Abstract
The transcription factor Myc is induced by mitogenic signals and regulates downstream cellular responses. If overexpressed, Myc promotes malignant transformation. Myc modulates expression of diverse genes in experimental systems, but few are proven direct targets. Here, we present a large-scale screen for genomic Myc-binding sites in live human cells. We used bioinformatics to select consensus DNA elements (CACGTG or E-boxes) situated in the 5′ regulatory region of genes and measured Myc binding to those sequences in vivo by quantitative chromatin immunoprecipitation. Strikingly, most promoter-associated E-boxes showed selective recovery with Myc, unlike non-E-box promoters or E-boxes in bulk genomic DNA. Promoter E-boxes were distributed in two groups bound by Myc at distinct frequencies. The high-affinity group included an estimated 11% of all cellular loci, was highly conserved among different cells, and was bound independently of Myc expression levels. Overexpressed Myc associated at increased frequency with low-affinity targets and, at extreme levels, also with other sequences, suggesting that some binding was not sequence-specific. The strongest DNA-sequence parameter defining high-affinity targets was the location of E-boxes within CpG islands, correlating with an open, preacetylated state of chromatin. Myc further enhanced histone acetylation, with or without accompanying induction of mRNA expression. Our findings point to a high regulatory and biological diversity among Myc-target genes.
Keywords: Human, genome, chromatin, transcription factor, Myc
Supplemental material is available at http://www.genesdev.org.
The behavior of mammalian cells is modulated by many extracellular stimuli, which trigger a variety of intracellular signaling pathways. Those signals induce expression of primary, or immediate early (IE) genes. Several IE genes encode transcription factors that, in turn, regulate secondary transcriptional responses (Winkles 1998). The c-myc proto-oncogene is such an IE gene. In normal cells, c-myc expression is generally induced by mitogens and suppressed by growth-inhibitory signals. Oncogenic activation of c-myc occurs by direct gene alterations, such as translocation or amplification, or by mutations in upstream signaling pathways. These accidents commonly result in deregulated and/or elevated expression of c-myc and its product, the Myc protein. c-myc is structurally related to two other genes, L- and N-myc, which are also overexpressed in specific tumor types (for reviews, see Garte 1993; Henriksson and Lüscher 1996; Grandori et al. 2000; Oster et al. 2002).
The changes in cellular behavior imparted by abnormal Myc expression have been studied extensively in cultured cells and transgenic animals. Myc has generally been associated with the promotion of cellular growth and proliferation, desensitization to growth-inhibitory stimuli, blockade of cell differentiation, cellular immortalization, and oncogenic transformation, as well as sensitization to apoptosis-inducing signals. Myc has also been linked to the regulation of various metabolic pathways, the induction of DNA damage, and genomic instability (for reviews, see Henriksson and Lüscher 1996; Amati et al. 1998; Grandori et al. 2000; Oster et al. 2002). In addition, Myc is likely to promote tumorigenesis in vivo through the induction of angiogenesis and tumor-cell invasiveness (Pelengaris et al. 1999, 2002; Brandvold et al. 2000; Ngo et al. 2000). Conversely, loss of Myc function has been associated with defects in cell growth, proliferation, and/or apoptosis, and is likely to profoundly impair the response of cells to their environment (e.g., Mateyak et al. 1997; Johnston et al. 1999; Bates et al. 2000; de Alboran et al. 2001; Douglas et al. 2001; Trumpp et al. 2001). In summary, the pathological effects of Myc overexpression are likely to result from multiple biological activities, reflecting the normal roles of Myc in coordinating cellular physiology with extracellular stimuli.
Myc is a transcription factor of the basic helix–loop–helix–leucine zipper (bHLH–LZ) family that can activate or repress gene expression. Activation occurs via dimerization with the bHLH–LZ partner Max and direct binding to the DNA sequence CACGTG, called the E-box (Blackwood and Eisenman 1991; Prendergast et al. 1991; Amati et al. 1992; Kretzner et al. 1992). Repression, instead, occurs through functional interference with transcription factors, such as Miz-1, that bind different DNA sequences (Oster et al. 2002). Both transcriptional activities of Myc appear to be critical for its biological function. Most relevant to the present work, dimerization with Max and binding to the E-box are essential for Myc to promote cell cycle progression, apoptosis, and cellular transformation (Amati et al. 1998). Thus, in order to understand the function of Myc, we need to identify its target genes, and in particular the E-box elements that Myc binds in the cellular genome.
Identification of Myc-regulated genes has generally relied on experimental activation of Myc followed by monitoring of changes in mRNA levels (Grandori et al. 2000; Oster et al. 2002). More than 10 investigative works reported the use of high-throughput screening based on cDNA microarrays or the SAGE assay, significantly expanding the list of genes that are up- or down-regulated by Myc (e.g., Schuldiner and Benvenisty 2001; Menssen and Hermeking 2002; Oster et al. 2002; Watson et al. 2002; O'Connell et al. 2003). Based on an updated online compilation (http://www.myc-cancer-gene.org/index.asp), this list now includes several hundred genes. It remains unclear, however, how many of these genes are direct targets of Myc. All studies based on mRNA expression have been hampered by the fact that a large fraction of Myc-target genes respond weakly, or even fail to respond to Myc activation, depending on the cell type or experimental conditions used. For example, several target genes that require Myc for induction by serum in Rat1 fibroblasts do not respond well to Myc alone in the same cells (Frank et al. 2001). Underlying this limitation, the lists of genes identified in high-throughput screens are only partially overlapping, and many genes were identified only once. Thus, we still possess a fragmentary picture of the loci that are directly targeted by Myc, and no accurate estimate of their numbers.
The primary criterion defining a direct target gene for any transcription factor is binding of the factor to regulatory DNA elements in cellular chromatin. This can be studied in live cells by chromatin immunoprecipitation (ChIP), which was used to demonstrate binding of Myc to several loci (Boyd and Farnham 1997; Bouchard et al. 2001; Frank et al. 2001; Xu et al. 2001; Zeller et al. 2001). Here, we have devised a large-scale, ChIP-based screen to identify E-box-containing genes that are bound by Myc in the human genome independently of their expression. The accompanying article (Orian et al. 2003) presents a large-scale analysis of Drosophila Myc-target loci. Both studies demonstrate the association of Myc with a very large population of genomic sites.
Results
Screening for Myc-binding sites in the human genome
We aimed to identify genes directly bound by Myc through the consensus E-box element CACGTG. Our strategy was based on the preselection of candidate sites with bioinformatic tools (Wang et al. 2001), followed by the experimental analysis of a large number of individual sites with a quantitative ChIP assay (Frank et al. 2001). From the scrutiny of known target genes, we knew that Myc-binding sites could be located in the promoter, first exon, or first intron. We therefore searched human genomic DNA sequences for CACGTG motifs located within a distance of 2 kb on either side of transcription start sites (henceforth labeled “promoter E-boxes”). Among a population of 6541 annotated GenBank entries, 1630 loci scored positive with one or more E-boxes within the predetermined boundaries, identifying a total of 2224 E-boxes. 654 of these sites were screened by ChIP, alongside 69 additional E-boxes that were outside the ±2-kb boundary. In total, we assayed Myc binding by ChIP at 723 genomic sites included into 533 different loci, listed in Supplementary Table A.
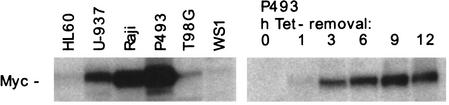
Our initial selection of cell lines was based on two criteria: (1) using cells growing in suspension to scale up the ChIP assay; and (2) comparing the distribution of Myc among cells expressing different Myc levels. We used the cell lines U-937 (monoblastic leukemia) and HL60 (myeloid leukemia), as well as P493-6 (or P493), a B-cell line in which expression of a c-myc transgene was repressed by tetracycline (Tet) and was reinduced following Tet removal (Fig. 1; Schuhmacher et al. 2001). Myc protein levels in these cell lines covered ∼2 orders of magnitude (Fig. 1). Our batch of HL60 cells, unlike others (data not shown), expressed low levels of Myc, comparable to those found in primary human fibroblasts after serum stimulation (WS1). Myc levels were the highest in P493 cells, slightly above those seen in Burkitt's lymphoma cells bearing a translocation of c-myc (e.g., Raji). U-937 cells expressed intermediate, although supraphysiological levels of Myc.
Figure 1.
