Abstract
The Caenorhabditis elegans coiled-coil protein LIN-5 mediates several processes in cell division that depend on spindle forces, including alignment and segregation of chromosomes and positioning of the spindle. Here, we describe two closely related proteins, GPR-1 and GPR-2 (Gprotein regulator), which associate with LIN-5 in vivo and in vitro and depend on LIN-5 for localization to the spindle and cell cortex. GPR-1/GPR-2 contain a GoLoco/GPR motif that mediates interaction with GDP-bound Gαi/o. Inactivation of lin-5, gpr-1/gpr-2, or the Gαi/o genes goa-1 and gpa-16 all cause highly similar chromosome segregation and spindle positioning defects, indicating a positive role for the LIN-5 and GPR proteins in G protein signaling. The lin-5 and gpr-1/gpr-2 genes appear to act downstream of the par polarity genes in the one- and two-cell stages and downstream of the tyrosine kinase-related genes mes-1 and src-1 at the four-cell stage. Together, these results indicate that GPR-1/GPR-2 in association with LIN-5 activate G protein signaling to affect spindle force. Polarity determinants may regulate LIN-5/GPR/Gα locally to create the asymmetric forces that drive spindle movement. Results in C. elegans and other species are consistent with a novel model for receptor-independent activation of Gαi/o signaling.
Keywords: LIN-5, GoLoco motif, G protein, chromosome segregation, spindle position
Supplemental material is available at http://www.genesdev.org.
Microtubule (MT)-based bipolar spindles are required for several critical processes in cell division and development, including segregation of chromosomes, maintenance of genetic stability, and specification of the cleavage plane (Straight and Field 2000; Wittmann et al. 2001). These multiple functions depend on correctly formed spindle structures and properly balanced spindle forces, which involve a complex interplay between MTs; MT-associated proteins (MAPs); and proteins at the spindle asters, kinetochores, and cell cortex (Wittmann et al. 2001). Although the roles of certain components have been elucidated, understanding the execution and coordination of the diverse spindle functions remains a major challenge.
Regulated positioning of the spindle is used during animal development, in particular during asymmetric cell divisions that generate daughter cells with different fates (Knoblich 2001). Because cell cleavage cuts the spindle perpendicularly and generally through the middle, changing the position of the spindle alters the axis and plane of cell division. Results from studies in Drosophila melanogaster and Caenorhabditis elegans have started to reveal conserved mechanisms used in asymmetric cell division (for review, see Knoblich 2001). Early in this process, the mother cell establishes an axis of polarity in coordination with the body plan. Cell fate determinants are subsequently localized asymmetrically in accordance with the polarity axis. Finally, spindle orientation is coordinated with the polarity axis during mitosis, so that cell cleavage generates daughter cells containing different concentrations of the determinants.
Several asymmetric divisions generate cells with different fates at defined positions in the early C. elegans embryo. During the first two divisions following fertilization, placement of the mitotic spindle is regulated by cell intrinsic polarity cues, whereas both intrinsic and extrinsic signals guide spindle positioning during subsequent divisions (for review, see Gönczy 2002). Anterior–posterior (A/P) polarity is established in the early embryo through asymmetric distribution of the PAR proteins. A sperm component, possibly the sperm aster, determines the posterior end of the fertilized oocyte and restricts the localization of the PDZ proteins PAR-3 and PAR-6 and the atypical protein kinase C PKC-3 to the anterior cortex (Tabuse et al. 1998; Wallenfang and Seydoux 2000; Cuenca et al. 2003). The RING-finger protein PAR-2 subsequently localizes to the posterior cortex and further limits PAR-3 localization because PAR-2 and PAR-3 localizations are mutually exclusive (Cuenca et al. 2003). PAR-2 is necessary for the posterior localization of PAR-1, a protein related to the MARK family of Ser/Thr kinases (Guo and Kemphues 1995; Drewes et al. 1997). PAR-2 and PAR-3 affect the net forces on the spindle asters in opposite ways (Grill et al. 2001). Thus, their asymmetric localizations create asymmetric spindle forces that induce spindle movement and ultimately determine the cleavage plane of the cell. However, the molecular mechanisms that translate asymmetric PAR localization into asymmetric forces and spindle positioning remain largely unknown.
Recent results have identified several candidate targets of polarity cues that influence the position of the spindle. Two genes encoding Gαi/o subunits of heterotrimeric G proteins, goa-1 and gpa-16, act redundantly to position the spindle in the early embryo, although they do not affect polarity and are not themselves asymmetrically localized (Miller and Rand 2000; Gotta and Ahringer 2001). In addition, the Gβ protein GPB-1 has a role in centrosome separation and rotation (Zwaal et al. 1996; Gotta and Ahringer 2001).
Another candidate downstream effector of PAR polarity is LIN-5. Embryos lacking lin-5 function do not properly position the mitotic spindles during the first embryonic divisions (Lorson et al. 2000). Moreover, subsequent cycles lack chromosome segregation and cytokinesis altogether, indicating that lin-5 is more generally required for spindle function. Similarly, following the depletion of maternal product, postembryonic cells in lin-5 null mutants form apparently normal spindles but fail chromosome alignment and segregation as well as cytokinesis (Albertson et al. 1978; Horvitz and Sulston 1980; Lorson et al. 2000). Despite the absence of chromosome segregation and cytokinesis, lin-5(null) cells exit from mitosis with normal timing and enter subsequent rounds of DNA synthesis and centrosome duplication. The different aspects of the Lin-5 phenotype are consistent with lin-5 acting in spindle force generation. The lin-5 gene has no obvious orthologs in other species and encodes a coiled-coil protein that localizes to the cell cortex and to meiotic and mitotic spindles (Lorson et al. 2000).
