TAP-Spd1; Ub indicates the monoubiquitinated form. (B) Ubiquitinated TAP-Spd1 forms are evident in mts2 mutant cells, and these are lost in mts2 csn1-d double mutants. Wild-type, csn1-d, mts2, and two csn1-d mts2 double-mutant isolates were used to express His6-ubiquitin. Soluble extracts were prepared and His6-ubiquitin was purified on Ni2+ beads. The resulting beads were boiled, and the eluate was resolved by 8% SDS-PAGE and probed for the TAP-tag. As before, nUb indicates the multiple-ubiquitinated forms of TAP-Spd1; Ub indicates the monoubiquitinated form. A control aliquot of TAP-Spd1 extract is also run to show the size of unmodified TAP-Spd1 and demonstrate that nonubiquitinated Spd1 is not purified by IMAC chromatography. The star indicates a nonspecific band.
Spd1 degradation requires Pcu4, which copurifies with the signalosome
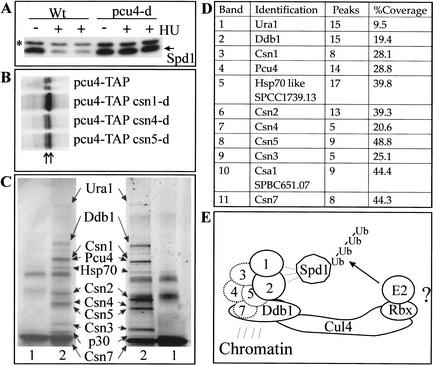
Identifying the mechanism of Spd1 degradation should provide insight into the function of the signalosome. The CSN complex is required for the deneddylation of the cullins, and the subunits have been identified as cullin-binding proteins (Lyapina et al. 2001). Mutants in both pcu3 and pcu4, which encode two of the three S. pombe cullin homologs, have been reported to exhibit slow growth and DNA damage sensitivity, phenotypes that would be consistent with a role in Spd1 degradation (Kominami et al. 1998). We therefore ascertained if Spd1 degradation was dependent on pcu3 or pcu4. First, we tagged Spd1 with a 3 × HA tag to simplify its identification (see Materials and Methods). In pcu4-d mutants (but not pcu3-d mutants, data not shown), loss of 3HA-Spd1 upon treatment of cells with HU is significantly attenuated (Fig. 7A), suggesting that Pcu4 plays a major role in Spd1 degradation.
Figure 7.
Analysis of Spd1 degradation. (A) HA-tagged Spd1 (Spd1) was analyzed by SDS-PAGE and Western blot of 50 μg of pcu+ and pcu4-d cell extracts either before (−) or after (+) HU treatment. The star indicates a nonspecific band. (B) C-terminal TAP-tagged Pcu4 detected by Western blot in extracts derived from csn+, csn1-d, csn4-d, and csn5-d cells. The left arrow indicates the unmodified form, the right arrow indicates the modified form. (C) TAP-purification of the signalosome from 30 L of S. pombe culture. (Left) Coomassie-stained SDS-PAGE from which the indicated bands were excised, subjected to trypsin digestion, and analyzed by mass spectroscopy. (Right) An aliquot of the same preparation stained with silver. (Lane 1) Untagged control. (Lane 2) csn2-TAP strain (Csn2-TAP is expressed from its own promoter from the normal genomic locus). (D) The number of identified peaks for each sample and the percentage sequence coverage. (E) A possible model for the function of the signalosome. Ddb1, based on its association with DET1, histones, and histone deacetylases (Benvenuto et al. 2002; Martinez et al. 2001; Schroeder et al. 2002), may anchor the signalosome and Pcu4 to specific sites on the chromatin to allow association of target proteins, such as Spd1, with Pcu4 via interactions with signalosome subunits.
Because the neddylation status of Pcu4 has not been reported, we used a strain in which the 3′ region of the pcu4 gene of S. pombe was modified to encode the TAP-tag (see Materials and Methods). Although not fully functional, Pcu4-TAP separates on SDS-PAGE as two major bands, consistent with Nedd8 modification (Fig. 7B). Surprisingly, in csn4-d and csn5-d mutant backgrounds, these two bands were unaltered, whereas in a csn1-d background the upper band was overrepresented compared with the lower band (Fig. 7B). These data suggest that there may be a more direct relationship between Pcu4 and the Csn1/Csn2 subunits of the signalosome than exists between the signalosome and either Pcu1 or Pcu3.