Myc protein expression levels in the cell lines used in this study. Fifty micrograms of whole cell lysates were analyzed by immunoblotting.
ChIP analysis was performed either in proliferating cells (U-937, HL60) or 8 h following Myc induction (P493). Cells were cross-linked with formaldehyde, chromatin was fragmented by sonication, and protein–DNA complexes were immunoprecipitated with antibodies directed against Myc. The recovered DNA was analyzed by real-time PCR with primer pairs spanning individual E-boxes (or occasionally multiple adjacent E-boxes). Each sequence was amplified in parallel reactions with either of three template DNAs purified from (1) Myc IPs, (2) control precipitation, and (3) input chromatin. The amount of DNA recovered in each IP was expressed as a percentage of the input. This assay is highly specific for Myc, because Myc IPs from myc−/− cells yielded no enrichment over the control (Frank et al. 2001). The data are given in Supplementary Table A.
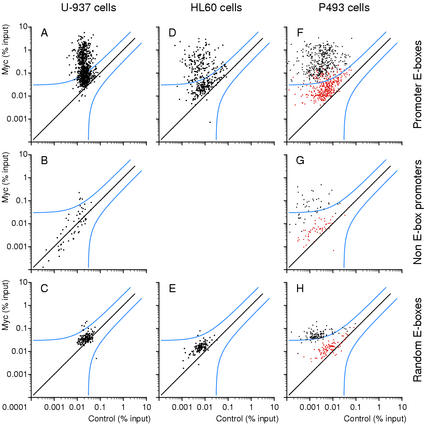
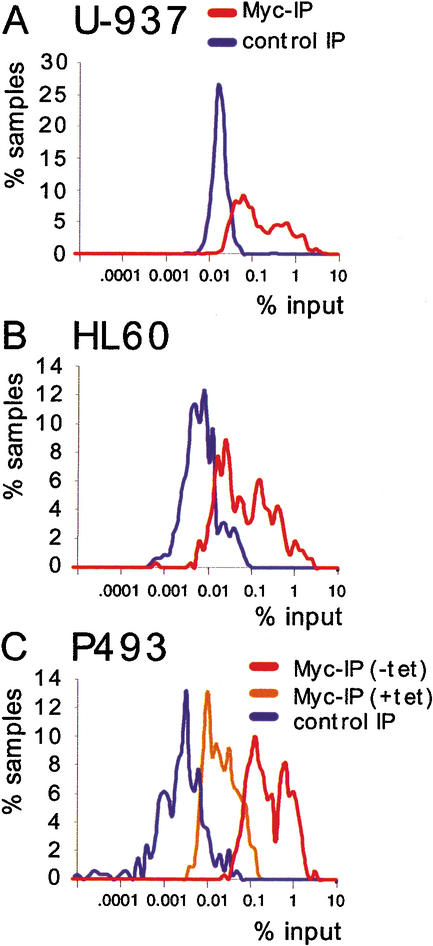
Figure 2A shows a scatter plot of the results from U-937 cells. Each dot represents an amplified genomic sequence. The amounts of DNA recovered in Myc and control precipitates are displayed on the Y- and X-axes, respectively, on a logarithmic scale. On the basis of the statistical analysis of our data (see Materials and Methods), a site had a >90% chance to be Myc-associated if it was at least the triple of its control value, plus an absolute threshold of 0.03 (in percentage of input). This threshold of confidence is shown in the graph (blue line): 482 out of 805 data points including duplicates, and thus 418 out of 720 tested E-boxes (58%), were above this line in U-937 cells. It was evident however, that the entire population of candidate sites was shifted above the diagonal in Myc IPs relative to control precipitates. As a further control in the same ChIPs, we analyzed genomic promoters that do not contain E-boxes (“non-E-box promoters”; Supplementary Table B): of these, only 4 out of 59 (6.7%) fell above our threshold of confidence for Myc-association (Fig. 2B). Most importantly, the bulk population of non-E-box promoters did not show any significant shift toward higher Myc values, demonstrating the specificity of the shift seen with promoter-associated E-boxes (Fig. 2A). Thus, although 58% of the promoter-associated E-boxes were associated with Myc in a statistically meaningful manner, it appears that all of these sequences showed enriched binding relative to non-E-box promoters. Upon closer inspection, it became evident that promoter E-boxes distributed in two clusters with different Myc-binding efficiencies, the lowest one being intersected by our threshold of confidence (Fig. 2A). These clusters are visualized better in the distribution curves shown in Figure 3A. In HL60 cells, with very low Myc levels, the data points for promoter-associated E-boxes also formed two clusters (Figs. 2D, 3B). As before, the whole population was shifted above the diagonal, with 167 of 357 data points above the statistical threshold (46.8%, i.e., 45.2% of different E-boxes).
Figure 2.
Myc associates with specific genomic sites. ChIP analysis of Myc binding in U-937 (A–C), HL60 (D,E), and P493 (F–H) cells. The sequences amplified were promoter-associated E-boxes (A,D,F), promoters that do not contain E-boxes (B,G), and random E-boxes (C,E,H) on Chromosome 21. The accession numbers, primers, and data are given in Supplementary Tables A–C. Each data point represents recovery of a given DNA site in the Myc IP (Y-axis) and control precipitation (X-axis). (% input) DNA recovery for each site was quantified as the percentage of input chromatin (Frank et al. 2001). P493 cells were analyzed in the presence of tetracycline (red dots) or 8 h after its removal (black dots).
Figure 3.
Myc-binding sites cluster in two groups of distinct affinities. Distribution curves show the percentage of data points for promoter E-boxes (% samples) that were within a narrow range around a given value (% input). ChIP was from U-937 (A), HL60 (B), and P493 (C) cells.
The amount of a given DNA site recovered by ChIP does not directly measure its affinity for Myc, but rather the fraction of cells in the population in which this site is bound at the time of analysis. Nonetheless, differences in recovery among various sites can be taken as an indication of their relative affinities for Myc in vivo, which account for chromatin accessibility and/or modifications. Thus, our data allow us to formally identify two populations of promoter-associated E-boxes, which we operationally label as “low-affinity” and “high-affinity” sites (Supplementary Table A).
As an additional control, we analyzed a series of random (i.e., not promoter-associated) E-boxes from Chromosome 21 (Fig. 2C; Supplementary Table C). In U937 cells, this population of E-boxes showed a marginal shift above the diagonal. Only 6 out of 137 (4.4%) fell above the statistical threshold, and remained bound at low efficiency. Similar data were obtained in HL60 cells (2/113; Fig. 2E). Thus, by selecting candidate E-boxes in the 5′ region of genes, we dramatically enriched for bona fide Myc-binding sites.
Acute overexpression of Myc enhances its association with low-affinity E-boxes and induces binding to nontarget chromatin
In P493 cells, which express Myc at very high levels (Fig. 1), 99% of promoter-associated E-boxes fell above our threshold of confidence (Fig. 2F, black dots). Although somewhat compressed by enhanced recovery of low-affinity sites, the two clusters of different affinities could still be distinguished (Fig. 3C). In cells kept in the presence of Tet, DNA recovery with Myc was reduced by ∼1 order of magnitude, confirming the specificity of our assay (Figs. 2F, red dots, 3C). Myc overexpression also caused its association with other populations of sites, because 32 of 51 random E-boxes (62.7%; Fig. 2G) and 104 of 118 non-E-box promoters (88%; Fig. 2H) were recovered above the threshold (whereas none was in the presence of Tet, red dots). In summary, the high levels of Myc expression in P493 cells led to (1) enhanced association with virtually all low-affinity E-boxes in promoters and (2) widespread association with sites that were not targeted in other cells. However, these sites were still bound at low efficiency relative to promoter E-boxes (Fig. 2F–H).
Myc-binding sites are conserved among different cell lines and primary cells
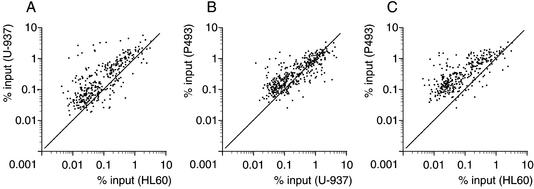
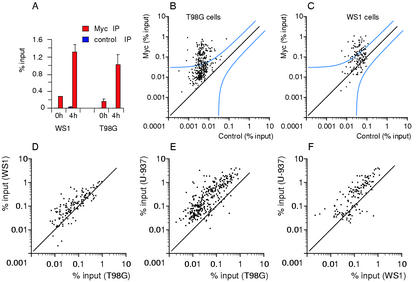
To address whether Myc bound overlapping populations of target sites in the three cell lines, the ChIP data for Myc IPs were plotted pairwise (Fig. 4A–C). In all combinations, most of the high-affinity sites clustered together, as did the low-affinity sites. Thus, the relative Myc-binding efficiencies of promoter E-boxes were strikingly conserved among different cells.
