Abstract
The Myc/Max/Mad transcription factor network is critically involved in cell behavior; however, there is relatively little information on its genomic binding sites. We have employed the DamID method to carry out global genomic mapping of the Drosophila Myc, Max, and Mad/Mnt proteins. Each protein was tethered to Escherichia coli DNA adenine-methyltransferase (Dam) permitting methylation proximal to in vivo binding sites in Kc cells. Microarray analyses of methylated DNA fragments reveals binding to multiple loci on all major Drosophila chromosomes. This approach also reveals dynamic interactions among network members as we find that increased levels of dMax influence the extent of dMyc, but not dMnt, binding. Computer analysis using the REDUCE algorithm demonstrates that binding regions correlate with the presence of E-boxes, CG repeats, and other sequence motifs. The surprisingly large number of directly bound loci (∼15% of coding regions) suggests that the network interacts widely with the genome. Furthermore, we employ microarray expression analysis to demonstrate that hundreds of DamID-binding loci correspond to genes whose expression is directly regulated by dMyc in larvae. These results suggest that a fundamental aspect of Max network function involves widespread binding and regulation of gene expression.
Keywords: myc, mad, Drosophila, target genes, transcription
Supplemental material is available at http://parma.fhcrc.org/AOryan.
The Myc, Mad, Max network comprises a group of widely expressed transcription factors, which function in cell proliferation and differentiation. This network includes members of the Myc and Mad families, the Mad-related protein Mnt, and Mga. All of these proteins possess basic helix–loop–helix zipper domains (bHLHZ), which mediate dimerization with the small bHLHZ protein Max, thereby forming heterodimers capable of recognizing the E-box sequence CACGTG (Grandori et al. 2000). Although Max homodimers lack transcriptional activity, recent evidence suggests that the transcriptional activities of Max heterodimers derive from their ability to recruit chromatin-modifying complexes to DNA (Amati et al. 2001; Eisenman 2001). For example, Mad family proteins directly bind the mSin3–histone deactylase corepressor complex permitting Mad–Max heterodimers to target histone deacetylation at E-box binding sites (Laherty et al. 1997; Bouchard et al. 2001). Myc proteins associate with the TRRAP coactivator, which in turn binds the histone acetyltransferase, GCN5, permitting Myc–Max dimers to direct acetylation of histones at E-box binding sites (McMahon et al. 2000; Bouchard et al. 2001; Frank et al. 2001). Furthermore, Myc is known to bind and inhibit the function of the transcription factor Miz-1, leading to E-box-independent repression of Miz-1 activated targets such as p15INK4b and other cyclin-dependent kinase inhibitors (Staller et al. 2001; Ziegelbauer et al. 2001; Seoane et al. 2002).
The varied activities of Max network proteins are manifested during proliferation and differentiation. Mad family proteins are generally induced during terminal differentiation and act to limit cell proliferation (Zhou and Hurlin 2001). Myc proteins are induced in response to a large number of growth factors and cytokines and may serve to integrate these external signals to sustain growth and proliferation. Targeted deletion of either N-myc or c-myc leads to early embryonic lethality in the mouse (Stanton et al. 1992; Davis et al. 1993) and recent studies demonstrate that myc gene function is essential for hematopoiesis and organogenesis (de Alboran et al. 2001; Douglas et al. 2001; Trumpp et al. 2001; Knoepfler et al. 2002). Deregulated overexpression of myc genes leads to malignant transformation, genetic instability, and apoptosis. In contrast to Myc and Mad, Max is a stable, constitutively expressed protein acting primarily as an obligate dimerization partner permitting Myc and Mad proteins to associate with DNA.
The Max network is highly conserved and Drosophila orthologs of vertebrate Myc and Max (dMyc and dMax) have been identified and characterized previously (Gallant et al. 1996; Schreiber-Agus et al. 1997; Johnston et al. 1999). More recently, a fly ortholog of mammalian Mad/Mnt was identified (termed dMnt; Bourbon et al. 2002; L.W.M. Loo and R.N. Eisenman, in prep.). Whereas vertebrates possess families of myc and mad genes, Drosophila myc, max, and mnt have no paralogs. Both dMyc and dMnt form heterodimers with dMax that specifically bind CACGTG. Whereas dMyc–dMax heterodimers activate transcription, dMnt–dMax associates with Drosophila Sin3 and represses transcription in an E-box-dependent manner (Gallant et al. 1996; L.W.M. Loo and R.N. Eisenman, in prep.). Moreover, similar to mammalian myc, dmyc is capable of cotransforming primary mammalian cells and rescuing the proliferation defect in c-myc null fibroblasts (Schreiber-Agus et al. 1997; Trumpp et al. 2001). Furthermore, both vertebrate and Drosophila myc regulate cell growth. The molecular and biological similarities between Drosophila and vertebrate Myc, Max, Mad, Mnt proteins coupled with the availability of Drosophila genetic tools and the complete genome sequence (Rubin et al. 2000) make Drosophila an attractive system to carry out a systematic analysis of genomic binding by the network. In this study, we have employed the recently devised DamID method (van Steensel and Henikoff 2000; van Steensel et al. 2001) in which a bacterial DNA methylase, fused to dMax network transcription factors, is used to “mark” DNA-binding sites in living cells.
Results
To identify genomic binding regions for Max network transcription factors we prepared fusion proteins consisting of the bacterial DNA adenine methylase (Dam) linked to the C termini of full-length dMax, dMnt, and dMyc as well as to the N terminus of dMyc. In each case, the transcription factor sequence was separated from Dam by a 9E10 (human Myc tag) peptide, permitting detection of the fusion protein with 9E10 antibody (see Materials and Methods for details). All of these fusion proteins were found to localize to the cell nucleus and activated (dMyc–Dam and Dam–dMyc) or repressed (dMnt–Dam) transcription from a synthetic reporter gene similar to their vertebrate orthologs (Grandori et al. 2000). In contrast, the tagged Dam protein alone, or a Myc mutant (amino acids 1–624) lacking the bHLHZ domain, had no effect on E-box-dependent transcription (data not shown; see Materials and Methods). These results indicate that fusion with Dam does not impair the functions of dMyc and dMnt. In order to ensure low levels of expression, the chimeric proteins were expressed separately in Drosophila Kc cells under control of the heat-shock promoter, but in the absence of heat-shock (van Steensel and Henikoff 2000). At 24 h posttransfection, genomic DNA was extracted. Subsequently, 0.1–2-kb fragments generated by digestion with DpnI (which cuts only at Gm6ATC) were purified by sucrose gradient centrifugation, labeled with the fluorochromes Cy5 (chimeric proteins) and Cy3 (Dam alone, serving as a reference for nonspecific binding/accessibility) and cohybridized to a Drosophila cDNA array that contains 6255 cDNAs and ESTs, representing roughly half of Drosophila coding sequences. Targeted sequences were identified based on the Cy5:Cy3 fluorescence ratio (van Steensel and Henikoff 2000; van Steensel et al. 2001; see Materials and Methods). The binding data for each protein on the entire array (chromatin profile) were generated as described previously (van Steensel et al. 2001). A set of statistical tests, as well as a “self on self” experimental set (dMnt–Dam vs. dMnt–Dam), were used to establish statistically significant targets and assess experimental noise (see Materials and Methods; Supplemental Material S3 for access to raw DamID binding data).