In this study, we applied a combination of approaches to further dissect the mechanisms governing spindle function and position. We describe two closely related proteins, GPR-1 and GPR-2 (G protein regulator), that associate with LIN-5 and contain GoLoco/GPR Gα interaction motifs. Importantly, inactivation of lin-5, gpr-1/gpr-2, or the Gαi/o subunits goa-1 and gpa-16 all cause highly similar defects in chromosome segregation and spindle positioning. Our results support a model in which GPR-1/GPR-2 are recruited to the spindle and cell cortex by LIN-5 and activate Gα protein signaling. These interactions appear necessary to generate the forces required in spindle and chromosome movements during mitosis. Polarity cues at the cell cortex act genetically upstream of lin-5 and gpr-1/gpr-2 and may locally regulate LIN-5/GPR/Gα to create asymmetric forces that alter spindle position during mitosis.
Results
LIN-5 and GPR-1/GPR-2 form part of a large-molecular-weight protein complex
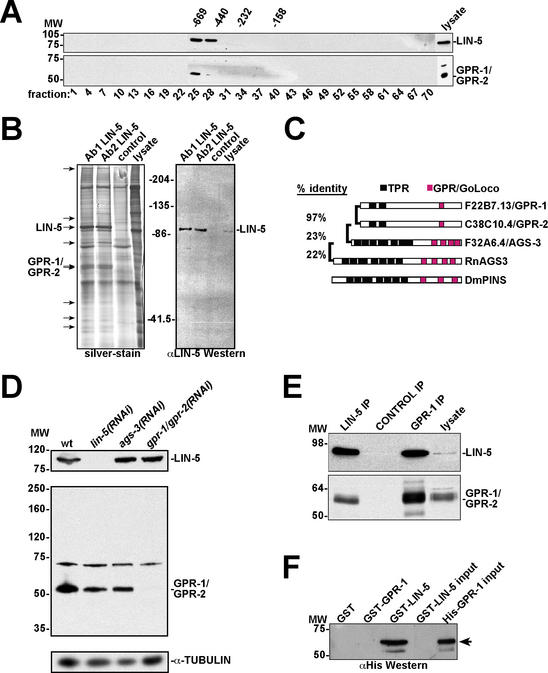
We set out to identify proteins that associate with LIN-5 in order to discover critical components of the spindle apparatus. Gel filtration chromatography revealed that LIN-5 forms part of a protein complex of ∼700 kD (Fig. 1A, top). To isolate proteins of this complex, we used anti-LIN-5 monoclonal antibodies for immunoprecipitation (IP) under low stringency conditions. Silver-stained SDS PAGE gels showed several proteins that were precipitated with two independent anti-LIN-5 monoclonal antibodies and not with control antibodies (Fig. 1B).
Figure 1.
A LIN-5 protein complex contains the GoLoco/GPR motif proteins GPR-1 and GPR-2. (A) Anti-LIN-5 (top panel) and anti-GPR-1 (bottom panel) Western blots of gel filtration chromatography fractions from C. elegans embryo lysates. LIN-5 and GPR-1/GPR-2 coelute in the early fractions. Numbers above panels are molecular weight markers in kilodaltons. Numbers below panels refer to fraction number. (B) Large-scale immunoprecipitation of LIN-5 for identification of interacting proteins. (Left) Silver-stained gel containing two LIN-5 IPs, the control (SD15 mAb) IP lane, and 50 μg lysate as indicated. Arrows point to protein bands in the LIN-5 IP lanes not observed in the control lane. Using tandem mass spectrometry, the ∼90-kD protein was identified as LIN-5 and the ∼60-kD doublet as GPR-1 and/or GPR-2. (Right) LIN-5 detected by immunoblotting, using 10% of the IPs. (C) Comparison between GPR-1, GPR-2, and other proteins containing TPR and GoLoco/GPR motifs. Rn, Rattus norvegicus; Dm, Drosophila melanogaster. C. elegans proteins are identified by their open reading frame designations and genetic names. (D) Anti-GPR antibody reactivity and specific elimination by RNAi. Western blots of RNAi embryo lysates using antibodies recognizing LIN-5 (top), GPR-1/GPR-2 (middle), and α-tubulin (loading control, bottom). (E) IP-Western blot demonstrating coprecipitation of LIN-5 and GPR-1/GPR-2 from C. elegans lysate. Antibodies indicated above the lanes were used in IPs followed by immunoblotting with anti-LIN-5 (top) and anti-GPR (bottom) antibodies. Input lysate (5%) was loaded on the right. (F) LIN-5 and GPR-1 interact directly in vitro. GST pulldowns with indicated proteins followed by immunoblotting with anti-His epitope antibody. GST-LIN-5 and His-GPR-1 input (5%) are loaded on the right.
Proteins eluted from the gels were subjected to tandem mass spectrometry. The major 90-kD band in the LIN-5 IP lanes was confirmed as LIN-5. In addition, a doublet of 60 kD apparent molecular weight (MW) was identified as a product of the predicted genes F22B7.13 and C38C10.4 (Fig. 1B). These genes are 96% identical at the nucleotide level and encode proteins that share 97% amino acid identity. The N-terminal part of the predicted proteins contains two domains weakly similar to tetratricopeptide (TPR) motifs, which are involved in protein–protein interactions (Fig. 1C). In addition, the C-terminal part contains a GoLoco/GPR (G protein regulatory) motif (Siderovski et al. 1999). The F22B7.13 and C38C10.4 products are not closely related to any specific GoLoco/GPR motif protein, and these genes have been named gpr-1 and gpr-2, respectively. In the following, gpr-1/gpr-2 will be referred to simultaneously when the close sequence identity of these genes prevented distinguishing between them in our experiments. A number of proteins in other metazoans contain both TPR and GoLoco motifs in various numbers, and several GoLoco motif proteins, including mammalian AGS3 and Drosophila Pins (Fig. 1C), have been shown to interact with Gαi/o subunits of heterotrimeric G proteins (Takesono et al. 1999; Schaefer et al. 2001). The closest C. elegans homolog of mammalian AGS3 is F32A6.4, which has been named ags-3.