Figure 4.
Pairwise comparisons of Myc-binding data among the indicated cells lines. Each data point represents recovery of a given DNA site in Myc immunoprecipitations (% input) from each of the indicated cells lines.
We next studied the population of genes bound by Myc following mitogenic stimulation of human glioblastoma cells (T98G) and primary human fibroblasts (WS1), which express Myc at low, physiological levels (Fig. 1) and in a serum-inducible manner (data not shown). Myc binding assayed by ChIP at NUC intron 1 was also serum-inducible (Fig. 5A). As assayed by large-scale ChIP (Fig. 5B,C), 110 of 273 tested sites (40.3%) in T98G and 51 out of 125 (40.8%) in WS1 cells fell above the statistical threshold. Pairwise comparisons showed that these populations were conserved among T98G, WS1, and U-937 cells (Fig. 5D–F). Furthermore, although the two affinity clusters were less readily visible in T98G and WS1 cells, the binding data were strongly consistent with the clustering in the other cell lines (Supplementary Table A). Thus, Myc binds a similar population of high-affinity targets in all the tumor and primary cells (WS1) tested here.
Figure 5.
Genomic Myc-target genes are largely shared between cancer and primary cells. (A) ChIP analysis of Myc binding to NUC intron 1 before and after serum stimulation. (B,C) ChIP analysis of Myc binding to promoter-associated E-boxes in T98G (B) and WS1 (C) cells 4 h following serum stimulation. The plots are as defined in Figure 2. The accession numbers, primers, and data are given in Supplementary Table A. (D–F) Pairwise comparisons of Myc-binding data among the indicated cell lines, as in Figure 4.
In all pairwise combinations, there were a minority of “outliers,” that is, sites that were bound efficiently, or excluded, only in a given cell line (Figs. 4A–C, 5D–F; Table 1; Supplementary Table A). These differences might be due to tissue-specific accessibility of chromatin or, in tumor cell lines, exclusion of Myc binding through de novo methylation of selected CpG islands (see below and Discussion).
Table 1.
Myc-target genes belong to diverse functional categories
| Adhesion/matrix/tissue remodeling β integrin (ITGB1)f Carcinoembryonic antigen (CEACAM5) Collagen type IV (COL4A1/COL4A2)ce Integrin α 6 (ITGA6)de Intercellular adhesion molecule 1 (ICAM1) MUC5Bd Mucin 1 (MUC1) Nidogen (NID)bc Plakophilin (PKP1)c Plasminogen activator inhibitor-1 (SERPINE1)f Tartrate-resistant acid phosphatase type 5 (ACP)cf Ligand/receptor Bone morphogenetic protein 4 (BMP4) CD30 CD63/Me491 CD79B/lg β/B29c Chemokine HCC-1 (CCL14)c Complement receptor 2 (CR2, CD21)a EMMPRIN (BSG) FGFR4 G-protein-coupled receptor (GPR4) Galanin (GAL)a Gastrin receptor (CCKBR) Hepatocyte growth factor receptor (MET) Hepatocyte growth factor-like protein homolog (MST1) IFRD2 Insulin receptor (INSR) Interleukin-2 (IL2) Interleukin-11 receptor α chain (IL11RA) Interleukin-13 (IL13)c Lymphotoxin-β (LTB)c Melanoma growth stimulatory activity β (CXCL2)b MHC class II HLA-SB-β-1c Muscle nicotinic acetylcholine receptor (CHRNB1) Netrin-2 like protein (NTN2L) NOTCH4 P protein (OCA2) Prepro-8-arginine-vasopressin-neurophysin II (AVP) Prostaglandin E2 receptor EP2 subtype (PTGER2) Transferrin receptor (TFRC)f Transforming growth factor β-1(TGFB1)f Transforming growth factor β-2 (TGFB2)b Transforming growth factor β-3 (TGFB3)c Type I σ receptor (σ R1)f Structural β-tropomyosin (TPM2) Cytoskeletal γ-actin (ACTG1) Erythrocyte membrane protein 4.2 (EPB42)c Lamin A/C (LMNA) Lamin B2 (LAMB2) Moesin (MSN)f Neurofilament subunit M (NEF3) Oncoprotein 18, stathmin, Pr22 (STMN1) Channels/components AE2 anion exchanger (SLC4A2) CLCN6 CLNS1A Na,K-ATPase subunit α 2 (ATP1A2)c TIRC7 protein (TCIRG1) Chaperone/protein folding Hsc70 (HSPA8)f Hsp10 (HSPE1)f Hsp60 (HSPD1)f Hsp90 α (HSPCAL3)f Prohibitin (PHB)f Translation/ribosomal protein Initiation factor 4AI (EIF4A1) Initiation factor 4E (EIF4E)f Initiation factor 5AII (EIF5A2)f Poly(A)-binding protein (PABP) RPL13A RPL19f RPL22f RPL27Af RPS19f RPS6 Translational inhibitor p14.5 Vesicle protein/trafficking ADP-ribosylation factor 1 (ARF1) ADP-ribosylation factor 4 (ARF4) Pex3 Synaptobrevin 2 (VAMP2) Synaptogyrin 2 (SYNGR2) Metabolism Amino acid AMP deaminase isoform L (AMPD2) Cystathionine β-synthase (CBS) Dihydropteridine reductase (QDPR) FAH Folylpolyglutamate synthetase (FPGS) Methylenetetrahydrofolate reductase (MTHFR) Amine S-adenosylmethionine decarboxylase (AMD1) Spermidine synthase (SRM)f Carbohydrate Aldehyde dehydrogenase 2 (ALDH2) Aldehyde reductase (AKR1A1) α(1,2)Fucosyltransferase (FUT1)c α-Galactosidase A (GLA) Galactose-1-phosphate uridyl transferase (GALT) Glucose transporter (SLC2A4) Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Energy metabolism Uncoupling protein 1 (UCP1)c Uncoupling protein 3 (UCP3) Heme α-Globin (HBA1) α-Globin (HBA2) β-Globin (HBB) Ferrochelatase (FECH)c Heme oxygenase-1 (HMOX1)cd Hydroxymethylbilane synthase (HMBS) Uroporphyrinogen decarboxylase (UROD) Iron Frataxin (FRDA)f Melanotransferrin (MF12) |
| Lipid Acyl-CoA oxidase (ACOX1) Fatty acid synthase (FASN)f Medium chain acyl CoA dehydrogenase (ACADM) Prosaposin (PSAP)c Nucleotide AIRC (PAICS)f AMP deaminase (AMPD3) Cadf Deoxycytidine kinase (DCK) GPAT (PPAT)f Platelet-derived endothelial cell growth factor (ECGF1)bc Steroid 11 β-hydroxysteroid dehydrogenase 2 (HSD11B2) Steroid 5-α-reductase (SRD5A1) Other Aconitase (ACO2) α-Mannosidase IIX, MIIX (MAN2A2) Epoxide hydrolase (EPHX1)e Glutathione S-transferase (GSTP1)f Inosine monophosphatase 2 (IMPA2) Microsomal glutathione transferase (MGST1)f Nitrilase homolog 1 (NIT1) Pyruvate dehydrogenase α (PDHA1) Reduced folate carrier (SLC19A1) Miscellaneous Angiotensin-I converting enzyme (ACE)c bA18I14.4a CFDP1/BCNT dbi/acbp (diazepam-binding inhibitor/acyl-CoA binding protein)f FCI-12 Growth arrest specific 11 (GAS11) Human homolog of rat insulinoma gene (rig) ISG20 JM26 protein MAGE-3 antigen (MAGEA3) Myeloperoxidase (MPO) Outer membrane receptor Tom20 (TOM20) Prostatic acid phosphatase (ACPP)c QM Ring zinc-finger protein ZNF127-Xp (MKRN4)c SLC9A3R2 Proteolysis Amyloid precursor protein (APP)f Arylsulfatase B (ARSB) Cathepsin F (CTSF) Cystatin B (CSTB)f Cystatin D (CST5)d Elafin (PI3)e Mif1/KIAA0025 (HERPUD1)f Nexin-1 (PN1)d Proteasome subunit HC5 (PSMB1) Ubiquitin-binding protein p62 Ubiquitin–52-amino-acid fusion protein (UbA52) Von Hippel-Lindau tumor suppressor (VHL)a Wegener's granulomatosis autoanitgen proteinase 3 (PRTN3) Signal transduction Acetylcholinesterase (ACHE)a Adenomatous polyposis coli (APC) |