Comparative chromatin profiling of Max network proteins
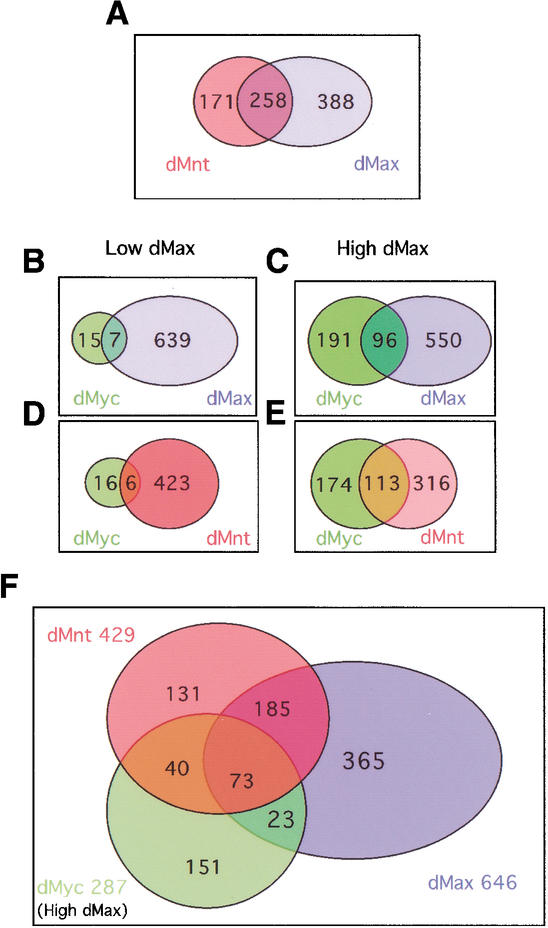
Having established chromatin profiles and a statistically significant set of binding targets for each of the network proteins, we compared the profiles derived from experimental sets for each of the network proteins to examine binding overlap among the different members. Figure 1A is a scatter plot comparison between the chromatin profiles of dMnt and dMax in which we compare separate binding data sets for each protein. These profiles show a high degree of similarity (correlation coefficient, r = 0.49) with both proteins displaying statistically significant binding to the same 258 genes (Fig. 1A, purple points). Many genes are not bound by either protein (Fig. 1A, gray points). We also found that increasing dMax levels had no effect on dMnt genomic binding (data not shown). The Venn diagrams in Figure 2 depict the number of overlapping and nonoverlapping genes. Overlapping genes represent 60% of the total genes occupied by dMnt and 40% of dMax bound genes (Fig. 2A). DamID analysis of binding by GAF, a transcription factor unrelated to dMax network proteins (van Steensel et al. 2003), shows only minimal overlap with dMnt bound loci (r = 0.11; Fig. 2F), further supporting the idea that the overlap detected among dMax network proteins is specific.
Figure 1.
Comparison of chromatin profiles for the Drosophila Max network. Scatter plot comparisons of chromatin binding profile data sets of the dMax network proteins derived from the entire cDNA array. The average binding is presented as Cy5:Cy3 log ratios between dMax–Dam:Dam vs. dMnt–Dam:Dam, r = 0.49 (A); dMax–Dam:Dam vs. dMyc–Dam:Dam with dMax expressed at low levels, r = −0.43 (B); dMax–Dam:Dam vs. dMyc–Dam:Dam with dMax expressed at high levels, r = 0.17 (C); dMnt–Dam:Dam vs. dMyc–Dam:Dam where dMyc–Dam is coexpressed with low levels of dMax, r = 0.26 (D); dMnt–Dam:Dam vs. dMyc–Dam:Dam where dMyc–Dam is coexpressed with high dMax levels, r = 0.71 (E); dMnt–Dam:Dam vs. dGAF–Dam, r = 0.11 (F). Ratios were calculated using a full set of experiments as described in Materials and Methods. Pearson correlation coefficients (r) were calculated for each set. Statistically significant binding targets are indicated by colored solid dots. Blue, dMax–Dam; green, dMyc–Dam; red, dMnt–Dam; brown, dGAF–Dam; purple, overlapping targets. The bicaudal (bic) gene is marked by open black circle and is printed twice on the array.
Figure 2.
Venn diagrams depicting the number of shared genes within the dMax network. Criteria for calculating statistically significant targets are described in Materials and Methods. (A) Comparison of dMnt and dMax targets. (B,D) dMyc data sets generated in the presence of low dMax levels. (C,E,F) dMyc data sets generated in the presence of high dMax levels.
A similar comparison of data sets derived from dMax and dMyc produced a surprising result (Fig. 1B). First, dMyc was recruited to a considerably smaller number of genes (22) than either dMax or dMnt (646 and 429 respectively; see Fig. 2B). Second, the dMax binding profile displayed little overlap with binding by either the C-terminal or N-terminal fusions of dMyc to Dam (Fig. 2B; data not shown). We asked whether dMax might be limiting for dMyc binding under our experimental conditions and therefore we expressed relatively higher levels of nonfused dMax along with the N-Dam–dMyc fusion (see Materials and Methods). As shown in Figures 1C and 2C, dMyc recruitment to targets and the binding profile overlap between dMyc and dMax were dramatically enhanced when dMax levels were augmented. Under these conditions the dMyc fusion associated with 287 targets, 96 shared with dMax (33.4%) and 73 (25.4%) shared with both dMnt and dMax (cf. Fig. 1B,C and the Venn diagrams in Fig. 2B,C,F). Augmented expression of dMax also resulted in increased overlap between dMyc and dMnt binding profiles, resulting in an overlap of 113 genes (39%; Figs. 1D,E, 2D–F). Myc recruitment to the majority of these target genes depends on functional dMax as the vast majority of the targets were not bound by dMyc when dMax lacking its leucine zipper domain (residues 91–119 deleted) was coexpressed (data not shown). Because the zipper region is essential for Max's ability to heterodimerize this result demonstrates that association between Myc and Max proteins is most likely responsible for the increased overlap between identified target genes. Nonetheless, although dMax levels were limiting for Dam–dMyc binding, a substantial number of dMyc targets did not overlap with either dMax or dMnt in the presence of either low or high dMax levels (17 and 152 unique Myc targets respectively; Fig. 2B,F; see Discussion). These findings suggest that dMax is limiting for dMyc binding to certain targets in Kc cells. Furthermore, the fact that dMax binds a subset of sites distinct from those bound by dMnt and dMyc implies that dMax heterodimerizes with an as-yet-unidentified factor (see Discussion).
Distribution of dMax network binding loci
Of 6255 genes represented on the array, 968 (15.4%) were found to associate with one or more dMax network proteins. The distribution and overlap is depicted in the Venn diagram in Figure 2F. Previous work showed that significant and specific methylation by tethered Dam occurs within 1.5–2 kb of the DNA-binding site (van Steensel and Henikoff 2000) permitting us to link binding regions detected on the array with a map of Drosophila chromosomes (Fig. 3A; see Discussion). Extensive binding of all three proteins was observed on the four major Drosophila chromosomes (Fig. 3A); however, repetitive DNA elements displayed relatively few binding sites despite the presence of E-boxes (e.g., Tirant; Fig. 3B). Even the low level of binding to repeats was not reproducible on identical repeats spotted multiple times on the array (e.g., the 297 and 412 repeats) and may therefore represent false positive hits. A previous DamID analysis showed that many of these repetitive elements are bound by Drosophila HP1 (heterochromatin protein 1), suggesting the lack of dMax network protein binding is likely to be specific, and not attributable to technical limitations or a generalized exclusion of chromatin-binding factors from this region (van Steensel et al. 2001). Therefore, although dMax network protein binding to chromatin is widespread, it appears to exclude specific regions, at least some of which are silenced regions associated with heterochromatin.
Figure 3.
Map of dMax network binding across Drosophila chromosomes and repetitive elements. Annotation of binding targets was preformed using the Drosophila gene collection subset (Rubin et al. 2000). Individual genes are depicted as vertical gray bars. dMyc, dMnt, and dMax binding sites are denoted by green, red, and blue solid circles, respectively. Color intensity is directly linked to binding P values as depicted at the bottom. (A) dMax network binding to the four Drosophila chromosomes. A partial list of genes found to be orthologs of direct targets of c-Myc (Fernandez et al. 2003) are indicated by black triangles and gene names. dMyc-regulated genes in the larval expression study that were detected in the the DamID analysis are denoted by purple (activated) and pink (repressed) arrows. (B) dMax network binding to repetitive elements.