We generated an affinity-purified rabbit antiserum against His-tagged full-length GPR-1 protein isolated from bacteria. This antiserum recognized proteins of ∼60 kD and 70 kD molecular weight in Western blotting experiments (Fig. 1D, middle). The faster migrating protein matches the predicted MW (60.6 kD) and apparent MW (Fig. 1B) of GPR-1/GPR-2, and this protein band was eliminated specifically by gpr-1/gpr-2 RNAi (Fig. 1D, middle). In contrast, RNAi of the GoLoco/GPR motif gene ags-3 did not affect the reactivity of anti-GPR-1 antiserum in Western blotting experiments. RNAi of lin-5 eliminated detection of LIN-5 protein but did not affect GPR-1 levels (Fig. 1D, top). These results confirm the specificity of the anti-GPR-1 antiserum.
To confirm that GPR-1/GPR-2 are in a complex with LIN-5, we performed gel filtration chromatography followed by Western blotting and observed that the majority of GPR-1/GPR-2 elutes in the same fractions as LIN-5 (Fig. 1A, bottom). Moreover, LIN-5 and GPR-1/GPR-2 reciprocally coprecipitated in IP-Western blotting experiments (Fig. 1E). Finally, bacterially expressed GST-LIN-5 protein, but not GST-GPR-1 or GST alone, was able to directly bind purified His-tagged GPR-1 (Fig. 1F). Together, these experiments establish that LIN-5 and GPR-1/GPR-2 form part of a protein complex and can bind each other directly.
LIN-5 and GPR-1/GPR-2 function together
If GPR-1/GPR-2 and LIN-5 function together in vivo, inactivation of either gpr-1/gpr-2 or lin-5 could cause a similar phenotype. In a large-scale RNAi screen, Gönczy et al. (2000) revealed essential roles for F22B7.13 (gpr-1) and C38C10.4 (gpr-2) in positioning the mitotic spindle in the early embryo. To determine functional similarity, we examined the lin-5 and gpr-1/gpr-2 RNAi phenotypes in detail and compared these phenotypes to the wild type.
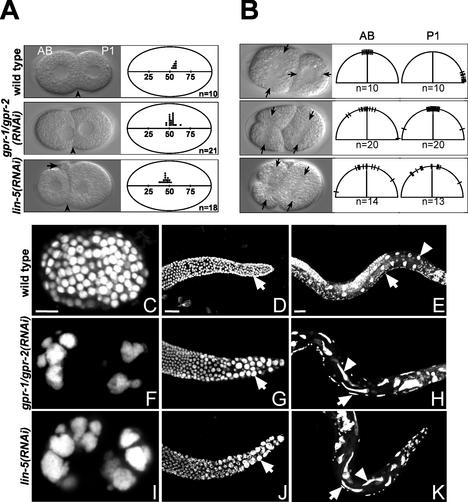
During wild-type C. elegans development, the oocyte-derived nucleus completes meiosis I and II after fertilization, and one polar body is extruded after each meiotic telophase (Albertson and Thomson 1993). Subsequently, a maternal pronucleus is formed in the anterior, which migrates toward the paternal pronucleus and meets it in the posterior (White and Strome 1996). After duplication of the paternally derived centrosomes, the two pronuclei and associated centrosomes move to the center of the egg and rotate together (centration). The first mitotic spindle is set up along the A/P axis in the center of the zygote, but is pulled toward the posterior of the embryo during anaphase. The posterior spindle pole oscillates and flattens during this process, and the anterior spindle pole remains relatively fixed in position. The asymmetric position of the spindle causes the first cleavage to produce two unequal cells: a larger anterior cell, AB, and a smaller posterior cell, P1 (Fig. 2A, top). Following this division, the centrosomes duplicate in both daughters and migrate to opposite sides of the nucleus, transverse to the A/P axis. In the AB cell, the spindle is formed in this direction and a transverse division follows. In the P1 cell, the nucleocentrosomal complex rotates 90° and the subsequent division is longitudinal (Fig. 2B, top). The fixed shape of the eggshell further helps to position the blastomeres correctly.
Figure 2.
Close similarity between gpr-1/gpr-2 and lin-5 loss-of-function phenotypes. (A,B) Defects in spindle movements. (Left) Representative DIC images. AB and P1 cells are marked. (A) Spindle positioning defects in the first mitotic division. Arrowheads mark the position of the cleavage plane. Note the abnormally large polar body in the lin-5(RNAi) embryo (arrow). (Right) Each dot represents the cleavage plane position relative to egg length in a single embryo. (B) Failure in nucleocentrosomal rotation. Arrows mark the position of the spindle. (Right) The spindle axes in AB and P1 are denoted by hatches on the half circles. n, number of embryos examined. (C–K). gpr-1/gpr-2 and lin-5 are required for all cell divisions in C. elegans. DNA stained with propidium iodide in wild-type (C–E), gpr-1/gpr-2(RNAi) (F–H), and lin-5(RNAi) (I–K) specimens. (C,F,I) Each image shows a single embryo. (D,G,J) Mitotic region of gonads from adults subjected to 3 d of RNAi feeding. Arrows point to single nuclei. (E,H,K) rrf-3(pk1426) animals after 3 d of RNAi feeding. Arrowheads point to nuclei of intestinal (In) cells and arrows mark nuclei of ventral cord precursor (P) cells that failed to undergo the normal postembryonic division in these lineages (E). Severely affected gpr-1/gpr-2(RNAi) animals have an average of 20.4 In nuclei and 36.6 P cells (P2–P10; n = 10). Similarly aged wild-type animals have 33 or 34 In nuclei and 48 P2–P10 cells (Sulston and Horvitz 1977). Bars, 10 μm.
Inactivation of gpr-1/gpr-2 using RNAi resulted in defects as early as the one-cell stage. Formation, meeting position, and migration of the pronuclei were normal, but in 4/19 embryos, the pronuclei and associated centrosomes rotated incompletely before the onset of mitosis. During anaphase, chromosome segregation was initiated, but oscillation and flattening of the posterior spindle pole were completely absent in 19 of 20 embryos (Table 1). Anterior and posterior spindle pole movement remained limited during anaphase B. Consequently, cell division resulted in AB and P1 cells of equal size in approximately half of the embryos (Fig. 2A, middle). The reformed nuclei in AB and P1 remained close to the cleavage plane for an abnormally long period of time (Fig. 2A, middle; data not shown). During the completion of cytokinesis, cytoplasmic blebbing at the cleavage plane was frequently observed.