| AHSGc AKAP1 blkc Cyclophilin 40 (PPID)f Cytosolic phospholipase A2 (PLA2G4A) Glycogen synthase kinase 3 β (GSK3B) Helix–loop–helix basic phosphoprotein (G0S8) hper 1 (PER1) mdm2 NRASf Protein phosphatase-1 regulatory subunit 7 (PPP1R7) PTEN RCC1 RhoG (ARHG) src-Like adaptor protein (SLAP)c Thioredoxin (TXN)f TSC2 Anti/proapoptosis baxf BBC3/PUMA/JFY1 BCL2 BCL2L12 Caspase 8 (CASP8) Caspase 9 (CASP9)c Familial Alzheimer's disease (STM2) MCL1 and MCL1 delta S/TM NOD1 Nuclear regulatory factors BN51f Histone deacetylase 3 (HDAC3) Lamin B receptor (LBR) Menin (MEN1)ab Nonhistone chromosomal protein HMG-17 (HMGN2)f OZF (ZNF146) Poly(ADP-ribose) polymerase (PPOL) SIRT1 (Sir2-like protein) Nucleolus/RNA-binding protein DKC1 Histone stem–loop-binding protein (SLBP) hnRNPA1 (HNRPA1)f hnRNPA2/B1 (HNRPA2B1) hPOP1 JKTBP2, JKTBP1 (HNRPDL) Nucleolin (NCL)f Proliferating cell nucleolar protein P120 (NOL1) Putative RNA-binding protein 3 (RBM 3) Surf-6 (SURF6) Tis11d U50′ snoRNA, U50 snoRNA: nonprotein coding (U50HG) Transcription factors Achaete-scute homolog 2 (ASCL2) Activator of transcription 6 (STAT6) ATF4 ATFa B-myb (MYBL2)b c-jun proto oncogene (JUN) CCAAT/enhancer binding protein α (CEBPA)f Chronic lymphatic leukemia protein (BCL3) E2F1f ear1 (NR1D1) EGR3 |
| elk1 (ELK1) Erythroid Kruppel-like factor (ELKF) ets2 (ETS2)f fra-1 (FOSL1)f HB9 homeobox (HLXB9) Helix–loop–helix protein (TCF12) HOXD13d HRY (Hairy, HHL, HES-1) Hypoxia-inducible factor 1 α (HIF1A) Id2df Id3 Insulinoma-associated (INSM1)b Interferon regulatory factor 2 (IRF2) Interferon regulatory factor 3 (IRF3) junB (JUNB) L-myc (MYCL1) MBP 1 (ENO1)f nm23-H1 (NME1)f Retinoic acid receptor α (RARA)f Retinoic X receptor B (RARB) |
| Single-minded 2 (SIM2)d Smad7 (MADH7) TRIDENT/HFM11 (FOXM1) Wilms tumor (WT1) DNA maintenance/repair Apurinic/apyrimidinic endonuclease (APEX)f β-Polymerase (POLB) brca2 DNA polymerase δ small subunit (POLD2)f DNA-PKcs (PRKDC) H4 histone (H4/b) H4 histone (H4/e) Histone (H2AZ)f MCM7 NBS1 NTHL1 Prothymosin α (PTMA)f Telomerase reverse transcriptase (TERT)f Topoisomerase (TOP1)f |
The 257 genes identified within the high-affinity group of Myc-targets (see Results) are listed and assigned to a functional category. The genes qualify as high-affinity targets in all cell lines tested, unless selectively indicated: aU-937, bHL60, cP493, dT98G, eWS1. Where applicable, the HUGO nomenclature is used (http://www.gene.ucl.ac.uk/nomenclature). fPreviously identified Myc-regulated genes (http://www.myc-cancer-gene.org/index.asp).
Sequence analysis of Myc-target sites reveals a preponderance of CpG islands
The above data confirm that recognition of the E-box motif by Myc/Max is a primary determinant of target gene selectivity in vivo (Amati et al. 1992; Kretzner et al. 1992; Boyd and Farnham 1997; Frank et al. 2001; Zeller et al. 2001; Orian et al. 2003). Most importantly, they also reveal that promoter-associated E-boxes have different and characteristic affinities for Myc. Therefore, there must be parameters besides the E-box that modulate DNA binding. These might include primary DNA sequence, DNA methylation, chromatin structure and modifications, or other DNA-binding proteins. To identify possible distinctive (and hence predictive) features of Myc-target genes, we compared two groups of 199 DNA sequences, one consisting of the highest- and the other of the lowest-affinity E-boxes in U-937 cells.
From in vitro binding-site selection and in vivo analysis of transfected DNA, it was proposed that T preceding and/or A following the core sequence (TcacgtgA) negatively restricted binding of Myc/Max, whereas CcacgtgG was a preferred site (Solomon et al. 1993; Boyd and Farnham 1997; O'Hagan et al. 2000). However, our analysis showed that flanking bases have no strong predictive value for Myc binding to cellular chromatin (data not shown; but Supplementary Table A gives the three bases on either side of each E-box). For each of the 16 NcacgtgN combinations—including TcacgtgA—we found sites that were efficiently bound by Myc and others that were not, and no single combination could a priori preclude or predict Myc/Max binding. This was also true for bases farther away from E-boxes, which, apart for a preponderance of G and C (see below), did not allow us to derive a consensus motif of general predictive value. We conclude that flanking nucleotides may influence, but are not a major determinant of Myc/Max selectivity in cellular chromatin.
We then considered the possibility that accessory DNA-binding protein(s) may be involved in enhancing or precluding binding of Myc to E-boxes. If such protein existed, its cognate binding sequence should be enriched selectively in either high- or low-affinity sites. We could identify no sequence that fulfilled this criterion, suggesting that Myc/Max binding to DNA does not systematically depend on another factor. This notwithstanding, Myc-target genes possess a variety of putative transcription factor-binding sites, which may influence Myc/Max binding and/or transcriptional activity at selective loci.
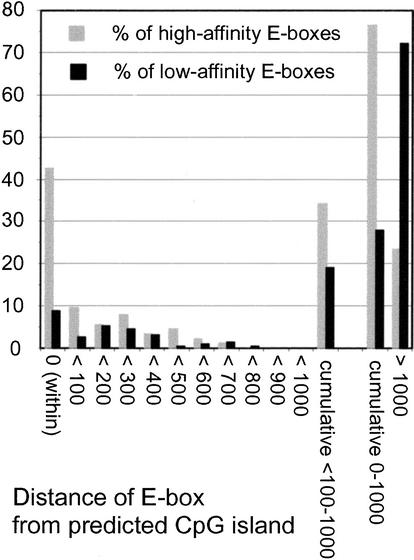
Cytosines in CpG dinucleotides are methylated in mammalian genomes, except when they occur within CpG islands (Bird 1987). Methylation of the central CpG in CACGTG prevents Myc/Max binding in vitro (Prendergast et al. 1991), which led to the suggestion that Myc-target sites should occur preferentially within CpG islands (Greasley et al. 2000). To address this hypothesis, each E-box in our collection was given a score that indicated its distance from a CpG island (Fig. 6; see Materials and Methods). Of the low-affinity E-boxes, 8.8% were within (score 0) and 19.1% were near E-boxes (<100–1000), yielding a total of 27.9% “CpG-island-associated” E-boxes (0–1000). This was expected, because all our sequences were selected a priori to be promoter-proximal, as CpG islands are (Bird 1987; Gardiner-Garden and Frommer 1987). Yet these proportions were further increased in the high-affinity population, with 42.5% of E-boxes within a CpG island and 34.1% nearby, for a total of 76.6% (Fig. 6). The remaining E-boxes were at much greater distances (>1000), generally meaning that no CpG island was found in the locus and/or available sequence. In summary, Myc binding markedly enriched for CpG islands independently from the initial conditions of our search (i.e., promoter association). As a corollary, any E-box that occurs within a CpG island has a high probability to be a high-affinity Myc-target site in vivo. This is a stronger predictor than simple GC content, which averaged 60.1% ± 8.2% and 51.7% ± 8.5% for the 500 bp encompassing E-boxes in the high- and low-affinity groups, respectively, both ranking significantly above the average for genomic DNA (41%). These observations support the notion that CpG methylation antagonizes Myc/Max binding in vivo (see Discussion).