Binding regions correlate with E-box sequences as well as other motifs
The E-box sequence CACGTG and several related sequences are binding sequences for vertebrate and Drosophila Myc, Max, and Mad/Mnt proteins (Blackwell et al. 1990, 1993; Blackwood and Eisenman 1991; Prendergast and Ziff 1991; Solomon et al. 1993; Gallant et al. 1996; James and Eisenman 2002; L.W.M. Loo and R.N. Eisenman, in prep.). To determine whether such sites are represented in our binding regions and to substantiate our in vivo-binding results, we applied an unbiased bioinformatics method, the REDUCE algorithm. REDUCE identifies putative regulatory sequences of 8 bp or less whose presence correlates with binding (Bussemaker et al. 2001; for review, see Li 2002). The REDUCE algorithm employs the binding profiles of the entire array without any preclustering or cut-off data restrictions. The readout of the REDUCE search is a list of distinct motifs with their statistical significance and (positive) regression coefficients quantifying their contribution to binding to the locus in which they occur. As shown in Table 1, binding sites for the canonical E-box CACGTG display significant correlation with the dMnt binding log-ratio (p < 10−12; Table 1A). However, the CACGTG correlated with dMyc binding only in the presence of high levels of coexpressed dMax (p < 10−12; Table 1C). A high correlation with short CG repeats may reflect a previously noted preference for binding by Myc and Mad to E-boxes that contain CG in flanking regions (Prendergast et al. 1991; Solomon et al. 1993; James and Eisenman 2002). When exogenous dMax levels are low, only AT-rich motifs correlate with binding (see Supplemental Material S5).
Table 1.
Highest scoring motifs detected by REDUCE analysis
| Motif
|
R2
|
P value
|
F
|
Matches
|
Loci
|
Consensus
|
|---|---|---|---|---|---|---|
| A dMnt | ||||||
| CGCG | 0.051 | 0.0E + 00 | 0.01 | 66,217 | 4366 | CG-repeat |
| GCGC | 0.048 | 0.0E + 00 | 0.01 | 97,925 | 4367 | CG-repeat |
| CGCGC | 0.047 | 0.0E + 00 | 0.02 | 16,661 | 4121 | CG-repeat |
| GCGCG | 0.042 | 0.0E + 00 | 0.02 | 17,392 | 4129 | CG-repeat |
| TATCGATA | 0.026 | 0.0E + 00 | 0.06 | 1618 | 1258 | TATCGATA |
| ATCGATA | 0.024 | 0.0E + 00 | 0.04 | 3863 | 2367 | TATCGATA |
| TCGATA | 0.015 | 1.0E − 12 | 0.02 | 8262 | 3541 | TATCGATA |
| TATCGAT | 0.015 | 5.0E − 12 | 0.03 | 3765 | 2369 | TATCGATA |
| GGTCACAC | 0.024 | 0.0E + 00 | 0.09 | 788 | 706 | GGTCACACT |
| GTCACACT | 0.017 | 0.0E + 00 | 0.08 | 691 | 633 | GGTCACACT |
| CACGTG | 0.019 | 0.0E + 00 | 0.03 | 4214 | 2558 | CACGTG |
| GCACGTG | 0.016 | 0.0E + 00 | 0.05 | 1370 | 1139 | CACGTG |
| GCACGTGT | 0.012 | 9.8E − 09 | 0.10 | 319 | 301 | CACGTG |
| B dMax | ||||||
| CGCGC | 0.025 | 0.0E + 00 | 0.02 | 16,401 | 4058 | CG-repeat |
| CGCG | 0.022 | 0.0E + 00 | 0.01 | 65,098 | 4301 | CG-repeat |
| GCGCG | 0.020 | 0.0E + 00 | 0.02 | 17,086 | 4069 | CG-repeat |
| GCGC | 0.014 | 2.0E − 12 | 0.00 | 96,364 | 4302 | CG-repeat |
| TATCGATA | 0.024 | 0.0E + 00 | 0.07 | 1600 | 1247 | TATCGATA |
| ATCGATA | 0.018 | 0.0E + 00 | 0.04 | 3797 | 2330 | TATCGATA |
| TATCGAT | 0.016 | 2.0E − 12 | 0.04 | 3705 | 2334 | TATCGATA |
| GGTCACAC | 0.013 | 2.0E − 09 | 0.08 | 776 | 693 | GGTCACACT |
| GTCACACT | 0.009 | 1.1E − 05 | 0.07 | 681 | 624 | GGTCACACT |
| C dMyc (high dMax) | ||||||
| CACGTG | 0.032 | 0.0E + 00 | 0.03 | 4203 | 2555 | CACGTG |
| ACGTG | 0.024 | 0.0E + 00 | 0.01 | 18,323 | 4228 | CACGTG |
| CACGT | 0.022 | 0.0E + 00 | 0.01 | 17,082 | 4221 | CACGTG |
| GCACGTG | 0.021 | 0.0E + 00 | 0.05 | 1365 | 1134 | CACGTG |
| ATCGATA | 0.022 | 0.0E + 00 | 0.03 | 3853 | 2362 | TATCGATA |
| TCGATA | 0.022 | 0.0E + 00 | 0.02 | 8255 | 3535 | TATCGATA |
| TATCGATA | 0.020 | 0.0E + 00 | 0.04 | 1616 | 1256 | TATCGATA |
| ATCGAT | 0.014 | 2.2E − 11 | 0.01 | 13,336 | 4037 | TATCGATA |
| TATCGAT | 0.013 | 2.1E − 10 | 0.02 | 3751 | 2362 | TATCGATA |
| CGCGC | 0.027 | 0.0E + 00 | 0.01 | 16,646 | 4116 | CG-repeat |
| CGCG | 0.026 | 0.0E + 00 | 0.00 | 66,118 | 4361 | CG-repeat |
| GCGC | 0.024 | 0.0E + 00 | 0.00 | 97,752 | 4362 | CG-repeat |
| GCGCG | 0.019 | 0.0E + 00 | 0.01 | 17,348 | 4124 | CG-repeat |
For each spot on the microarray the Berkeley Drosophila Genome Sequence Release 2 was used to generate an associated DNA region comprising the sequence of the array probe plus 2 kb flanking region. The Matches column indicates the total number of matches to the motif in the combined sequences. The Loci column indicates the number of sequences with at least one match. F represents the regression coefficient for a given motif. R2 equals the square of the Pearson correlation between the binding log ratio and the motif count (see Materials and Methods). All oligonucleotides of up to 8 bp in length were tested in parallel. Only the most significant motifs (p < 10−4) with a positive correlation are shown. E-box and DRE sequences are depicted in bold. Complete REDUCE results available in Supplemental Material S5.
Interestingly, REDUCE analysis also identified other sequence motifs that correlate with binding. Because of their high degree of association with Max network binding sites these motifs may serve as binding sites for factors that cooperate with dMyc and dMnt. One such putative binding site is the palindromic sequence TATC GATA (DNA replication element, DRE), a sequence reported near genes involved in cell proliferation and growth (Hirose et al. 1993, 2001; Jasper et al. 2002; Table 1, bold). The DRE was shown to be a consensus binding element for several factors: DREF (DNA replication element factor, a factor associated with the TRF2 complex; Hochheimer et al. 2002), Cut (a repression component within the Notch pathway; Jackson and Blochlinger 1997; Nepveu 2001), and BEAF32 (a boundary element associated binding factor; Zhao et al. 1995). Interestingly, REDUCE analysis demonstrates that the DRE sequence was the most significant motif among dMax targets (Table 1), suggesting that dMax is involved in as yet unidentified complexes (see Discussion).