Table 1.
gpr-1/gpr-2 and lin-5 are required for embryonic spindle movements in mitosis
|
|
P0 spindle position
|
Anterior aster oscillation/flattening
|
Posterior aster oscillation/flattening
|
Spindle rotation
|
|---|---|---|---|---|
| N2 | + | − | + | + |
| par-2a | − | − | − | − |
| gpr-1/gpr-2(RNAi) | − | − | − | − |
| lin-5(ev571ts) or lin-5(RNAi) | − | − | − | − |
| goa-1 gpa-16(RNAi) | − | − | − | − |
| par-3 or par-3;par-2a | − | + | + | +/+b |
| lin-5(RNAi);par-3(it71) | − | − | − | − |
| par-3(it71) gpr-1/gpr-2(RNAi) | − | − | − | − |
See text for numbers.
Spindle rotation is observed in both AB and P1. In wild-type embryos, only the P1 spindle rotates.
On reappearance at the two-cell stage, the duplicated centrosomes were often positioned incorrectly. The spindles in AB and P1 were formed without active rotation of the nucleocentrosomal complex (Fig. 2B, middle). Mitosis in AB and P1 was initiated somewhat more concurrently than usual, with P1 lagging AB by only 82.4 sec ± 5.5 sec (S.E.M.) compared with 126 sec ± 6.0 sec (S.E.M.) in wild-type embryos. Again, the movement of chromosomes and poles during anaphase B was always limited. Using a strain expressing green fluorescent protein fused to the β-tubulin protein (TBB-2∷GFP), we observed that the cleavage plane frequently displaces the midzone MTs toward the cortex, apparently breaking the midspindle and dragging the reforming nuclei toward the cleavage plane (data not shown). After cell cleavage, the nuclei remained in close proximity of each other and the cell cortex. Chromosome segregation and cytokinesis failed completely during one of the next divisions. As DNA replication continued, gpr-1/gpr-2(RNAi) embryos arrested with just a few polyploid nuclei (Fig. 2, cf. C and F).
All defects described earlier for gpr-1/gpr-2(RNAi) embryos were also seen in lin-5(RNAi) embryos and lin-5(ev571ts) embryos at the nonpermissive temperature (Fig. 2A,B,I; Table 1; Lorson et al. 2000). However, lin-5 embryos displayed two additional defects. Meiosis was generally abnormal in lin-5(RNAi) embryos, as evidenced either by abnormally large polar bodies (Fig. 2A arrow, bottom left) or the presence of multiple maternal pronuclei (data not shown). These extra maternal pronuclei also migrated to the posterior and fused with the paternal pronucleus. The lin-5 and gpr-1/gpr-2 loss-of-function phenotypes also deviated during anaphase of the first mitosis. While the posterior spindle pole failed to migrate and oscillate in both types of embryos, the anterior pole slowly migrated toward the anterior cortex late in anaphase in 18 of 19 lin-5(RNAi) embryos (Fig. 2A, bottom). This shift in spindle position toward the anterior resulted in embryos containing an AB blastomere smaller than P1. Double RNAi for lin-5 and gpr-1/gpr-2 resulted in embryos with the lin-5-associated anterior aster migration phenotype (data not shown), suggesting that lin-5 normally antagonizes this movement independent of gpr-1/gpr-2 function. All other aspects of early cell division were indistinguishable between lin-5(RNAi) and gpr-1/gpr-2(RNAi) embryos.
The similarities between lin-5 and gpr-1/gpr-2 loss-of-function phenotypes were seen in postembryonic as well as embryonic divisions. In adult animals, mitosis occurs only in the distal ends of the gonad arms. Feeding-induced lin-5 or gpr-1/gpr-2 RNAi of adult hermaphrodites resulted in fewer nuclei in the distal gonad with polyploid DNA and increased numbers of associated centrosomes (Fig. 2D,G,J; data not shown). In addition, following feeding RNAi, lin-5(RNAi) and gpr-1/gpr-2(RNAi) larvae showed division defects and polyploidy in all postembryonic lineages, with some lineage-dependent variation (Fig. 2E,H,K, legend). The reduced cell numbers and polyploidy seen in gpr-1/gpr-2 animals match the Lin-5 phenotype and indicate continued rounds of DNA replication in the absence of completed mitoses (Fig. 2I; Lorson et al. 2000). Thus, gpr-1/gpr-2 not only act in spindle positioning but also mediate chromosome segregation, pointing toward a more general spindle function. The close similarity in phenotype, combined with the observed co-IP, strongly suggests that the spindle functions of LIN-5 and GPR-1/GPR-2 depend on their direct interaction.
The localizations of GPR-1/GPR-2 overlap with and depend on LIN-5
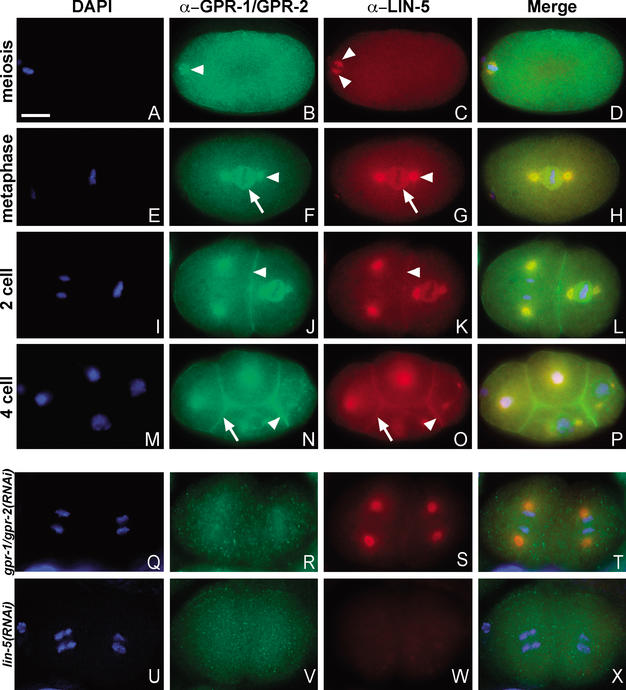
To further examine the interaction between LIN-5 and GPR-1/GPR-2 in vivo, we determined their subcellular localizations. The affinity-purified GPR-1 antiserum showed a distinct staining pattern that was eliminated by gpr-1/gpr-2 RNAi. Therefore, we believe that this staining reflects the localization of GPR-1/GPR-2 (Fig. 3).