Figure 6.
Myc-binding sites are strongly enriched for CpG islands. Two groups of 199 Myc-target and nontarget E-boxes each (from data in U-937 cells) were analyzed informatically for the presence of CpG islands. Each E-box was assigned one of the scores indicated below the graph according to its distance from a CpG island (see Materials and Methods). The plot indicates the percentage of high-affinity (gray bars) and low-affinity (black bars) E-boxes that fit each score.
Effects of Myc on histone acetylation and target-gene expression
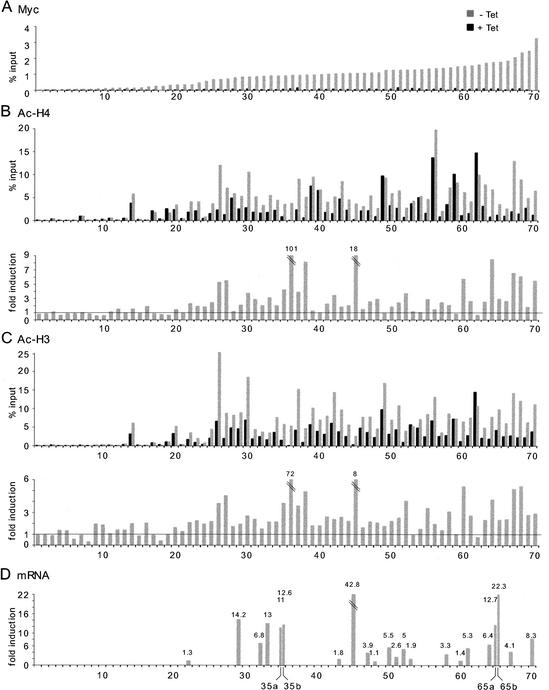
Myc has been shown to regulate acetylation of histones H3 and H4 at several chromosomal loci (Bouchard et al. 2001; Frank et al. 2001; Nikiforov et al. 2002), most likely through the direct recruitment of various acetyltransferases (Oster et al. 2002). To address the generality of this phenomenon, we selected a series of sites with diverse levels of Myc binding in P493 cells (Fig. 7A; genes listed in Supplementary Table D) and used ChIP to analyze their state of acetylation on histones H4 and H3 (Fig. 7B,C). For accurate analysis, we show both the net values (% input) and fold induction of acetylation after Myc binding. The data revealed an unexpected feature: prior to Myc induction, high-affinity sites showed higher basal acetylation of both H4 and H3 (Fig. 7B,C, black bars), relative to the lowest-affinity sites. This finding is discussed below. Myc binding (Fig. 7A) further increased acetylation of both histones at a majority of high-affinity sites (Fig. 7B,C, gray bars), but not at low-affinity sites (<23). Even though the fold increases induced by Myc at some target sites were modest, the net increases in acetylation were substantial (e.g., #34, #41, #65, #66). Our data also confirmed previous observations for several loci in other cell types (#65: HSP10/60; #35: GPAT/AIRC; #70: NUC; #45: TERT; Frank et al. 2001; Nikiforov et al. 2002). A subset of sites at which Myc binding did not enhance acetylation showed elevated net values prior to Myc induction (e.g., #39, #40, #49 for H4; #53, #54, #59, #62 for H3/H4; note that in other cells Myc induced acetylation at #59, or cyclin D2: Bouchard et al. 2001). This is reminiscent of the situation for H3 in Rat1 fibroblasts, which was constitutively hyperacetylated, and seemed therefore not to be regulated by Myc (Frank et al. 2001). It is important to note that the antibodies used here do not distinguish between different acetyl-lysines in the N-terminal tails of H3 and H4. Thus, we do not yet know which residues in the H3 and H4 tails are targeted by Myc, and distinct acetylation events might mask Myc-specific changes. This notwithstanding, we can conclude that Myc modulates acetylation of histones H3 and H4 at a majority of its high-affinity target loci.
Figure 7.
Analysis of Myc binding, histone acetylation, and gene expression in P493 cells. The list of 70 loci and the corresponding ChIP data are given in Supplementary Tables D and A, respectively. (A) Myc binding to selected loci before (black bars) and after (gray bars) Tet removal. Aligned below this graph is shown ChIP data on acetylation of histone H4 (B) and histone H3 (C). (Upper graphs) Net values (% input). (Lower graphs) Fold induction in acetylation following Myc activation. (D) mRNA induction for selected genes following Myc activation. The maximal induction level over a time course of 12 h is given for each gene. Genes that were equally induced by seeding in fresh Tet-containing medium were removed from this analysis. Several control genes showed constant mRNA levels in all the samples (data not shown). The primers for mRNA analysis are listed in Supplementary Table D.
To address whether Myc regulated expression of the target genes identified by ChIP, the levels of various mRNAs were monitored over a time course between 0 and 12 h following Tet removal. Figure 7D shows the maximal induction point for each gene (those without bars were either not tested, or gave no conclusive results). The effect of Myc on target-gene expression was variable, ranging from none (#48, #60) to large increases in relative mRNA levels (e.g., #45, hTERT). No gene among those analyzed here was repressed upon Myc binding. It is important to note that even genes that are efficiently bound by Myc and become hyperacetylated may be induced weakly, if at all (e.g., #60). Thus, as seen previously (Frank et al. 2001), Myc binding and acetylation do not systematically correlate with gene activation. Furthermore, genes that are activated by Myc in a given cell type and/or condition may not respond in others. These observations emphasize the need to avoid generalizations based on the unresponsiveness of a given mRNA in a particular cell type or experimental setting.
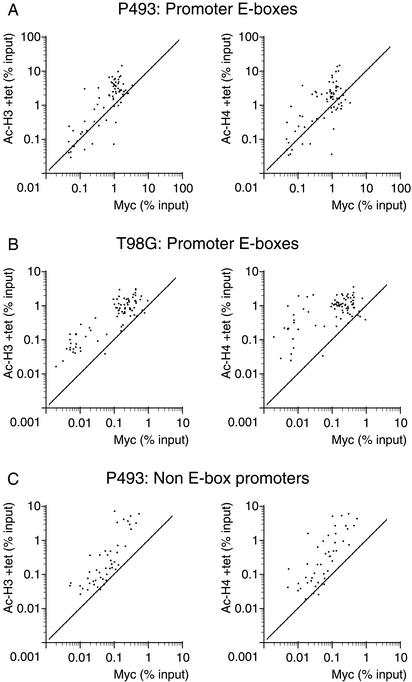
Myc may associate preferentially with preacetylated chromatin
As noted above (Fig. 7), we observed a higher level of pre-existing H3 and H4 acetylation at Myc-target E-boxes relative to nontarget sites, prior to induction of Myc. This is visualized better in Figure 8A, in which we compare H3 and H4 acetylation (in the presence of Tet) with Myc binding (after Tet removal). Although this might be caused by a small leakiness in Myc expression and/or defects in the dynamics of acetylation/deacetylation in this cell line, similar observations were made in quiescent T98G cells prior to Myc induction by serum (Fig. 8B). Thus, a basal level of histone acetylation might be a determinant of Myc binding to E-boxes in chromatin. This hypothesis is further supported by the enrichment of Myc-binding sites in CpG islands (see above; Fig. 6), which are nonmethylated and expected to be associated with an open chromatin state (Richards and Elgin 2002; see Discussion).
Figure 8.
A preacetylated state of chromatin correlates with Myc binding. Each data point represents recovery of a given DNA site in the acetyl-H3 or acetyl-H4 IPs as indicated (Y-axes) and Myc IPs (X-axes). The cells for ChIPs and sequences amplified are indicated on top. In P943 and T98G cells, Ac-H3 and Ac-H4 ChIPs were performed before Myc induction by Tet withdrawal and serum induction, respectively, whereas Myc ChIPs were performed after those treatments.