Binding regions are proximal to genes implicated in Max network functions as well as genes regulated by dMyc
We sorted dMax network binding sites into functional groups using the Drosophila Gene Ontology database (GO; http://www.godatabase.org), a gene annotation system (Ashburner et al. 2000). Table 2 is a partial list of annotated gene targets. Our screen reveals both novel and previously described pathways and target genes associated with dMax network function (see Discussion). The complete binding profiles and partial annotation can be found as Supplemental Material (S1, S2).
Table 2.
Partial list of the dMax network target loci
| Biological pathway/molecular function
|
dMnt targets
|
dMax targets
|
dMyc targets
|
|---|---|---|---|
| Actin/cytoskeleton/cell migration | Dmn, nup44A, Tm1, Sop2, CG6546, CG25C, Dlc90F, nop5, bif | nup44A, Tm1, Arp11, bif, CG15669, puc, Klp3A | Tm1 |
| Cell cycle regulators | Myt1, ial, slbp | ial, cdk4, cdc2, cdc2c, Rbf, CycJ, CycC, CycD, stg | cdk4, CycA, CycB, CycB3, Rbf |
| Cell death | Dredd | Dredd, Dcp-1, Ice, debcl | Reaper L |
| Channels/transports | CG10444, icln | CG10444, Nrv1, Nhe1, icln | rpk, icln |
| Chromatin | HP1b, HmgD, Df31, Nap1, Caf1 | HP1b, HmgD, Caf1 | HP1b, fs (1) Ya |
| Immunity Mitochondria biogenesis, structure, function | FK506bp1, Dredd, Rel, PGRP-LE Tim10, Tim9, mRSp30, mRp19, mRpS7, TFAM, Ferrochelatase | FK506 bp1, Myd88, Rel, PGRP-LE, CG10535, CG3829 Tim10, mRpS30, mRp19, mRpS7, TFAM, colt | Rel Tim10, mRpL10, mRpS7, TFAM, CG3476, mge |
| Nucleolus | CG12909, CG8939, Surf6 | CG1135 | CG1135, CG12909, Rpp30, Surf6 |
| Peptidyl prolyl cis-transisomerase | Fk560bp1 | CG3511, FK506bp1, FKBP59 | CG5482, cyp33 |
| Protein synthesis/Ribosome | Aast-g1n, eIF6, mRpL19, mRpS30, mRpS7, RpL1, RpS5, RpS9, Nmd3, Surf6, CG1475 | bonsai, cg1475, mRpL19, mRpS7, RpL1, eIF6, pelo | RpL1, RpL19, pelo, mRpS7, mRpL1, RpS12, RpS13, RpS9, Surf6 |
| Replication/DNA repair | RnrS, CG8142, pms2 | RnrS, Rfc40, Mcm2, Mcm6, SPT4, Orc1, mus209, CG8142 | RnrS, UbcD6 |
| RNA binding | CG11738, CG12909, CG14230, CG5589, CG8636, CG8862, mod, Nmd3, Rbp1, SC35, SF2, Slbp, tra2, U2af38 | BcDNA: GH01073, CG11738, CG14230, CG1704, CG5589, CG8862, Hrb98DE, mxc, Nmd3, qkr58E-1, Rbp1, SC35, snf, Srp54 | CG10214, CG11738, CG12909, CG8862, cyp33, mod |
| RNA methyltransferase | CG11837, CG5220, CG7319, CG7818 | CG5220, CG7319 | CG1837, CG5220 |
| Signaling | aay, CG1815, CG4527, CG6805, LD08534 | loco, pak3, spz, grk, PP2-AB, puc, stg, cg66805, LD08534, aay | aay, LD08534 |
| Small GTPases GTP binding proteins | Rab14, RhoL | Rab17, Ran, Rap21, Ras85D, RhoL, Arf84F | Rab11 |
| Splicosome | CG10754, CG4980, CG6876, SC35, SF1, SF2, SmB, U2af38 | SC35, Snf, Srp54 | |
| Synaptic vesicles traffic | Syx13, Syx16, Syx17, Syx18, Syndapin, usnp, Rop, KdelR | Syx17, Syx18, Arf84D, Syndapin, Rop, KdelR | Syx13, CG8862 |
| Transcription factors | bic, Max, DREF, Pan, Ell, TFAM, Rel, Jra, Med6 | bic, DREF, bigmax, myb, Xbp1, Rpll33, Rbf, TFAM | bic, bigmax, E2F2, Rel, Rbf, TFAM |
| Transcription initiation and elongation | Taf55, Tfb2, Cyp1 | Taf60-2, Cyp1 | Taf60-2, Cyp1 |
| Transferase transferring glycosyl groups | CG18012, CG4802, CG5537, CG6437, CG9249 | CG5220, CG7319, CG9249 | CG3434, CG4802, CG5537, CG9249 |
| Translation factors | EfTuM, Adam, CG5705 | EfTuM, Adam, eif6, eif4A | Adam |
| Ub-proteasome | CG5384, CG5505, smt3, UfD1-like, Rpn5, Rpn7, Pros25, Pros26 | CG5384, smt3, Ufd1-like, Rpn5, Rpn7, Pros45, Tbp1, Uba1 | UbcD6, smt3 |
Standardized gene annotation was applied from the Drosophila Genome Ontology database (release February 2002). Only annotations in the biological process and molecular function categories with three or more targets are listed. A list of the network target genes can be found in Supplemental Material S2. Shared targets between all three proteins are in italic.
We next asked whether the binding loci defined by DamID in Kc cells could be identified as dMyc-responsive genes in the rapidly growing third instar larvae. Using the heat-shock Flp/Gal4 method (Pignoni and Zipursky 1997; Neufeld et al. 1998), we have generated transgenic flies expressing a marker GFP and dMyc, or GFP alone, both under UAS control. Total RNAs collected from either Myc-expressing or control larvae 7 and 14 h after heat-shock induction were Cy3;Cy5 labeled and hybridized to the same Drosophila microarrays used for the DamID experiments (see Materials and Methods). Statistically significant mRNA expression changes were defined for 845 genes: 544 genes activated and 301 repressed (see Supplemental Material S1, S4). Comparison of the dMax network DamID binding profile with the gene expression profile indicates a high degree of overlap (Fig. 3, purple and pink arrows; Table 3). The highest degree of correlation between binding and expression was observed for loci that are bound by both dMyc and dMnt (60%) and by all three Max network proteins (48%; Table 3). Furthermore, applying the REDUCE algorithm to the gene expression profile revealed a significant correlation with E-box and DRE sequences, supporting the notion that our binding loci correspond to transcriptionally regulated genes (see Supplemental Material S5). Within the group of genes bound by both dMyc and dMnt no genes were found to be repressed by dMyc, reinforcing the notion that the distinct biological effects of dMyc and dMnt derives from their opposing transcriptional functions (see below). Of the total dMax network targets identified by DamID and whose expression is dMyc regulated (253), the majority (232) were activated and only 21 repressed. Thus, the other 280 repressed genes are likely to be due to indirect or downstream effects of dMyc.
Table 3.
Comparison of DamID binding loci with dMyc-induced larval gene expression targets
| A Loci bound by
|
B No. of binding loci identified by DamID
|
C No. of dMyc regulated genes also identified by DamID
|
D Percent of overlap
|
E Activated genes
|
F Repressed genes
|
|---|---|---|---|---|---|
| dMyc + dMax + dMnt | 73 | 35 | 47.9% | 34 | 1 |
| dMyc + dMnt | 40 | 24 | 60.0% | 24 | 0 |
| dMyc + dMax | 23 | 6 | 26.0% | 5 | 1 |
| dMnt + dMax | 185 | 64 | 34.5% | 60 | 4 |
| dMyc | 151 | 24 | 15.8% | 24 | 0 |
| dMax | 365 | 56 | 15.3% | 42 | 14 |
| dMnt | 131 | 44 | 33.5% | 43 | 1 |
| Total targets | 968 | 253 | 26.1% | 232 | 21 |
Column A subgroups: targets were sorted according to subgroup specification as reflected in Figure 2F. Column B: Number of loci detected as statistically significant DamID targets. Column C: Number of genes that are depicted in column B that also show dMyc-regulated gene expression changes.