Figure 3.
GPR-1/GPR-2 localize to the mitotic spindle and cell cortex in a LIN-5-dependent manner. DNA staining with DAPI (blue) and immunostaining of GPR-1/GPR-2 (green) and LIN-5 (red). Merged images are shown in D, H, L, P, T, and X. (A–D) Fertilized embryo in meiosis II. LIN-5, but not GPR-1/GPR-2, showed specific localization to the meiotic spindle (arrowheads, B,C). (E–H) One-cell embryo in metaphase. GPR-1/GPR-2 (F) and LIN-5 (G) colocalize to the spindle asters (arrowheads) and kinetochore MTs (arrows). (I–L) Two-cell embryo. GPR-1/GPR-2 (J) and LIN-5 (K) colocalize to the spindle apparatus as well as the cell cortex (arrowheads). (M–P) Four-cell embryo. In addition to spindle staining, both proteins showed stronger staining at the P2/EMS boundary (arrowhead) than at other cell membranes (arrow). (Q–X) Localization in RNAi embryos. (Q–T) GPR-1/GPR-2 staining is strongly reduced but not fully eliminated in gpr-1/gpr-2(RNAi) embryos. (S) LIN-5 staining remained at the spindle apparatus but appeared reduced at the cell periphery. (U–X) lin-5 RNAi eliminated LIN-5 staining (W) and disrupted GPR-1/GPR-2 localization (V). Bar, 10 μm.
During all mitotic divisions examined, GPR-1/GPR-2 and LIN-5 were present at the cell cortex and spindle asters (Fig. 3E–P; Lorson et al. 2000). GPR-1/GPR-2 and LIN-5 were detected at the spindle asters and at the membranes between germ-precursor nuclei in the distal gonad arms (data not shown). During the formation and maturation of oocytes, GPR-1/GPR-2 and LIN-5 localized diffusely to the cytoplasm and more prominently to the nuclear and cytoplasmic membranes, in particular to membranes between adjacent oocytes. Following fertilization and meiosis, GPR-1/GPR-2 and LIN-5 appeared at the duplicated centrosomes associated with the sperm pronucleus (data not shown). GPR-1/GPR-2 and LIN-5 both became progressively more abundant at the spindle asters during the formation of the first mitotic spindle (Fig. 3E–H). Both proteins also localized diffusely around the kinetochore MTs in metaphase (Fig. 3E–H). Although the latter localization disappeared in early anaphase, GPR-1/GPR-2 and LIN-5 persisted at the spindle asters until chromosome decondensation in telophase. On completion of cell cleavage, GPR-1/GPR-2 and LIN-5 were detected only at the cell cortex and cytoplasm. This pattern of spindle-associated localizations was repeated during subsequent mitotic divisions (Fig. 3I–L). The cortical localization appeared enriched between blastomeres, especially the cortical staining of LIN-5 (Fig. 3K). We did not detect other asymmetries in localization until the four-cell stage. At that stage, both LIN-5 and GPR-1/GPR-2 showed significant accumulation at the boundary between the EMS and P2 blastomeres (Fig. 3N,O; see below). Similar asymmetries were detected during some of the subsequent divisions (data not shown). The overlapping localizations of LIN-5 and GPR-1/GPR-2 further support their concerted mitotic function.
Unlike LIN-5, GPR-1/GPR-2 were not detected at the meiotic spindle. In 15/18 embryos, GPR-1/GPR-2 antiserum diffusely stained the maternal pronucleus or condensed meiotic chromosomes (Fig. 3B). However, this staining is likely not specific, as similar staining was seen in 9/16 gpr-1/gpr-2(RNAi) embryos. In contrast, LIN-5 was abundantly present at the polar regions of the meiotic spindle (Fig. 3C). These different localizations correspond to the different meiotic requirements for GPR-1/GPR-2 and LIN-5.
In addition to the overlap in LIN-5 and GPR-1/GPR-2 localizations, GPR protein localization required LIN-5. Following gpr-1/gpr-2 RNAi, GPR staining was mostly eliminated from the spindle and cortex, but LIN-5 was still present at the spindle asters (Fig. 3Q–T). LIN-5 also remained detectable at the cell periphery, although the level appeared reduced. In the reverse experiment, lin-5 RNAi not only eliminated LIN-5 detection, but it also eliminated the spindle and cortical localizations of GPR-1/GPR-2 (Fig. 3U–X). Western blotting experiments demonstrated that GPR-1/GPR-2 protein levels are similar in the presence or absence of LIN-5 (Fig. 1D), excluding effects on protein stability. Together, our results indicate that the coiled-coil protein LIN-5 acts to recruit or anchor the GPR-1/GPR-2 proteins to the spindle and cortex.
LIN-5 and GPR-1/GPR-2 likely act downstream of cell polarity cues in spindle positioning
LIN-5 and GPR-1/GPR-2 are required for the proper positioning of the mitotic spindle during the early embryonic cell divisions. Because polarity defects also lead to abnormal spindle positioning, LIN-5 and GPR-1/GPR-2 could act to establish or maintain cell polarity, or they could respond to cell polarity established by the par genes in positioning the spindle. To distinguish between these possibilities, we examined whether lin-5 and gpr-1/gpr-2 act upstream or downstream of the par genes.