To explore this further, we generated a TAP-tagged csn2 allele (integrated at the endogenous locus and expressed from its own promoter) and purified the signalosome to homogeneity (Fig. 7C; Tasto et al. 2001). Purified proteins were separated by SDS-PAGE and samples were stained with Coomassie blue (left) or silver (right). Individual Coomassie bands were excised and identified by trypsin cleavage and mass fingerprinting using mass spectrometry. Eleven bands generated fingerprinting data compatible with annotated ORFs in the S. pombe genome database (Fig. 7D). In addition to the identification of Csn1, Csn2, Csn3, Csn4, Csn5, and Csn7, we also identified Ura1, Ddb1, Pcu4, an Hsp70-like protein, and an unknown ORF (SPBC651.07) encoding a protein of 30.48 kD that we have named Csa1 (for COP9/signalosome-associated protein 1). We did not identify the putative Csn6 homolog or the second ORF in S. pombe with homology to Csn7 subunits (SPAC1751.03). This is consistent with the fact that, unlike the other csn genes, csn6 and SPAC1751.03 (annotated as csn7b) are both essential, suggesting functions distinct from the signalosome (C. Zhou and D.A. Wolf, pers. comm). Although deneddylation of Pcu1 and Pcu3 is signalosome-dependent, we did not find Pcu1 or Pcu3 association with the signalosome in these preparations, suggesting that these interactions (Lyapina et al. 2001; Zhou et al. 2001) are unstable. It is possible that the 11 proteins identified are present in a single complex. An alternative explanation would be that two or more independent complexes containing TAP-Csn2 are copurifying. To date, we have not distinguished between these two possibilities.
Discussion
Although mutations in csn1, csn2, csn3, csn4, and csn5 each cause constitutive Nedd8 modification of the cullins Pcu1 and Pcu3, only two subunits of the S. pombe signalosome, Csn1 and Csn2, are important for proper progression through S phase and efficient survival of DNA damage (Mundt et al. 1999, 2002; Zhou et al. 2001). Our data strongly suggest that the failure to degrade Spd1 in a Csn1/Csn2-dependent manner is the reason for both the S-phase delay and the repair defect. Spd1 acts to keep the small RNR subunit Suc22 in the nucleus, thereby regulating RNR activity. Because the large RNR subunit, Cdc22, is constitutively dispersed throughout the cell, we suggest: (1) There is a nuclear RNR pool that can be activated in the absence of signalosome function, because the complete loss of RNR would be a lethal event. (2) To increase the supply of deoxyribonucleotides during DNA replication and repair, Suc22 delocalizes from the nucleus to the cytoplasm in a signalosome-dependent manner to form additional active complexes with Cdc22. In cells deleted for spd1, Suc22 is constitutively cytoplasmic and most probably associated with Cdc22 (the C-terminal epitope is masked; cf. Figs. 3, 4 and Lycksell et al. 1994) and therefore constitutively active. Thus, our data suggest that the Csn1 and Csn2 subunits of the signalosome are required to degrade Spd1. This releases Suc22 from the nucleus so it can associate with Cdc22 and dNTPs can be produced.
This model is supported by the response of Spd1 and Suc22 after DNA damage: following irradiation of G2 cells, Csn1 and Csn2 are both required for degradation of Spd1 and Suc22 relocalization to the cytoplasm. In csn1-d cells, Suc22 is always nuclear and relocalization fails to occur after DNA damage. However, in cells deleted for both csn1 and spd1, Suc22 is constitutively cytoplasmic, which suppresses both the UV and ionizing radiation sensitivity, showing that DNA damage sensitivity is coincident with the inability to localize Suc22 to the cytoplasm after DNA damage. Thus, the sensitivity of csn1-d cells probably reflects inefficient repair caused by a lack of dNTPs. This hypothesis is supported by the observation that overexpression of suc22 in csn1-d cells rescues UV sensitivity as effectively as spd1 deletion.