Chromatin immunoprecipitation analysis of dMyc and dMnt binding and histone acetylation at the bic locus
We employed chromatin immunoprecipitation (ChIP) to assess whether dMyc and dMnt bind the same site proximal to a DamID-identified target gene and alter the status of histone tail acetylation. We chose Bicaudal (bic), as an example of a gene that displayed binding by dMyc, dMax, and dMnt (Fig. 1, bic is circled) and whose mRNA is also induced by dMyc in our expression analysis. bic is related to vertebrate BTF3, a general transcription factor whose expression was found to correlate with c-Myc in a global expression study (Zheng et al. 1987; Watson et al. 2002). bic possesses one E-box in the vicinity of its transcriptional start site (Fig. 4A). Using the ChIP assay we specifically detect dMyc and dMnt association with the bic E-box region in cells overexpressing each protein (Fig. 4B, cf. IgG control lanes 3,4 and dMyc binding lanes 5,6, and dMnt binding lanes 7,8). Importantly, binding by dMyc and dMnt alters the status of endogenous histone acetylation at this site: dMyc binding correlates with hyperacetylation of both histones H3 and H4 (Fig. 4B, lanes 11,13), whereas dMnt binding correlates with deacetylation (Fig. 4, lanes 12,14). These findings support the notion that dMyc and dMnt are capable of alternate occupation of the same binding site, as predicted from the DamID data, and promote opposing changes in chromatin modification.
Figure 4.
Occupancy of the bic promoter by dMyc and dMnt proteins correlates with changes in histone acetylation. (A) Diagram of the bic promoter, the CACGTG E-box is denoted as an open box. The PCR primer set (−50 left primer, +75 right primer) used is denoted by upper black arrows. (B) Promoter occupancy was determined using monoclonal antibodies to dMyc (lanes 5,6), dMnt (lanes 7,8), acetylated histone H3 (lanes 11,12), and acetylated histone H4 (lanes 13,14). ChIP using genomic extracts derived from dMyc-expressing cells (odd-numbered lanes) or dMnt-expressing cells (even-numbered lanes) is described in Materials and Methods. Input, 2 ng non-IP genomic DNA; bic, the bic E-box specific primers; kis, control primers for the kis gene promoter serving as a loading control; αIgGp and αIgGm, Anti-mouse polyclonal and monoclonal IgG antibodies. Enrichment is represented as the ratio between the bic and the kis PCR products as described in Materials and Methods. (C) Ecdysone treatment leads to endogenous dMnt recruitment to the bic promoter and correlates with deacetylation of histone H3 and H4. Ecdysone treatment and ChIP were as described in Materials and Methods. Amplification linearity was tested in the presence of ecdysone (lanes 2–4). All other abbreviations are as depicted in Figure 4B.
To determine whether the endogenous dMnt protein occupies the bic locus E-box, we induced Kc cell differentiation using ecdysone. Other work has shown that dMnt is induced during differentiation and by ecdysone in pupal stages (L.W.M. Loo and C. Thummel, unpubl.). Figure 4C shows that endogenous dMnt associates with the bic E-box only following ecdysone treatment (Fig. 4C, cf. lanes 6 and 7). Similar experiments with endogenous dMyc were not feasible because of the inability of our antibody to detect the low levels of endogenous protein. Importantly, dMnt binding correlates with deacetylation of histone H3 and H4 (Fig. 4C, lanes 8–12), consistent with the ability of dMnt to recruit a mSin3–HDAC complex to its binding site (L.W.M. Loo and R.N. Eisenman, in prep.).
Discussion
A significant gap in our understanding of the function of many transcriptional regulatory proteins has been the lack of comprehensive identification of their in vivo binding sites and the genes whose expression they regulate. This problem is especially pertinent for transcription factors such as Myc, Mad/Mnt, Max, and other members of the Max network that function as relatively weak transcriptional regulators, whose consensus binding site is ubiquitous, and whose expression elicits profound effects on cell growth and proliferation. Standard methods of target gene evaluation do not reliably differentiate between genes bound and directly regulated by Myc and Mad from genes whose expression is altered as a secondary or later consequence of Myc or Mad induction. In principle, it is important to know about both sets of genes, but it is also crucial to distinguish between them. The DamID method employed in this paper permits determination of transcription factor binding site regions in live cells and is not dependent on chemical cross-linkers or PCR primers. Because it involves “marking” of DNA in chromatin by a methyltransferase linked to a transcription factor, even transient or low affinity interactions with DNA, as well as proximity to regions distal to the binding site (through looping or higher-order folding), might be detected (van Steensel and Henikoff 2000; van Steensel et al. 2001). Because we used a cDNA array to detect targeted methylation regions, only binding sites within a few kb of transcription units are detected. Therefore, our enumeration of dMax network binding sites is likely to be an underestimate. The mapping resolution also does not permit precise pinpointing of the binding site within each probed locus, although the REDUCE analysis strongly suggests that E-box motifs within target loci mediate the protein recruitment (e.g., as for bic; Fig. 4).
The validity of our approach is strongly supported by several lines of evidence. First, the degree of overlap between dMyc, dMax, and dMnt binding regions (Fig. 2) is consistent with the relationship between E-box binding and heterodimerazation with Max established previously for the vertebrate proteins as well as for their orthologs in Drosophila (Blackwood and Eisenman 1991; Ayer et al. 1993; Zervos et al. 1993; Gallant et al. 1996; L.W.M. Loo and R.N. Eisenman, in prep.). Importantly, the GAGA factor, a ubiquitous transcription factor unrelated to the dMax network, displays only minimal overlap with dMnt binding sites (Fig. 1F), suggesting our results are specific for binding by dMax network transcription factors. Furthermore, studies in mammalian cells have shown both overlapping and nonoverlapping functions and target genes for Myc and Mad proteins (O'Hagan et al. 2000; Iritani et al. 2002; James and Eisenman 2002) in agreement with our DamID findings. Second, using a ChIP assay, the direct binding of dMyc and dMnt to a DamID-defined target gene, bic (bicaudal), was demonstrated (Fig. 4). In addition, the mammalian orthologs of at least 18 genes identified as binding targets for dMyc, dMax, and dMnt in our study have been demonstrated to be direct targets for vertebrate Myc using ChIP (Fernandez et al. 2003) and are indicated in Figure 3. Third, application of the REDUCE algorithm, which correlates binding with the occurrence of DNA sequence motifs (Bussemaker et al. 2001) reveals a statistically significant correlation between the E-box CACGTG and the presence of dMnt binding regions. CACGTG enrichment also correlated with dMyc binding in the presence of high dMax levels, for dMax binding in the presence of high dMyc levels, and for genes whose expression is modulated by dMyc. Fourth, a substantial set of target genes identified in our Drosophila gene expression microarray analysis, employing larvae overexpressing dMyc, correspond to target genes defined by DamID (Fig. 3; Table 3; Supplemental Material S1). In addition, target genes identified here are in accord with genes regulated by Myc and Mad as described in several recently published gene-expression studies in vertebrate systems (Coller et al. 2000; Guo et al. 2000; Boon et al. 2001; Neiman et al. 2001; Schuhmacher et al. 2001; Iritani et al. 2002).