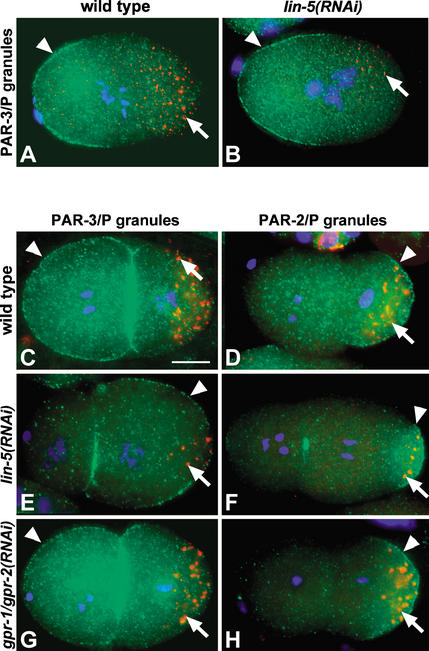
In immunostaining experiments, LIN-5 and GPR-1/GPR-2 were detected at the cortex and spindle in par-1(b274), par-2(it5), par-3(it71), par-4(it47), and par-6(zu222) mutant embryos (data not shown). Thus, these localizations do not depend on proper cell polarity. In lin-5 or gpr-1/gpr-2 RNAi embryos, PAR-1, PAR-2, PAR-3, PAR-6, P granules, PAR-2∷GFP, and PIE-1∷GFP all showed normal asymmetric localization patterns starting from female meiosis and becoming fully established by anaphase (Fig. 4A,B; data not shown; at least 20 embryos examined per experiment). Thus, the establishment of polarity does not require lin-5 or gpr-1/gpr-2. However, at the two-cell stage, PAR-3 and PAR-6 were abnormally localized to the posterior rather than the anterior blastomere in a subset of lin-5(RNAi) embryos. This abnormal localization correlated with the cleavage plane: 36/40 embryos with AB smaller than P1 showed PAR-3 localization in P1, compared with 0/28 embryos with AB equal to or larger than P1 (Fig. 4, cf. C and E; data not shown). In some embryos, PAR-2 and PAR-3/PAR-6 localizations still appeared mutually exclusive, whereas these PAR protein localizations appeared to overlap in other embryos (Fig. 4D,F; data not shown).
Figure 4.
gpr-1/gpr-2 and lin-5 do not determine polarity in the early embryo. Embryos are shown with P granules in red (arrows), DNA in blue (DAPI), and either PAR-3 (A–C,E,G) or PAR-2 (D,F,H) in green. (A,B) lin-5 is not required to establish polarity in the one-cell embryo. PAR-3 (arrowheads) localizes to the anterior cortex in wild-type (A) and in lin-5(RNAi) (B) one-cell embryos, and P granules localize to the posterior in both embryos. (C,D) Two-cell wild-type embryos. PAR-3 localizes to the entire cortex of the anterior AB cell and the anterior region of the posterior P1 cell (C, arrowhead). PAR-2 (D, arrowhead) and P granules (arrows) localize to the posterior in P1. (E,F) Two lin-5(RNAi) embryos in which AB is smaller than P1. PAR-3 (E, arrowhead) localized to the P1 cortex and posterior of AB. PAR-2 (F, arrowhead) localized to the posterior cortex of P1, although at times in a more restricted area. P granules show wild-type localization. The punctate PAR staining at the cortex between the blastomeres is not reproducible. (G,H) Two gpr-1/gpr-2(RNAi) embryos with AB and P1 of similar size. PAR-3 (G, arrowhead), PAR-2 (H, arrowhead), and P granules (arrows) localized like in wild type. Bar, 10 μm.
The mislocalizations of PAR-3 and PAR-6 could either reflect a polarity-maintenance function of lin-5 or be caused by the abnormal cell-division plane. The latter possibility is supported by the apparently normal polarity of lin-5(RNAi) embryos throughout the one-cell stage (Fig. 4A,B) and the normal PAR-3/PAR-6 localizations in all two-cell embryos in which AB was larger or similar in size to P1. In addition, PAR-3 was found at the cortex between all blastomeres at later stages, as in wild type, rather than segregating only to the periphery of the P cells like PAR-2. Moreover, PAR-3 and PAR-6 localizations were normal in gpr-1/gpr-2(RNAi) embryos, which rarely have P1 cells larger than AB (Fig. 4G,H). Based on these results, it is unlikely that lin-5 and gpr-1/gpr-2 act in spindle positioning by affecting cell polarity.
To further examine if lin-5 and gpr-1/gpr-2 act downstream of the par genes in spindle positioning, we carried out genetic epistasis experiments. In embryos lacking maternal par-3 function, or par-2 and par-3 together, both spindle asters oscillate and flatten at the one-cell stage and the spindle rotates in both AB and P1 at the two-cell stage (Table 1; Cheng et al. 1995). These results are consistent with a model in which spindle migration and rotation are the default state, and par-3 acts to inhibit spindle movement in the anterior (Cheng et al. 1995). In time-lapse differential interference contrast (DIC) recordings, we observed anterior migration of the spindle without oscillation or flattening in 6/7 lin-5(RNAi);par-3(it71) mutant embryos (Table 1). In addition, the spindle did not rotate in either AB or P1 in 7/7 lin-5(RNAi);par-3(it71) embryos. Similarly, the spindle in P0 failed to oscillate and flatten in 9/9 par-3(it71) gpr-1/gpr-2(RNAi) mutant embryos, and neither AB nor P1 showed spindle rotation in 12/17 of these embryos (compared with spindle rotation in both AB and P1 in 56% of par-3 embryos (Cheng et al. 1995)). In sum, the spindle movement defects in lin-5 and gpr-1/gpr-2 RNAi embryos are not affected by par gene function, and PAR proteins can localize normally in the absence of lin-5 or gpr-1/gpr-2 function. Thus, lin-5 and gpr-1/gpr-2 must act downstream of par-3, or in parallel, in positioning the mitotic spindle.
In contrast to spindle movement, several characteristics of the par-3 mutant phenotype were visible in the absence of lin-5 or gpr-1/gpr-2 function. AB and P1 divided completely synchronously in 8/8 lin-5(RNAi);par-3(it71) and 15/16 par-3(it71) gpr-1/gpr-2(RNAi) embryos, and P granules failed to localize to the posterior in all lin-5(RNAi);par-3(it71) and par-3(it71) gpr-1/gpr-2(RNAi) embryos examined. Thus, lin-5 and gpr-1/gpr-2 act downstream of the par genes exclusively in spindle positioning.