The signalosome and Pcu4 cooperate to degrade Spd1
Csn1 and Csn2, but not Csn3, Csn4, or Csn5, are required in vivo for the degradation of Spd1. The cullin Pcu4 is also required for Spd1 degradation (Fig. 7; data not shown). Pcu4 is most homologous to human cullins CUL4A and CUL4B and like these human cullins lacks the N-terminal Skp1-binding motif found in Pcu1 and its human homolog CUL1. Skp1 is the adaptor protein that links CUL1 to F-box proteins that, in turn, target the SCF to its substrates. The other S. pombe cullin, Pcu3, is most homologous to human CUL3, which associates with an Skp1-like protein and has an N-terminal domain bearing homology to the Skp1-binding site of CUL1. It is not known if S. pombe Pcu3 associates with Skp1, but it is not thought that Pcu4 associates with the adapter protein Skp1, and therefore it is unlikely to bind F-box proteins.
Our data suggest a close relationship between Pcu4 and the signalosome that is distinct from the relationship between the signalosome and either Pcu1 or Pcu3: the genetic dependency of the Nedd8 modification status of Pcu4 is apparently distinct from that seen with Pcu1 or Pcu3 and appears to mirror the biological phenotypes of null mutations in signalosome subunits. Furthermore, the purified S. pombe signalosome contained Pcu4, but not Pcu1 or Pcu3, and no F-box proteins were found to copurify. There is a strong prediction in the literature that Csn5 encodes the isopeptidase that cleaves Nedd8 from cullins (Lyapina et al. 2001). We observe that Pcu4 migrates as two bands (usually diagnostic for Nedd8 modification) in the csn5-d mutant, but that this pattern is altered to a single upper (Nedd8-modified) band in csn1-d and csn2-d mutants. This suggests that, for Pcu4 at least, Csn5 is not required for Nedd8-isopeptidase activity, although alternative explanations cannot be ruled out at this stage.
Each signalosome subunit (Csn1–8) contains either an MPN or a PCI domain. In the human and Arabidopsis signalosomes, only the Csn5 and Csn6 subunits are MPN-domain proteins. Csn5 is a member of the MPN+/JAMM motif subclass of MPN-domain proteins, whereas Csn6 contains a conventional MPN domain (Cope et al. 2002; Maytal-Kivity et al. 2002). The MPN+/JAMM motif contains five conserved polar residues embedded within the conventional MPN domain that may define a catalytic domain, perhaps coordinating a metal ion. Although it has been predicted that the MPN+/JAMM motif relates to a novel isopeptidase activity, active recombinant protein has not been reported, and thus formal proof of this prediction is unavailable (Cope et al. 2002; Maytal-Kivity et al. 2002). The purified S. pombe signalosome did not contain the putative Csn6 homolog, which in any case would not be predicted to contain a catalytic activity because this is only thought to be associated with the MPN+/JAMM subfamily of MPN domain proteins (Maytal-Kivity et al. 2002), but not with the conventional MPN domain proteins. Thus, if there is a Csn5-independent isopeptidase activity directed against Pcu4–Nedd8, it is most likely to be encoded by one of the classical cystine proteases, possibly recruited to the complex by association with the Csn1–Csn2 PCI-domain proteins, which themselves do not contain a known protease motif.
Interestingly, a homolog of the DDB1 protein was identified as a signalosome-associated protein. Human DDB1 is the p127 subunit of the heterodimeric damaged DNA binding complex. The second human DDB subunit, p48 (DDB2), is a WD-repeat protein mutated in Xeroderma pigmentosum group E patients (XPE). DDB has been implicated in NER (Tang et al. 2000), although DDB subunits clearly have other functions in the cell, possibly in the regulation of transcription (Martinez et al. 2001; Schroeder et al. 2002). Interestingly, Ddb1 has been found in human cells to be associated with CUL4A (Shiyanov et al. 1999), and Ddb2 (the WD-repeat subunit of DDB) is targeted for degradation by CUL4A (Nag et al. 2001). This suggests that our finding Ddb1 in the complex is not coincidental and reflects a role for Ddb1 in Cul4-dependent polyubiquitination (Fig. 7E).