Drosophila Max network binding sites define biological functions of the network
We have used the Drosophila Gene Ontology Database to derive an unbiased classification of genes associated with dMax network binding regions (see Table 2 for a partial list; for complete list, see Supplemental Material S2). Many of our dMax network targets identified are genes that fit well with the established biological functions of Myc and Mad. In addition, a significant number of targets point to new pathways likely to be regulated by the network. Our data demonstrate both binding to, and regulation of, genes encoding proteins broadly involved in biosynthetic processes, in accord with genetic and biochemical analyses demonstrating that Myc is involved in cell growth in Drosophila and vertebrates (Schmidt 1999; Schuhmacher et al. 1999; Kim et al. 2000; de Alboran et al. 2001; Douglas et al. 2001), and from earlier global gene expression studies (see http://www.myc-cancer-gene.org/index.asp for a compilation of candidate target genes; Guo et al. 2000; Boon et al. 2001; Neiman et al. 2001; Schuhmacher et al. 2001). Our DamID binding loci also include genes involved in cell cycle and DNA replication (Table 2). Our list of putative dMax network targets also reveals potential novel pathways such as mitochondrial biogenesis and function, as well as vesicular transport. Other pathways known to be linked to Myc such as apoptosis, proteolysis, and the immune response are also reflected in our list of dMax network target genes as are a number of transcription factors (Table 2).
Widespread genomic binding by the dMax network
Our findings demonstrate a surprisingly large number (968) of binding sites for proteins of the dMax network (Fig. 2). Considering that the array represents a random sampling of ∼50% of Drosophila coding regions, a conservative estimate is that dMax network proteins interact with ∼2000 genes, and, as mentioned above, this is likely to be an underestimate. It is important to note however that dMax network proteins do not bind profligately to DNA, as evidenced by the low degree of overlap with GAGA factor, the general correlation of E-box sequences with binding, and the lack of association with repeat elements linked to HP1 binding previously. HP1 is predominantly localized to pericentric heterochromatin, and its binding is associated with silenced chromatin structure (James et al. 1989). The lack of association of dMyc, dMax, or dMad with such elements may indicate that the network proteins are primarily associated with genes that are subject to ongoing transcriptional modulation. These findings are in accord with extensive ChIP assays carried out by Fernandez et al. (2003) in human cells. That study suggests that 8%–10% of cellular genes associate with Myc and in general display enhanced histone H3 and H4 acetylation.
The large number of binding sites and regulated target genes identified in this study contrasts with earlier ideas of Myc function that posited a small number of critical targets. However, not all binding sites necessarily result in direct transcriptional regulation by dMax network factors. This is evident from our dMyc-dependent gene expression data carried out in growing third instar larvae. At this developmental stage, 31% (89/287) of our Myc binding loci (as determined in Kc cells) displayed altered mRNA epression in larvae. Of genes that were detected as overlapping targets of all three proteins or of only dMyc and dMnt, 48.6% and 60.5% respectively, displayed concomitant changes in mRNA levels upon Myc induction (Fig. 3; Table 3). Interestingly, Fernandez et al. (2003) describe Myc binding and histone acetylation at mammalian genes whose expression does not appear to change in response to induction of Myc. One possible explanation is that Myc binding to a subset of genes, although not immediately affecting gene expression, confers a permissive state on chromatin allowing binding by other cis-acting factors at later times.
The many dMax targets detected that are shared with dMyc and dMnt most likely represent binding by dMyc–dMax and dMnt–dMax heterodimers. However, the extent of nonoverlap between binding sites for these proteins is more extensive than expected. For example, we found that dMax expressed at low levels binds to 365 genes that do not overlap with either dMnt or dMyc targets. However, 15% of these binding loci are regulated by dMyc in our larval expression analysis. Thus, the degree of overlap is probably influenced by the temporal pattern and levels of dMyc expression. This has implications for tumorigenesis where vertebrate Myc proteins are often dramatically overexpressed. Our work provides evidence that such overexpression may shift the spectrum of target genes relative to those expressed in normal cells.
Max homodimers bind E-boxes with relatively low affinity and in mammalian cells are inhibited by phosphorylation from binding DNA (Berberich and Cole 1992; Koskinen et al. 1994). Although we do not know whether dMax homodimers are similarly blocked from binding to DNA in vivo, we favor the idea that the large number (365) of unique dMax binding sites and the lack of correlation with E-boxes reflects dimerization and DNA binding by dMax with as-yet-unidentified interacting proteins. Interestingly, in mammalian cells Max has been found, in association with the bHLHZ protein Mga, in E2F6 repression complexes (Ogawa et al. 2002). Similarly, unique sites found for dMnt and dMyc may represent non-E-box DNA binding through formation of higher-order complexes. For mammalian Myc, interaction of Myc–Max heterodimers with the Miz-1 protein has been shown to direct Myc to non-E-box sites (Staller et al. 2001). It is likely that associations with other partners may redirect dMyc and dMnt to unique binding sites. If so, our findings indicate that such interactions may be extensive and are an important part of dMax network function.
The canonical E-box sequence alone is unlikely to be sufficient to determine specific binding by dMax network proteins and, indeed, many E-box-containing promoters are not associated with Max network proteins (Bouchard et al. 2001; Fernandez et al. 2003). One possibility is that other sequences in the vicinity of an E-box may play a role in target gene specificity. For example, the DRE, which correlated with binding of all three dMax network proteins is located within <1 kb of many of our E-box sequences. Therefore, it is tempting to hypothesize that the DRE operates in cis with adjacent E-boxes to recruit protein complexes that will either promote activation or repression. Alternatively, the proximity of DRE and E-box sites may reflect coordinate regulation of the same genes through distinct signaling pathways.
In addition, REDUCE analysis has revealed a number of unexpected correlations. For example, we found association between dMyc and AT-rich sequences when dMax levels are limiting (Supplemental Material S5). In several loci examined, these AT-rich regions occur in the vicinity of genes lacking E-boxes, perhaps reflecting dMyc association with as-yet-undefined binding proteins when dMax levels are limiting. REDUCE analysis of dMax binding regions failed to detect a binding correlation with CACGTG. However, when high levels of dMyc were expressed together with dMax–Dam, REDUCE analysis of dMax binding regions found the E-box significantly correlated with binding (data not shown). This is in accord with previous data that dMax homodimers bind only weakly to E-boxes, and that Max binding is largely directed by its heterodimeric partners (Berberich and Cole 1992; Fieber et al. 2001). Perhaps the AT- (and CG-) rich sequences influence architecture of the binding site or serve as binding motifs for factors that enhance dMax network protein association with DNA.
Taken together, our data suggest a rather more complex picture of the functioning of Max network transcription factors then has been considered previously. The results suggest extensive yet specific interaction with chromatin probably encompassing thousands of binding sites and directly affecting expression of hundreds of genes. In addition, the DamID results indicate the possibility of several different modes of Myc, Max, and Mad/Mnt interactions. These include binding to partner proteins yet to be identified as well as potential cooperation with other transcription factors. Earlier experiments have shown that Myc and Mad expression is under tight control by the cell. Such control is likely to be important in balancing the multiple protein–protein and DNA binding interactions inferred from our data.
Materials and methods
Plasmids and constructs
Generation of chimeric Dam∼proteins: Full-length cDNAs encoding Drosophila melanogaster Max, Mnt, Myc, and Myc Δct, (amino acids 1–624) were cloned in either pNDam-9E10 or p9E10-CDam vectors (van Steensel and Henikoff 2000) resulting in the formation of either N-terminal or C-terminal Dam-fusion proteins. GAF-Dam was described previously (van Steensel et al. 2001). For expression in Kc cells, the appropriate cDNAs were subcloned into heat shock (hs)-inducible pCasper vector or pMTV, a copper-inducible insect expression vector (Invitrogen). All constructs was verified by automated sequencing.