Asymmetric localization of GPR-1/GPR-2 correlates with spindle rotation in EMS
The asymmetric localization of the Pins GoLoco-motif protein has been implicated in the control of asymmetric division in D. melanogaster (for review, see Knoblich 2001). We did not observe apparent asymmetries in GPR-1/GPR-2 or LIN-5 localization at any point during the first two rounds of embryonic cell divisions, in which the spindle position is determined cell autonomously. Interestingly, both LIN-5 and GPR-1/GPR-2 were asymmetrically distributed at the cortex at later stages, as antibody staining revealed enrichment between the P2 and EMS blastomeres and between the P3 and E blastomeres (Fig. 5A,E; data not shown). At the four-cell stage, the posterior blastomere P2 induces spindle rotation in its anterior sister cell EMS, causing it to divide along the A/P axis and generate daughters with different fates. Similarly, cell–cell signaling between P3 and E also determines spindle position in E (Goldstein 1995). Thus, the accumulation of cortical LIN-5/GPR correlates with extrinsic control of spindle orientation.
Figure 5.
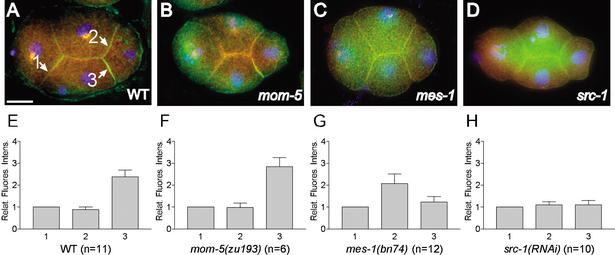
LIN-5 and GPR-1/GPR-2 enrichment at the P2/EMS boundary requires mes-1 and src-1. (A–D) Merged images, showing staining of LIN-5 (red), GPR-1/GPR-2 (green), and DNA (DAPI, blue). LIN-5 and GPR-1/GPR-2 are enriched between P2 and EMS (arrow marked 3) in wild-type (A) and mom-5(zu193) (B) embryos, but not in mes-1(bn74) (C) and src-1(RNAi) (D) embryos. (E–H) Measurements of GPR-1/GPR-2 relative fluorescence intensity for the AB.a/EMS (1), AB.p/P2 (2), and P2/EMS (3) cell boundaries. GPR-1/GPR-2 is enriched two- to threefold at the P2/EMS boundary (E). This enrichment is lost in mes-1(bn74) (G) and src-1(RNAi) (H) embryos but not in mom-5(zu193) (F) embryos. Error bars indicate standard error of the mean. Bar, 10 μm.
P2/EMS signaling involves two redundant pathways that act in parallel to rotate the spindle apparatus in EMS and to specify the endoderm fate: a mom-2/mom-5 Wnt ligand/Frizzled (Fz) receptor cascade and a second pathway consisting of the tyrosine-kinase related genes mes-1 and src-1 c-Src (Bei et al. 2002). To examine whether mom-2/mom-5 or mes-1/src-1 pathways promote the asymmetric localizations of GPR-1/GPR-2 and LIN-5, we examined embryos mutant for components of one of the two pathways. In mom-5(zu193) frizzled mutant embryos, both LIN-5 and GPR-1/GPR-2 were still enriched at the P2/EMS boundary (Fig. 5B,F). In contrast, mes-1(bn74) embryos showed either no enrichment or enrichment at both the P2/EMS and P2/AB.p boundaries (Fig. 5C,G). Inactivation of the src-1 gene by RNAi caused a similar loss of asymmetric distribution (Fig. 5D,H). These results indicate that the mes-1/src-1 pathway acts to enrich LIN-5 and GPR-1/GPR-2 between P2 and EMS, which may contribute to spindle positioning in EMS. Thus, both cell intrinsic and cell extrinsic polarity signals may act on LIN-5 and GPR-1/GPR-2 to position the spindle.
Functional and physical interactions between LIN-5, GPR, and heterotrimeric G proteins
The G protein regulatory motif, or GoLoco domain, predicted within GPR-1/GPR-2 was previously found in a number of proteins that regulate the Gαi/o subunit of heterotrimeric G proteins (Fig. 1C; Siderovski et al. 1999). G protein heterotrimers consist of Gβγ modules associated with GDP-bound Gα (Gilman 1987). GoLoco motif proteins interact with specific Gαi/o subunits in the GDP-bound state, inhibit GDP dissociation, and compete with Gβγ for binding (Bernard et al. 2001; Natochin et al. 2001; Schaefer et al. 2001; Kimple et al. 2002). Like gpr-1/gpr-2 RNAi, mutation of the C. elegans goa-1 Gαi/o gene disrupts spindle positioning in early embryos, albeit with partial penetrance (Miller and Rand 2000). Importantly, double RNAi of two related Gαi/o subunits, goa-1 and gpa-16, causes spindle defects that are indistinguishable from gpr-1/gpr-2(RNAi) (Fig. 6A,B; Gotta and Ahringer 2001). As in gpr-1/gpr-2(RNAi) embryos, meiosis and mitotic spindle assembly were apparently normal, but spindle movement to the posterior was reduced in all goa-1 gpa-16(RNAi) embryos, and oscillation and flattening of the posterior spindle aster were absent (Table 1, cf. Figs. 6A and 2A). During the second round of division, active rotation of the nucleocentrosomal complex was absent in all embryos examined (n = 9, Fig. 6B). Embryos lacking goa-1 and gpa-16 function failed in the next rounds of cell division and arrested with few and polyploid nuclei (Fig. 6C). Thus, the terminal phenotype of these embryos was also similar to gpr-1/gpr-2(RNAi), lin-5(ev571ts), and lin-5(RNAi) embryos (Fig. 2F,I; Lorson et al. 2000). Finally, nuclei in the distal ends of the gonads failed in mitosis, and polyploid nuclei accumulated in adult animals following goa-1 gpa-16 RNAi (cf. Figs. 6D and 2D). The close similarity of the gpr-1/gpr-2 and Gαi/o loss-of-function phenotypes and the presence of a Gαi/o binding domain in the GPR proteins suggest that GPR-1/GPR-2 act as positive regulators of G protein signaling.