As noted above, overexpressed Myc associated with non-E-box promoters in P493 cells. Here again, the most highly preacetylated sites were also the better bound by Myc (Fig. 8C). Similar data were obtained for nonpromoter E-boxes (data not shown). Thus, although elevated histone acetylation per se is insufficient for maximal Myc binding (Fig. 8A,C), it positively correlates with binding to both E-box and non-E-box promoters. It is worth reminding that the latter were bound much less efficiently and only upon acute overexpression of Myc. Altogether, these data are suggestive of a weak, nonspecific association of Myc with “open” promoter chromatin, which might precede and/or facilitate its specific binding to E-boxes.
Myc-associated genes belong to diverse functional categories
Among those screened in this work, 272 promoter-associated E-boxes belong to the “high-affinity” Myc-target group. These E-boxes identify 257 genes, listed in Table 1 by functional categories. Only 53 of these genes (∼20%) were previously identified as Myc-regulated genes (indicated with f) and have been compiled elsewhere (http://www.myc-cancer-gene.org/index.asp). It is important to point out that our screen did not saturate potential Myc-binding sites in the genome (see Discussion). Thus, our list is still partial and may not reflect all the biological processes targeted by Myc. In addition, Myc may regulate an even larger group of “low-affinity” target genes identified in our work, which are not included in Table 1 (but see Supplementary Table A). Most of the functional categories listed in Table 1 were expected, including apoptosis, cell cycle, nucleolar function, ribosomal proteins and translation factors, growth factors, receptors, signal transduction, and so on. In each category, new target genes were identified that may shed new light on how Myc regulates these biological processes. We identified a striking number of genes encoding metabolic enzymes, confirming and extending the possible involvement of Myc in diverse pathways, most noticeably energy, redox, and DNA metabolism. We also identified several new targets involved in DNA replication and repair. Finally, Myc targeted many genes encoding other transcription factors and nuclear regulators. Thus, Myc is likely to participate in an extensive reprogramming of gene expression, both directly and through the action of other transcriptional regulators.
Discussion
We have performed a large-scale analysis of Myc binding to promoters that contain the E-box consensus element CACGTG in live human cells. As in the accompanying study of Drosophila Myc (Orian et al. 2003), our data reveal that the protein associates with a strikingly large number of genomic loci, suggesting significant diversity in the ensuing transcriptional response.
Our ChIP analysis showed that most E-boxes in promoter regions showed enhanced Myc binding relative to E-boxes in bulk genomic DNA or to promoters without E-boxes. Therefore, the presence of an E-box in a regulatory domain is already an indication that Myc is likely to bind this element. Yet, promoter E-boxes were bound neither in a uniform manner, nor with a continuous range of efficiencies. Instead, they were distributed in two distinct clusters of high- and low-affinity Myc-binding sites (see Results). These clusters were conserved between cells expressing very different levels of Myc, and thus represented an intrinsic property of cellular chromatin. Depending on Myc expression levels, a variable proportion of the sites in the low-affinity group were enriched above the threshold of statistical significance: in P493 cells that acutely overexpressed Myc, this included virtually all sites. Most high-affinity sites, on the other hand, were generally Myc-associated in all cell lines. We identified 257 genes in this category, including 53 that were regulated by Myc in previous studies (Table 1). This group represents 44% of the E-box promoters tested in our work. Because ∼25% of the total promoter entries in our database (1630 of 6541) contained one or more E-boxes within the preset boundary (−2 to +2 kb from the transcription start site), the conserved core of high-affinity Myc-target genes represents ∼11% of all cellular promoters.
This number of target genes is most likely an underestimate, because our screen was based solely on the “canonical” E-box element CACGTG, whereas another E-box (CACATG or CATGTG) and variant sites (e.g., CACGCG) can also be bound by Myc/Max (Grandori et al. 2000; Oster et al. 2002). The frequency and efficiency of binding to these alternative sites in vivo remain to be determined, but individual cases have been confirmed by ChIP (Morrish et al. 2003; P.C. Fernandez, unpubl.). This is not counting an unknown number of repressed loci to which Myc is recruited via interaction with other proteins like Miz-1 (Oster et al. 2002). In addition, Myc can bind E-boxes farther away than −2 kb or +2 kb, as shown by several examples in our data set (Supplementary Table A). The direct binding of Myc to such a large number of sites both in humans and Drosophila (Orian et al. 2003) was not anticipated and is not a general feature of eukaryotic transcription factors. The Gal4 protein, for example, was detected on only 10 target promoters (Ren et al. 2000), and Rap1 bound ∼5% of all yeast genes (Lieb et al. 2001).
An unresolved question is whether the oncogenic function of Myc relies on “pathological” target genes that are not regulated in normal cells. As noted above, Myc overexpression results in enhanced binding to low-affinity E-boxes. In U-937 cells, which expressed Myc ∼10-fold over physiological levels, the fraction of all promoters bound at a significant level was 14.5% (i.e., 58% of E-box-containing promoters), compared with 11.3% in HL60 cells (i.e., 45.2%). In P493, an inducible B-cell line overexpressing Myc by at least 2 orders of magnitude, 25% of all promoters were bound (i.e., virtually all E-box promoters). Similar observations were made in Burkitt's lymphoma cells (Raji; data not shown). In P493 cells, Myc was also detectable on sequences that were not enriched in other cells, including random (i.e., nonpromoter-associated) E-boxes and non-E-box promoters. Yet these sites remained bound at relatively lower efficiencies. It remains to be investigated to what extent deregulation of either low-affinity target genes or aberrant nontarget genes contributes to tumorigenesis. On the other hand, it is reasonable to assume that deregulated Myc expression (and not always great overexpression) provokes permanent binding to, and hence potential deregulation of, bona fide target genes in tumor cells.
The Myc-binding patterns that we have observed in our cell lines suggest that target sites in chromatin compete for limiting Myc protein levels. The peak expression of Myc in serum-stimulated human fibroblasts has been estimated at 3–6000 molecules per cell (mpc), whereas cycling cells expressed 1–3000 mpc (Waters et al. 1991). Thus, there may rarely, if ever, be enough Myc in normal cells to bind all potential targets, assuming that 11% of cellular genes means >4000 target loci (Hogenesch et al. 2002), many of which possess multiple E-boxes. This may explain why high-affinity sites showed a relative independence, but the extent of binding to low-affinity sites was more dependent on Myc levels. In this context, it is important to point out that ChIP analysis does not provide a cell-based but, rather, a population-based readout. Myc-target sites in our assays were recovered at efficiencies ranging from 0.03% (the minimal significant value) to ∼6% of input chromatin. Although DNA recovery in ChIP may not be fully quantitative, these values imply that Myc is bound to any given site in a minority of cells at a time and most likely in a transient manner. Thus, there can be more sites bound in a cell population than Myc molecules per cell.
Although Myc bound E-box-containing promoters, E-boxes in bulk genomic DNA were not targeted at a significant frequency. Yeast Rap1 and SBF showed a similar tendency (Iyer et al. 2001; Lieb et al. 2001), suggesting that eukaryotic promoters possess unique structural features. These might include primary DNA sequence, accessory DNA-binding proteins, chromatin structure, histone modifications, and, in mammalian cells, DNA methylation. Our data revealed no secondary sequence motifs systematically associated with high-affinity E-boxes and showed that, contrary to previous expectations (Solomon et al. 1993; Boyd and Farnham 1997; O'Hagan et al. 2000), nucleotides flanking the CACGTG core have little predictive value. On the other hand, high-affinity Myc-target sites showed a dramatic enrichment for CpG islands. These islands are short stretches of genomic DNA, generally proximal to promoter regions, distinguished by a high GC content, normal occurrence of CpG dinucleotides, and lack of cytosine methylation (Bird 1987; Gardiner-Garden and Frommer 1987). The linkage of high-affinity E-boxes to CpG islands was not simply due to the study of promoter regions, because promoter-associated E-boxes with the lowest affinity for Myc were less frequently found within or near an island (see Results). Thus, our data formally identify CpG islands as a major determinant of Myc binding to E-box elements. As a corollary, DNA methylation and Myc binding must be mutually antagonistic in vivo.