Cell culture, transfections, and reporter assays
Reporter assays were performed as described previously using pCI-Neo to express the different proteins in 293T cells (Laherty et al. 1997). Kc167 Drosophila cells were maintained, transfected, and stained as described earlier (van Steensel et al. 1995; Henikoff et al. 2000). Cells were treated with 0.2 μM 20-hydroxy ecdysone (Sigma) for 18 h where indicated. Cell cultures for microarray analysis were as follows: Where indicated, pMTV–dMyc (Cu+2-inducible) was cotransfected in the presence of 0.5 mM CuSo4 (Sigma), along with hs-dMax–Dam. Alternatively, cells were transfected with a plasmid coding for either hs-dMax (low Max) or copper-induced pMTV–dMax (high Max), or pMTV–dMaxΔZip (Δ92–115) along with hs-Ndam–Myc. pMTV–dMax was also cotransfected with hs-dMnt–Dam. All assays were carried out in the absence of heat shock.
Microarray analysis and chromatin profiling
All chromatin profiles were performed as described (van Steensel et al. 2001) using spotted microarrays constructed from release 1 of the Drosophila Gene Collection (Rubin et al. 2000) and 430 additional cDNA and genomic sequences. Three independent experiments were performed for each of the proteins. In an additional experimental set the dye labeling was reversed for Dam and the fusion protein to exclude bias related to the dye. Microarray images were quantified using GenePix Pro v3.0 imaging software (Axon Instruments). Microarray results were analyzed using Cyber-T microarray analysis software (Baldi and Long 2001) and corresponding Bayesian-derived P values were adjusted for multiple hypotheses testing using a false discovery rate (FDR) method (Benjamini and Hochberg 1995) where FDR was set at 0.05. Additionally, dMnt–Dam versus dMnt–Dam control experiments were used to assess experimental array variation within the DamID assay. Accordingly, a lower-bound ratio threshold of log2 0.58 (i.e., 3 × S.D.) was also imposed. For access to DamID primary data, see Supplemental Material S3.
Larval dMyc expression profile experiments
Fly strains: All transgenes are P[+] in w− strains. w+;+;Act 5c > CD2 > GAL4 UAS–GFP (Neufeld et al. 1998); y w hs-FLP122; +; UAS–dMyc (Zaffran et al. 1998). y w hs-FLP122; +; +. Adult flies and larvae were raised in regular fly food consisting of cornmeal and molasses at 25°C. Larvae overexpressing either UAS-regulated dMyc;GFP or GFP alone transgenes were generated using the Flp/Gal4 method (Struhl and Basler 1993; Pignoni and Zipursky 1997; Neufeld et al. 1998). Larvae were staged from hatching and raised at a density of 50 per vial at 25°C. Third instar larvae (110 h after egg deposition, AED) were heat shocked at 37°C for 2 h, and larvae were collected 7 h after heat shock (∼120 h AED). Total RNA was isolated using TRIzol reagent (Invitrogen) as described by manufacturer followed by RNeasy (Qiagen) clear up. cRNA targets were generated using a standard amino-allyl labeling protocol, where 30 μg each of “experimental” (dMyc;GFP: hs-FLP122; Act–GAL4, UAS–GFP; UAS–dMyc) and “reference” (GFP only :ywhs–FLP122; Act–GAL4, UAS–GFP; +) total RNAs were coupled to either Cy3 or Cy5 fluorophores. Paired labeled targets were processed on microarrays using protocols described elsewhere (Fazzio et al. 2001). Posthybridized arrays were scanned using a GenePix 4000 scanner (Axon Instruments). Data were generated from five independent replicates (two with one dye orientation and three with the reversed dye orientation) at 7 h and four independent replicates (two with one dye orientation and two with the reversed dye orientation) at 14 h. Spot-level ratios were log2 transformed and a loess normalization (f = 0.33) strategy was applied using S-Plus (MathSoft, Cambridge, MA) to correct for observed intra-array intensity-dependent ratio biasing. Expression data analysis was performed as described for DamID. Accordingly, any gene/est with a P value that meets the FDR = 0.05 criterion and has a fold-change outside of the range ±1.5 were identified as differentially expressed. Access to gene expression data is available as Supplemental Material (S4).
Motif-based regression analysis of Max network proteins binding using REDUCE
To identify motifs responsible for the preferential binding of the transcription factors studied, we used REDUCE, a tool originally developed for the analysis of gene expression data but which can also be used to analyze binding data. To remove any remaining bias attributable to the varying number of matches with GATC (the restriction site used in the DamID procedure) in the probe sequences, we first fitted a linear model based on GATC. The corrected log-ratios were subsequently analyzed as described (Bussemaker et al. 2001) using software available at http://bussemaker.bio.columbia.edu/reduce. REDUCE relies on standard linear regression of the binding log-ratio for each probe on the number of matches of a given motif to the DNA sequence associated with the binding log-ratio (i.e., the probed region plus flanking sequence).
The regression coefficient (slope) associated with each motif is determined by performing a least-squares fit of this model to the data; it estimates the contribution to binding to a locus by each occurrence of the motif. The value of R2 for this fit is equal to the square of the Pearson correlation r between the binding log-ratio and the motif count; it measures the statistical significance of the regression coefficient. The P value associated with R2 has to be corrected to control the family-wise error rate in the parallel test of a large number of motifs. We used the Bonferroni correction, which amounts to multiplying the P value by the total number of motifs tested. The complete REDUCE search results can be found in Supplemental Material S5.
Chromatin immunoprecipitation
Chromatin fixation and purification procedures were as described (Schubeler et al. 2000; Takahashi et al. 2000; Sawado et al. 2001). Where indicated, cells were treated with 0.5 mM CuSO4. Immunoprecipitation of cross-linked DNA was carried out using the following antibodies: rabbit polyclonal αAcH3, αAcH4 (Upstate), αdMnt mouse monoclonal antibodies (b8, b11; L. Loo, in prep.), and αdMyc mouse monoclonal (b10; Prober and Edgar 2002). Duplex γ[P32]-dCTP-labeled PCR reactions were preformed and quantified using “Storm” PhosphorImager system (Molecular Dynamics). Specific protein binding was measured as a comparison between the relative ratio of intensity of the specific target “bic” PCR products to the reference. The reference gene kismet (kis) had negative binding values for both dMyc and dMnt, was not detected as a target in the larval myc expression experiment, had linear amplification profile under the various conditions, and, therefore, serves as a loading control. Primer sequences can be found in Supplemental Material.
Acknowledgments
We are grateful to Steve Henikoff, Dirk Schübeler, Harmit Malik, Fumiko Hirose, and Masamitsu Yamaguchi for valuable reagents, advice, and unpublished data. We are also grateful to Bruno Amati and Paula Fernandez for discussion and sharing unpublished data. We thank Steve Henikoff and Carla Grandori for critical readings of the manuscript and Grace Navaja for technical assistance. Support for this work was from the Human Frontiers Science Program (A.O.), NIH grants GM47852 (S.P.), CA80295-03S (E.W.), 1P20LM007276-01 (HJB), Uehara Memorial Foundation (T.S.), RO1CA57138 (R.N.E.), and NIH/NCI Cancer Center Support Grant P30 CA15704-29 (J.D.), and R01 GM51186 (B.A.E.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL eisenman@fhcrc.org; FAX (206) 667-6522.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1066903.