The S. pombe ddb1-d strain exhibits slow growth and modest DNA damage sensitivity (Zolezzi et al. 2002) and, in results to be presented elsewhere (A.M. Carr and C. Liu, unpubl.), we find that Ddb1 plays a role in Spd1 degradation. Whereas a Ddb1 homolog is present in S. pombe, no clear homolog of p48 (Ddb2) can be identified. Intriguingly, S. cerevisiae lacks both Ddb2 and Ddb1 homologs, and also does not have a highly conserved signalosome or a cullin with clear homology to CUL4A/B. An attractive explanation for our observations would be that the signalosome provides a targeting function for Pcu4-dependent E3 ubiquitin ligases, recruiting this polyubiquitination apparatus to nuclear proteins such as Spd1 (and possibly a range of transcription factors) through interactions with Ddb1 (Fig. 7E). In such a model, Csn1 and Csn2 could be required to target Pcu4-dependent E3 ligase to Spd1, whereas other subunits may target other as yet unidentified proteins. Alternatively, the regulation of Pcu4 by Csn1 and Csn2 through deneddylation may be more strict than the regulation of Pcu1 by Csn-dependent deneddylation (note that Pcu1 is an essential protein, but csn5-d mutants, which are defective in deneddylation of Pcu1, display no discernable phenotype in log-phase cells).
The association of Ddb1 with histone deacetylases in human cells (Martinez et al. 2001) and of Arabidopsis Ddb1 with Det1 (Schroeder et al. 2002), an H2A-binding protein required for photomorphogenic transcription (Benvenuto et al. 2002), suggests that there may be relationship between chromatin functions and Ddb1 that may involve Pcu4 and the signalosome in the regulation of transcriptional control.
Two distinct signalosome-dependent mechanisms regulate Spd1 degradation and Suc22 relocalization
We can clearly distinguish two separate inputs to the Spd1 degradation machinery: In unperturbed S phase, Spd1 degradation is csn1- and csn2-dependent but rad3-, chk1-, and cds1-independent. In G2 cells after DNA damage, Spd1 degradation requires both the signalosome subunits Csn1 and Csn2 plus the rad3- and chk1-dependent DNA damage checkpoint.
This suggests that Chk1 either directly or indirectly modifies Spd1 in response to DNA damage, or that it modifies an aspect of the protein degradation machinery to target Spd1. In S. cerevisiae, a similar small protein, Sml1, acts as a formal inhibitor of RNR activity (Chabes et al. 1999). Although it is unknown if this acts in vivo by regulating the subcellular localization of RNR subunits, it is known that Sml1 is targeted by the Mec1–Dun1 kinase pathway for phosphorylation (Zhao and Rothstein 2002) and that this phosphorylation correlates with Sml1 degradation and RNR induction both during S phase and in response to DNA damage.
We have seen no evidence of Spd1 modification in response to cell cycle progression or DNA damage in S. pombe (Fig. 5A,B; data not shown). We presently favor the probability that Csn1-dependent Spd1 degradation is controlled by modifications to the Pcu4 and signalosome-dependent protein-degradation machinery. These modifications could be directed by Chk1 in response to DNA damage and by alternative mechanisms during cell cycle progression (e.g., cyclin-dependent kinase activity).
Materials and methods
Elutriation and FACS analysis
Cell were synchronized using a JE-5.0 elutriating centrifuge (Beckman). Small G2 cells were collected, harvested, and resuspended in fresh medium at 3 × 106 cells/mL. Septation was determined by fixing cells in methanol and staining with DAPI and Calcofluor (Russell and Nurse 1986). Samples for protein extract were washed in ice-cold water, frozen in LN2, and stored at −80°C. Of this, 50 μg (total protein) was Western-blotted. Cells were prepared for FACS analysis as previously described (Moreno et al. 1991; Lindsay et al. 1998). FACS profiles of asynchronous cultures show the majority of cells in G2 (2n) and a population of <10% binucleate cells within S phase, indicated by the 2–4n shoulder.
Immunofluorescent microscopy
Staining was as described (Hagan and Hyams 1988). Cells were fixed in 3.7% paraformaldehyde for 10 min and stained with primary antibody (α-Suc22 at 1:50, α-GFP at 1:200) and secondary antibody (FITC-conjugated, DAKO, at 1:150). TAP-Tag was detected directly with FITC-conjugated rabbit anti-mouse (DAKO) at 1:150.