Two mechanisms may explain the effect of DNA methylation. First, methylation within the CACGTG motif prevented Myc/Max binding to naked DNA in vitro (Prendergast et al. 1991). Second, CpG methylation is generally associated with a repressed, heterochromatic state, characterized by hypoacetylation of histones H3 and H4, methylation of histone H3 on Lys 9, and recruitment of the HP1 protein (Richards and Elgin 2002). Of note, Myc- and HP1-binding sites were nonoverlapping in Drosophila (Orian et al. 2003), pointing to a possible conservation in the determinants of target selectivity. In our work, histones H3 and H4 at low-affinity E-boxes were generally hypoacetylated, whereas high-affinity sites showed significant basal acetylation prior to Myc induction. Similarly, the relatively weak and less frequent association of Myc with non-E-box promoters (detected only upon acute overexpression of Myc) preferentially occurred on preacetylated sequences. This might indicate the existence of a “scanning” mechanism, by which binding of Myc to open, preacetylated chromatin precedes and facilitates sequence-specific binding. Consistent with this idea, deacetylation of histone H4 interfered with binding of several transcription factors to chromatin in vitro (Vettese-Dadey et al. 1996). Additional features of open chromatin, such as nucleosome phasing or nonhistone proteins, are likely to modulate Myc binding: this may explain the existence of loci at which multiple E-boxes are targeted with different efficiencies (e.g., TERT, AKAP1, PUMA, CAD, CBS, STAT6, NOD1, HC8, NOTCH4, TGF-β1). Access to specific E-boxes might also be controlled by inducible or tissue-specific chromatin remodeling events, because some sites were either preferentially bound or excluded in a given cell line (Table 1; Supplementary Table A).
CpG methylation is a frequent mechanism of promoter silencing in tumor cells (Jones and Baylin 2002). Interestingly, among the Myc targets identified in our screen, we find tumor-suppressor genes that are silenced by methylation in human tumors (e.g., CASP8, VHL, PTEN, APC, BRCA2, GSTp, WT1; Teitz et al. 2000; Jones and Baylin 2002). Up-regulation of those genes by Myc may constitute a fail-safe mechanism against tumor progression, but may simultaneously set the selective pressure for their silencing. An interesting example is CASP8, whose silencing in human childhood neuroblastomas correlated with N-Myc overexpression (Teitz et al. 2000). Such a paradigm is further supported by experimental systems in which genetic blockade of Myc-induced apoptosis leads to a dramatic acceleration of tumorigenesis (e.g., Eischen et al. 2001; Pelengaris et al. 2002). We also speculate that Myc, perhaps as an abnormal consequence of overexpression, may play a transient role in initiating DNA methylation, as shown for the chimeric oncoprotein PML-RARα (Di Croce et al. 2002). However, given that the overall Myc-binding pattern was conserved between tumor and primary cells in our study, it seems unlikely that the former have suffered widespread methylation of Myc-target sites.
The abundance of Myc-binding sites in the genome supports the hypothesis that only a subset of target genes is induced in any given experimental system or condition. At a large subset—and possibly all—of its high-affinity target loci, Myc recruits enzymes that induce acetylation of histones H3 and H4 on lysine residues that yet have to be mapped (see Results; Bouchard et al. 2001; Frank et al. 2001; Nikiforov et al. 2002; S.R. Frank, unpubl.). This event appears to be required, but is insufficient for transcriptional activation (Frank et al. 2001). Among other effects, it may render chromatin permissive for binding and/or activation by other transcription factors. In addition, Myc may recruit other coactivators (Oster et al. 2002). In summary, the response of any target gene following Myc activation is likely to depend on a variety of other factors, and may change dramatically as a function of cell type and environment. Among the many thousands of loci bound by Myc, we must now identify those that are critically regulated in vivo during development and tumorigenesis.
A complete picture of transcription factor-binding sites has been achieved in the yeast genome by hybridizing DNA recovered by ChIP onto DNA microarrays (Ren et al. 2000; Iyer et al. 2001; Lieb et al. 2001; Simon et al. 2001). The same strategy has been used to identify target genes of human E2F (Ren et al. 2002; Weinmann et al. 2002) but, as in our present work, only a fraction of all possible binding sites was covered. The PCR-based approach used here possesses the drawback of a pre-established sequence bias (the E-box), but the advantage of enhanced sensitivity and resolution. This is illustrated by the distinction between low- and high-affinity Myc-binding sites made in this work. The advent of high-density robotized PCR and of more extensive genomic microarrays should improve both approaches, offering strong complementary tools to unravel chromatin modifications in mammalian cells at a large scale.
Materials and methods
Bioinformatic methods
The preselection of promoter-associated E-boxes (Supplementary Table A) and other sites (Supplementary Table B) for ChIP analysis was performed as previously described for p53 consensus sites (Wang et al. 2001). Briefly, the FindPatterns program (Wisconsin Sequence Analysis Package Version 10) was used to search a database of annotated human 5′ region sequences within a 2-kb boundary on either side of predicted transcription start sites. This sequence database was derived from GenBank release 120 (October 2000).
For further analysis of the Myc-target sites identified by ChIP and their comparison with nontarget sequences, two populations of 199 E-boxes each were generated, one positive and the other negative for Myc binding in U937 cells. To avoid ambiguities caused by adjacent Myc-binding sites, multiple E-boxes closer than 500 bp to each other were excluded. These subsets were derived from the data listed in Supplementary Table A. CpG islands were identified using criteria defined by Gardiner-Garden and Frommer (1987), using the GrailExp v1.0 software (Xu and Uberbacher 1997).
Statistical analysis of ChIP data
A Spearman's rank order correlation coefficient between random subsets of background values (∼0.64; data not shown) shows a strong consistency of the ordering of the background values, and thus the existence of a site-dependent background. An error range prediction model was built with matched pairs analysis on the logarithms of duplicate values for the same genomic site (with either same or different PCR primers, for a total of 891 pairs). This showed that the resolution of the system is affected by a small component of random noise (not dependent by the measured value itself) plus a scale error. These two noise components have been merged in an estimation of consistency of ordering between pairs with a given percentile of certainty in the form log A < (k log B + k/100). For k = 3, the above relationship is above the 90th percentile. Thus, if for two readings, A and B, log A < (3 log B + 3/100), we can assert with 90% confidence that the real value of B is greater than A.
Distribution analysis of the values of ChIP showed the existence of two separate clusters of sites in promoter E-boxes (Fig. 3). To identify the members of the two clusters, we used the K-means method in a five-dimensional space, where the input variables to the model were the logarithms of Myc/control ratios for the five cell lines used. Input data were previously normalized toward a common mean and variance to avoid biases arising from differences in size of the data sets, and missing values in the input were handled by a dynamic substitution model. Bootstrap analysis was done with randomly chosen initial attractors for ∼200 runs. In all the cases, the model converged within 78 cycles, always resulting in the same clusters. The sites belonging to each of the two clusters (high and low overall affinity) are marked in Supplementary Table A (available online at http://www.genesdev.org). In addition, because some sites may be selectively targeted or excluded by Myc in a given cell line, we also identified sites classified by the overall clustering as low affinity, but which for at least one cell line could be considered as high affinity as those having the variable corresponding to that cell line twice as distant from the low-affinity centroid of the K-means model than from the high-affinity one.
Cell culture
U-937 and HL60 cells were grown in RPMI supplemented with 10% fetal calf serum (FCS). For analysis of Myc by ChIP, 1.5 L of logarithmically growing cells was split to 2 to 3 × 105 cells/mL a day before harvesting. P493 cells were grown in RPMI supplemented with 10% FCS, NEAA (BioWhyttaker), and 2 mM L-glutamine (BioWhittaker). Repression and re-expression of Myc was as described (Schuhmacher et al. 2001). For ChIP, 2 L of logarithmically growing cells was split to 3 × 105 cells/mL, and 0.1 μg/mL tetracycline (Sigma) was added for 72 h. To reinduce expression of Myc, cells were washed three times in prewarmed RPMI containing 10% FCS before culturing for the indicated period of time. T98G and WS1 were purchased from ATCC and grown in D-MEM supplemented with 10% FCS. Cells were rendered quiescent by growth to confluent density followed by incubation for 3 d in serum-free medium. To induce cell cycle entry, cells were harvested by trypsinization and reseeded 1:4 onto plates containing D-MEM/10% FCS. For ChIP, 15 confluent 150-mm dishes, or the equivalent amount of cells following splitting, were used.