References
- Amati B, Frank SR, Donjerkovic D, Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and trasncription. Biochim Biophys Acta. 2001;1471:M135–M145. doi: 10.1016/s0304-419x(01)00020-8. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer DE, Kretzner L, Eisenman RN. Mad: A heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: Regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. JR Statistical Soc Ser B-Methodological. 1995;57:289–300. [Google Scholar]
- Berberich SJ, Cole MD. Casein kinase II inihibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers. Genes & Dev. 1992;6:166–176. doi: 10.1101/gad.6.2.166. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA-binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, Weintraub H. Binding of Myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN. Max: A helix–loop–helix zipper protein that forms a sequence-specific DNA binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus M-C, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes & Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon HM, Gonzy-Treboul G, Peronnet F, Alin MF, Ardourel C, Benassayag C, Cribbs D, Deutsch J, Ferrer P, Haenlin M, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygous and reduced fertility in heterozygous female mice. Genes & Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene- targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- Douglas NC, Jacobs H, Bothwell AL, Hayday AC. Defining the specific physiological requirements for c-Myc in T cell development. Nat Immunol. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- Eisenman RN. Deconstructing myc. Genes & Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread collaboration of Isw2 and Sin3–Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, P.C., Frank, S.R., Wang, L., Schroeder, M., Liu, S., Greene, J., Cocito, A., and Amati, B. 2003. Genomic targets of the human c-Myc protein. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Fieber W, Schneider ML, Matt T, Krautler B, Konrat R, Bister K. Structure, function, and dynamics of the dimerization and dna-binding domain of oncogenic transcription factor v-myc. J Mol Biol. 2001;307:1395–1410. doi: 10.1006/jmbi.2001.4537. [DOI] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced actylation of histone H4 and gene activation. Genes & Dev. 2001;15:2069. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P, Shiio Y, Cheng PF, Parkhurst S, Eisenman RN. Myc and Max homologs in Drosophila. Science. 1996;274:1523–1527. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The MYC/MAX/MAD network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET. Identification of c-myc responsive genes using rat cdna microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose F, Yamaguchi M, Handa H, Inomata Y, Matsukage A. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J Biol Chem. 1993;268:2092–2099. [PubMed] [Google Scholar]
- Hirose F, Ohshima N, Shiraki M, Inoue YH, Taguchi O, Nishi Y, Matsukage A, Yamaguchi M. Ectopic expression of DREF induces DNA synthesis, apoptosis, and unusual morphogenesis in the Drosophila eye imaginal disc: Possible interaction with Polycomb and trithorax group proteins. Mol Cell Biol. 2001;21:7231–7242. doi: 10.1128/MCB.21.21.7231-7242.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- Iritani BM, Delrow J, Grandori C, Gomez I, Klacking M, Carlos LS, Eisenman RN. Modulation of T lymphocyte development, growth, and cell size by the Myc-antagonist Mad1 transcriptional repressor. EMBO J. 2002;21:4820–4830. doi: 10.1093/emboj/cdf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SM, Blochlinger K. cut interacts with Notch and protein kinase A to regulate egg chamber formation and to maintain germline cyst integrity during Drosophila oogenesis. Development. 1997;124:3663–3672. doi: 10.1242/dev.124.18.3663. [DOI] [PubMed] [Google Scholar]
- James L, Eisenman RN. Myc and Mad bHLHZ domains possess identical DNA-binding specificities but only partially overlapping functions in vivo. Proc Natl Acad Sci. 2002;99:10429–10434. doi: 10.1073/pnas.162369299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- Jasper H, Benes V, Atzberger A, Sauer S, Ansorge W, Bohmann D. A genomic switch at the transition from cell proliferation to terminal differentiation in the Drosophila eye. Dev Cell. 2002;3:511–521. doi: 10.1016/s1534-5807(02)00297-6. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Li Q, Dang CV, Lee LA. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc Natl Acad Sci. 2000;97:11198–11202. doi: 10.1073/pnas.200372597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation . Genes & Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen PJ, Västrik I, Mäkelä TP, Eisenman RN, Alitalo K. Max activity is affected by phosphorylation at two NH2-terminal sites. Cell Growth Diff. 1994;5:313–320. [PubMed] [Google Scholar]
- Laherty CD, Yang W-M, Sun J-M, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Li H. Computational approaches to identifying transcription factor binding sites in the yestad genome. Methods Enzymol. 2002;350:484–495. doi: 10.1016/s0076-6879(02)50980-0. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman PE, Ruddell A, Jasoni C, Loring G, Thomas SJ, Brandvold KA, Lee Rm, Burnside J, Delrow J. Analysis of gene expression during myc oncogene-induced lymphomagenesis in the bursa of Fabricius. Proc Natl Acad Sci. 2001;98:6378–6383. doi: 10.1073/pnas.111144898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AFA, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc- responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- O'Hagan RC, Schreiber-Agus N, Chen K, David G, Engelman JA, Schwab R, Alland L, Thomson C, Ronning DR, Sacchettini JC, et al. Gene-target recognition among members of the Myc superfamily and implications for oncogenesis. Nat Genet. 2000;24:113–119. doi: 10.1038/72761. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of Max, with c-Myc stimulates methylation-sensitive DNA binding and Ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prober DA, Edgar BA. Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing. Genes & Dev. 2002;16:2286–2299. doi: 10.1101/gad.991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Hong L, Brokstein P, Evans-Holm M, Frise E, Stapleton M, Harvey DA. A Drosophila complementary DNA resource. Science. 2000;287:2222–2224. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- Sawado T, Igarashi K, Groudine M. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc Natl Acad Sci. 2001;98:10226–10231. doi: 10.1073/pnas.181344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EV. The role of c-myc in cellular growth control. Oncogene. 1999;18:2988–2996. doi: 10.1038/sj.onc.1202751. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, Stein D, Chen K, Goltz JS, Stevens L, DePinho RA. Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutive phenotype. Proc Natl Acad Sci. 1997;94:1235–1240. doi: 10.1073/pnas.94.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, Francastel C, Cimbora DM, Reik A, Martin DI, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes & Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, Eick S, Kohlhuber F. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. The transcriptional program of a human B cell line in response to myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- Solomon DLC, Amati B, Land H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 1993;21:5372–5376. doi: 10.1093/nar/21.23.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Möröy T, Bartek J, Massague J, Hänel F, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes & Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes & Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nat Biotech. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Brink M, van der Meulen K, van Binnendijk EP, Wansink DG, de Jong L, de Kloet ER, van Driel R. Localization of the glucocorticoid receptor in discrete clusters in the cell nucleus. J Cell Sci. 1995;108:3003–3011. doi: 10.1242/jcs.108.9.3003. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Bussemaker HJ. Genome-wide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc Natl Acad Sci. 2003;100:2580–2585. doi: 10.1073/pnas.0438000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Oster SK, Shago M, Khosravi F, Penn LZ. Identifying genes regulated in a Myc-dependent manner. J Biol Chem. 2002;277:36921–36930. doi: 10.1074/jbc.M201493200. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Chartier A, Gallant P, Astier M, Arquier N, Doherty D, Gratecos D, Semeriva M. A Drosophila RNA helicase gene, pitchoune, is required for cell growth and proliferation and is a potential target of d-Myc. Development. 1998;125:3571–3584. doi: 10.1242/dev.125.18.3571. [DOI] [PubMed] [Google Scholar]
- Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc–Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- Zheng XM, Moncollin V, Egly JM, Chambon P. A general transcription factor forms a stable complex with RNA polymerase B (II) Cell. 1987;50:361–368. doi: 10.1016/0092-8674(87)90490-9. [DOI] [PubMed] [Google Scholar]
- Zhou ZQ, Hurlin PJ. The interplay between Mad and Myc in proliferation and differentiation. Trends Cell Biol. 2001;11:S10–14. doi: 10.1016/s0962-8924(01)02121-3. [DOI] [PubMed] [Google Scholar]
- Ziegelbauer J, Shan B, Yager D, Larabell C, Hoffmann B, Tjian R. Transcription factor MIZ-1 is regulated via microtubule association. Mol Cell. 2001;8:339–349. doi: 10.1016/s1097-2765(01)00313-6. [DOI] [PubMed] [Google Scholar]