Genetics, cell biology, protein extracts, and Cds1 kinase assay
Strain construction, gene deletion, and epitope tagging was performed by standard genetic techniques (Moreno et al. 1991; Bahler et al. 1998; Werler et al. 2003). TAP-cdc22, TAP-suc22, and csn2-TAP alleles were fully functional, as judged by a lack of phenotype. The TAP-spd1 and 3HA-spd1 alleles had intermediate phenotypes as judged by their ability to prevent lethality in rad3-ts csn1-d rescue experiments. However, degradation of tagged Spd1 appeared normal after HU treatment, was slightly delayed after irradiation of G2 cells, and showed the same genetic dependencies. Cells harboring the pcu4-TAP allele were viable but slightly UV-sensitive and cells were elongated (a gift from D.A. Wolf). The phenotype was similar to the null allele. Using the same methodology, the coding region of Suc22 driven by the adh1 constitutive promoter (Russell and Hall 1983) was integrated at the ura4 locus. In the resulting strain, Suc22 was expressed ∼10-fold higher than in wild-type cells. The protocols for checkpoint measurements, cell scoring, and irradiation are described in Edwards and Carr (1997). Protein extracts for Western blotting were prepared by TCA extraction as described in Caspari et al. (2000). Soluble extracts were prepared by grinding cells in liquid nitrogen as described in Caspari et al. (2000). Cds1 kinase assays were performed, as previously described (Lindsay et al. 1998), using MBP as an in vitro substrate. Spd1 ubiquitination was assayed as described (Shiozaki and Russell 1997). Probing for Spd1-TAP, we found that the presence of multiubiquitinated bands was dependent on the presence of the TAP-tagged Spd1 and on the expression of H6-ubiquitin as previously described.
Acknowledgments
This work was supported by HFSP grant RGO178/2000-M and MRC grant E160/106. L.W. is a recipient of a predoctoral fellowship from the Ministry of Education, Taiwan. We thank A. Brown and D. Stead of the COGEME facility for mass spectrometry analysis, and C. Zhou and D.A. Wolf for communicating unpublished results and valuable discussion. Finally we thank numerous colleagues for constructive criticism, ideas, and support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL a.m.carr@sussex.ac.uk; FAX 44-1273-678121.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1090803.
References
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C. The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr Biol. 2002;12:1529–1534. doi: 10.1016/s0960-9822(02)01105-3. [DOI] [PubMed] [Google Scholar]
- Borgne A, Nurse P. The Spd1p S phase inhibitor can activate the DNA replication checkpoint pathway in fission yeast. J Cell Sci. 2000;113:4341–4350. doi: 10.1242/jcs.113.23.4341. [DOI] [PubMed] [Google Scholar]
- Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A, Domkin V, Thelander L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J Biol Chem. 1999;274:36679–36683. doi: 10.1074/jbc.274.51.36679. [DOI] [PubMed] [Google Scholar]
- Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng XW. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell. 1996;86:115–121. doi: 10.1016/s0092-8674(00)80082-3. [DOI] [PubMed] [Google Scholar]
- Christensen PU, Bentley NJ, Martinho RG, Nielsen O, Carr AM. Mik1 levels accumulate in S phase and may mediate an intrinsic link between S phase and mitosis. Proc Natl Acad Sci. 2000;97:2579–2584. doi: 10.1073/pnas.97.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of NEDD8 from CUL1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Dubiel W, Wei N, Hofmann K, Mundt K, Colicelli J, Kato J, Naumann M, Segal D, Seeger M, et al. Unified nomenclature for the COP9 signalosome and its subunits: An essential regulator of development. Trends Genet. 2000;16:202–203. doi: 10.1016/s0168-9525(00)01982-x. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes & Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RJ, Carr AM. Analysis of radiation-sensitive mutants of fission yeast. Meth Enzymol. 1997;283:471–494. doi: 10.1016/s0076-6879(97)83038-8. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Gordon C, McGurk G, Wallace M, Hastie ND. A conditional lethal mutant in the fission yeast 26 S protease subunit mts3+ is defective in metaphase to anaphase transition. J Biol Chem. 1997;271:5704–5711. doi: 10.1074/jbc.271.10.5704. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the role of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Harris P, Kersey PJ, McInerny CJ, Fantes PA. Cell cycle, DNA damage and heat shock regulate suc22+ expression in fission yeast. Mol Gen Genet. 1996;252:284–291. doi: 10.1007/BF02173774. [DOI] [PubMed] [Google Scholar]
- Hellmann H, Estelle M. Plant development: Regulation by protein degradation. Science. 2002;297:792–797. doi: 10.1126/science.1072831. [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes & Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp D, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kominami K, Ochotorena I, Toda T. Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1-cullin-1-F-box) ubiquitin ligase. Genes Cells. 1998;3:721–735. doi: 10.1046/j.1365-2443.1998.00225.x. [DOI] [PubMed] [Google Scholar]