Chromatin immunoprecipitation (ChIP) assay
Our ChIP protocol and quantification have been described (Frank et al. 2001). The following modifications were made: 1.5–3.3 × 108 fixed cells were sonicated in 6 mL of SDS buffer. The lysate was diluted with 3 mL of Triton Dilution Buffer (100 mM Tris at pH 8.6, 100 mM NaCl, 5 mM EDTA, 5% Triton X-100). IPs were from 9 mL of lysate, with either 50 μg of polyclonal antibodies specific for c-Myc (N262, Santa Cruz Cat. #SC764) or 500 μL of blocked protein A beads (50% slurry protein A-Sepharose; Amersham). For large-scale experiments, DNA preparations from three independent ChIPs were pooled and diluted in 6 mL of water. For analysis of histone acetylation, we used polyclonal antibodies directed against acetylated H3 (Upstate Biotech, Cat. #06-599) and acetylated H4 (Upstate Biotech, Cat. # 06-866). PCR was performed with 4 μL of DNA and 800 nM primers diluted in a final volume of 20 μL in SYBR Green Reaction Mix (Perkin Elmer). Accumulation of fluorescent products was monitored by real-time PCR using a GeneAmp 5700 Sequence Detector (Perkin Elmer). Primers for ChIP analysis are indicated in Supplementary Tables A–C.
mRNA and protein analysis
For total RNA extraction, 1.5 × 107 P493 cells were used, before and after Myc induction. RNA extraction, cDNA synthesis, and real-time PCR were carried out as described (Frank et al. 2001). The PCR primers are listed in Supplementary Table D. For immunoblotting, whole cell lysates were made from exponentially growing cells using TX-100 lysis buffer (1% Triton X-100, 50 mM Tris at pH 8.1, 150 mM NaCl, 1 mM DTT, 20 mM NaF, 0.5 mM PMSF, and protein inhibitor cocktail; Roche). Then 50 μg of total cellular protein was separated in a 10% SDS-polyacrylamide gel and blotted onto PVDF membranes. The monoclonal antibody 9E10 was used for identification of Myc.
Acknowledgments
We thank Tiziana Parisi, Stefan Taubert, Dubravka Donjerkovic, Dave Parry, Martin Oft, Emma Lees, Fernando Bazan, and Kostis Alevizopoulos for insightful discussions at various stages of this project; and Carla Grandori and Dirk Eick for P493 cells. We are grateful to Robert Eisenman and Amir Orian for sharing their data prior to publication. Dirk Dobbelaere is thanked for helpful comments and his generous support to P.C.F. during the revision of this work. DNAX Research Institute is supported by Schering-Plough.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bruno.amati@ieo-research.it; FAX 39-02-57-489-851.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1067003.
References
- Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- Amati B, Alevizopoulos K, Vlach J. Myc and the cell cycle. Front Biosci. 1998;3:D250–D268. doi: 10.2741/a239. [DOI] [PubMed] [Google Scholar]
- Bates CM, Kharzai S, Erwin T, Rossant J, Parada LF. Role of N-myc in the developing mouse kidney. Dev Biol. 2000;222:317–325. doi: 10.1006/dbio.2000.9716. [DOI] [PubMed] [Google Scholar]
- Bird A. CpG islands as gene markers in the vertebrate nucleus. TIGS. 1987;3:342–347. [Google Scholar]
- Blackwood EM, Eisenman RN. Max: A helix–loop–helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Lüscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes & Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Farnham PJ. Myc versus USF: Discrimination at the cad gene is determined by core promoter elements. Mol Cell Biol. 1997;17:2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvold KA, Neiman P, Ruddell A. Angiogenesis is an early event in the generation of myc-induced lymphomas. Oncogene. 2000;19:2780–2785. doi: 10.1038/sj.onc.1203589. [DOI] [PubMed] [Google Scholar]
- de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- Douglas NC, Jacobs H, Bothwell AL, Hayday AC. Defining the specific physiological requirements for c-Myc in T cell development. Nat Immunol. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Roussel MF, Korsmeyer SJ, Cleveland JL. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol Cell Biol. 2001;21:7653–7662. doi: 10.1128/MCB.21.22.7653-7662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes & Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Garte S. The c-myc oncogene in tumor progression. Crit Rev Oncogenesis. 1993;4:435–449. [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Greasley PJ, Bonnard C, Amati B. Myc induces the nucleolin and BN51 genes: Possible implications in ribosome biogenesis. Nucleic Acids Res. 2000;28:446–453. doi: 10.1093/nar/28.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson M, Lüscher B. Proteins of the Myc network: Essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Ching KA, Batalov S, Su AI, Walker JR, Zhou Y, Kay SA, Schultz PG, Cooke MP. A comparison of the Celera and Ensembl predicted gene sets reveals little overlap in novel genes. Cell. 2002;106:413–415. doi: 10.1016/s0092-8674(01)00467-6. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Kretzner L, Blackwood EM, Eisenman RN. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992;359:426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proc Natl Acad Sci. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Giedt C, Hockenbery D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes & Dev. 2003;17:240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo CV, Gee M, Akhtar N, Yu D, Volpert O, Auerbach R, Thomas-Tikhonenko A. An in vivo function for the transforming Myc protein: Elicitation of the angiogenic phenotype. Cell Growth Differ. 2000;11:201–210. [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, Park J, Kotenko I, Matheos D, Johnsson A, McMahon SB, Cole MD. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol Cell Biol. 2002;22:5054–5063. doi: 10.1128/MCB.22.14.5054-5063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, B.C., Cheung, A.F., Simkevich, C.P., Tam, W., Ren, X., Mateyak, M.K., and Sedivy, J.M. 2003. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. (In press). [DOI] [PubMed]
- O'Hagan RC, Schreiber-Agus N, Chen K, David G, Engelman JA, Schwab R, Alland L, Thomson C, Ronning DR, Sacchettini JC, et al. Gene-target recognition among members of the myc superfamily and implications for oncogenesis. Nat Genet. 2000;24:113–119. doi: 10.1038/72761. [DOI] [PubMed] [Google Scholar]
- Orian, A., van Steensel, B., Delrow, J., Bussemaker, H.J., Li, L., Sawado, T., Williams, E., Loo, L.W.M., Cowley, S.M., Yost, C., et al. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: Induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in β cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of Max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes & Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O, Benvenisty N. A DNA microarray screen for genes involved in c-MYC and N-MYC oncogenesis in human tumors. Oncogene. 2001;20:4984–4994. doi: 10.1038/sj.onc.1204459. [DOI] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Solomon DLC, Amati B, Land H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 1993;21:5372–5376. doi: 10.1093/nar/21.23.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, Behm FG, Look AT, Lahti JM, Kidd VJ. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Vettese-Dadey M, Grant PA, Hebbes TR, Crane- Robinson C, Allis CD, Workman JL. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wu Q, Qiu P, Mirza A, McGuirk M, Kirschmeier P, Greene JR, Wang Y, Pickett CB, Liu S. Analyses of P53 target genes in the human genome by bioinformatic and microarray approaches. J Biol Chem. 2001;276:43604–43610. doi: 10.1074/jbc.M106570200. [DOI] [PubMed] [Google Scholar]
- Waters CM, Littlewood TD, Hancock DC, Moore JP, Evan GI. c-myc protein expression in untransformed fibroblasts. Oncogene. 1991;6:797–805. [PubMed] [Google Scholar]
- Watson JD, Oster SK, Shago M, Khosravi F, Penn LZ. Identifying genes regulated in a Myc-dependent manner. J Biol Chem. 2002;277:36921–36930. doi: 10.1074/jbc.M201493200. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes & Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkles JA. Serum- and polypeptide growth factor-inducible gene expression in mouse fibroblasts. Prog Nucleic Acid Res Mol Biol. 1998;58:41–78. doi: 10.1016/s0079-6603(08)60033-1. [DOI] [PubMed] [Google Scholar]
- Xu D, Popov N, Hou M, Wang Q, Bjorkholm M, Gruber A, Menkel AR, Henriksson M. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc Natl Acad Sci. 2001;98:3826–3831. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Uberbacher EC. Automated gene identification in large-scale genomic sequences. J Comput Biol. 1997;4:325–338. doi: 10.1089/cmb.1997.4.325. [DOI] [PubMed] [Google Scholar]
- Zeller KI, Haggerty TJ, Barrett JF, Guo Q, Wonsey DR, Dang CV. Characterization of nucleophosmin (B23) as a Myc target by scanning chromatin immunoprecipitation. J Biol Chem. 2001;276:48285–48291. doi: 10.1074/jbc.M108506200. [DOI] [PubMed] [Google Scholar]