- Lindsay HD, Griffiths DJF, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes & Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Lycksell P, Ingemarson R, Davis R, Gruslund A, Thelander L. 1H NMR studies of mouse ribonucleotide reductase: The R2 protein carboxy-terminal tail, essential for subunit interaction, is highly flexible but becomes rigid in the presence of protein R1. Biochemistry. 1994;33:2838–2842. doi: 10.1021/bi00176a013. [DOI] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinho RG, Lindsay HD, Flaggs G, DeMaggio A, Hoekstra M, Carr AM, Bentley NJ. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth Enzymol. 1991;194:795–826. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Mundt KE, Porte J, Murray JM, Brikos C, Christensen PU, Caspari T, Hagan IM, Millar JBA, Simanis V, Hofmann K, et al. The COP9/signalosome is conserved in fission yeast and has a role in S-phase. Curr Biol. 1999;9:1427–1430. doi: 10.1016/s0960-9822(00)80091-3. [DOI] [PubMed] [Google Scholar]
- Mundt KE, Liu C, Carr AM. Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol Biol Cell. 2002;13:493–502. doi: 10.1091/mbc.01-10-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Bondar T, Shiv S, Raychaudhuri P. The Xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol Cell Biol. 2001;21:6738–6747. doi: 10.1128/MCB.21.20.6738-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F, Saeki M, Katayama S, Aida N, Toh-e A, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng X-W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:662–666. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Russell PR, Hall BD. The primary structure of the alcohol dehydrogenase gene from the fission yeast Schizosaccharomyces pombe. J Biol Chem. 1983;258:143–149. [PubMed] [Google Scholar]
- Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol. 2002;12:1462–1472. doi: 10.1016/s0960-9822(02)01106-5. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Deng XW. COP9 signalosome revisited: A novel mediator of protein degradation. Trends Cell Biol. 2001;11:420–426. doi: 10.1016/s0962-8924(01)02091-8. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Deng XW. Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell. 2002;14:2553–2563. doi: 10.1105/tpc.003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, Gordon C, Naumann M, Dubiel W. A novel protein complex involved in signal transduction possessing similarities to 26S proteosome subunits. FASEB J. 1998;12:469–478. [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Stress-activated protein kinase pathway in cell cycle control of fission yeast. Methods Enzymol. 1997;283:506–520. doi: 10.1016/s0076-6879(97)83040-6. [DOI] [PubMed] [Google Scholar]
- Shiyanov P, Nag A, Raychaudhuri P. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J Biol Chem. 1999;274:35309–35312. doi: 10.1074/jbc.274.50.35309. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig K, Tyers M, Elledge SJ, Harper JW. F-Box proteins are components of E3 complexes and act as receptors to recruit phosphorylated substrates for ubiquitination. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp D, Kamura T, Conrad M, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Yanagawa Y, Kwok SF, Matsui M, Deng XW. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes & Dev. 2002;16:554–559. doi: 10.1101/gad.964602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasto JJ, Carnahan RH, McDonald WH, Gould KL. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Osterlund MT, Kwok SF, Deng XW. Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol. 1997;114:779–788. doi: 10.1104/pp.114.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S, Hetfeld B, Dubiel W, Wolf DA. Conservation of the COP9/signalosome in budding yeast. BMC Genet. 2002;3:15. doi: 10.1186/1471-2156-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werler P, Hartsuiker E, Carr AM. A simple Cre-loxP method for chromosomal N-terminal tagging of essential and inessential S. pombe genes. Gene. 2003;304:133–141. doi: 10.1016/s0378-1119(03)00402-5. [DOI] [PubMed] [Google Scholar]
- Woollard A, Basi G, Nurse P. A novel S phase inhibitor in fission yeast. EMBO J. 1996;15:4603–4612. [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- Zhou C, Seibert V, Geyer R, Rhee E, Lyapina S, Cope G, Deshaies RJ, Wolf DA. The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem. 2001;2:7–12. doi: 10.1186/1471-2091-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolezzi F, Fuss F, Uzawa S, Linn S. Characterization of a Schizosaccharomyces pombe strain deleted for a sequence homologue of the human damaged DNA binding 1 (DDB1) gene. J Biol Chem. 2002;277:41183–41191. doi: 10.1074/jbc.M207890200. [DOI] [PubMed] [Google Scholar]