Figure 6.
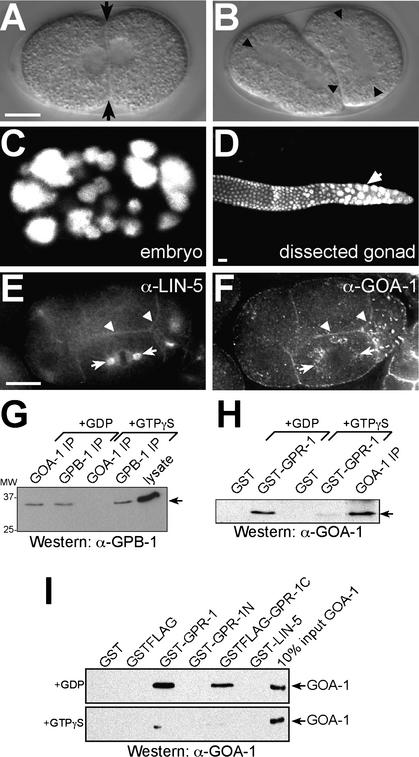
The Gαi/o subunits goa-1 and gpa-16 act with gpr-1/gpr-2 and lin-5 in spindle and chromosome movements. (A–D) The goa-1 gpa-16 double RNAi phenotype closely resembles the lin-5 and gpr-1/gpr-2 phenotype (cf. Fig. 2). DIC images illustrating symmetric first division (A) and lack of spindle rotation in P1 (B). (C,D) goa-1 and gpa-16 are required for cell divisions in C. elegans. DNA was stained with propidium iodide. (C) goa-1 gpa-16(RNAi) embryo containing polyploid nuclei and few cells. (D) Mitotic germ nuclei in distal gonad arm of adults treated with goa-1 gpa-16 RNAi fail in mitosis and become polyploid. (E,F) GOA-1 (F) colocalizes with LIN-5 (E) and GPR-1/GPR-2 (see Fig. 3N) at the cell membrane (arrowheads). Punctate staining of GOA-1 peripheral to spindle asters is observed (F, arrows), but this staining does not overlap with the spindle apparatus (E, arrows). (G–I) GOA-1 interacts with GPB-1 Gβ and GPR-1 in a GDP-dependent manner. (G) GPB-1 Western blot of IPs from embryo lysates containing either 100 μM GDP or GTPγS. Input (10%) is shown on the right. (H) GOA-1 Western blot of GST pulldowns from lysates containing GDP or GTPγS. A GOA-1 IP is shown on the right. (I) Direct association between GPR-1 and GOA-1·GDP. GOA-1 Western blot of GST pulldowns from equal amounts of pure GOA-1 preloaded with GDP (top panel) or GTPγS (bottom panel). GST-GPR-1 and the C terminus of GPR-1 containing the GoLoco motif (GSTFlag-GPR-1C) interact more efficiently with GOA-1·GDP than with GOA-1·GTPγS. (GST-GPR-1N) N terminus of GPR-1. Input (10%) is shown on the right. Bars, 10 μm.
As previously proposed, GPR/GoLoco motif proteins could activate G protein signaling by releasing Gβγ proteins from the heterotrimer (Natochin et al. 2001; Schaefer et al. 2001). This implies that loss of gpr function should mimic loss of Gβγ function. However, the previously reported loss of Gβγ function phenotype was clearly distinct from loss of gpr-1/gpr-2, lin-5, or goa-1 gpa-16 (Zwaal et al. 1996; Gotta and Ahringer 2001). In our experiments, gpb-1 Gβ RNAi, either alone or in combination with RNAi or mutational inactivation of gpb-2 Gβ5, resulted in incomplete centration (gpb-1, 5/5 embryos; gpb-2;gpb-1, 10/13 embryos) and abnormal nuclear rocking (gpb-1, 5/5 embryos; gpb-2;gpb-1, 10/12 embryos). In addition, the anterior aster oscillated slightly without flattening during anaphase (gpb-1, 5/5 embryos; gpb-2;gpb-1, 10/15 embryos). However, posterior aster oscillation, flattening, and migration were normal, in contrast to the phenotype of gpr-1/gpr-2, lin-5, or goa-1 gpa-16 embryos. Division of P0 was still asymmetric and spindle rotation in P1 still occurred, albeit later than normal (data not shown; Zwaal et al. 1996; Gotta and Ahringer 2001). Additionally, triple RNAi of goa-1, gpa-16, and gpb-1 yielded both Gαi/o and Gβ phenotypes in the first two divisions (data not shown). Together, these results indicate that lin-5 and gpr-1/gpr-2 function in G protein signaling primarily through Gα activation rather than Gβγ release.
In immunostaining experiments, we observed overlapping localizations of GPR-1/GPR-2, LIN-5, and GOA-1 at the cell cortex. In addition, diffuse GOA-1 staining was observed surrounding the spindle asters, but not at the spindle asters themselves (Fig. 6E,F). The localizations of LIN-5 and GPR-1/GPR-2 were unaffected by goa-1 gpa-16 RNAi, and GOA-1 localization did not depend on the LIN-5 or GPR proteins (data not shown).
The partially overlapping localizations could allow interactions between the Gαi/o and GPR proteins. To examine whether GPR-1/GPR-2 interact preferentially with GDP-bound GOA-1, we added GDP or the slowly hydrolyzable GTP-analog GTPγS to lysates. As expected, the Gβ subunit GPB-1 coprecipitated with GOA-1 from lysates containing excess GDP but not GTPγS (Fig. 6G). Likewise, GST-GPR-1 precipitated endogenous GOA-1 in the presence or absence of excess GDP, but not from lysates containing GTPγS (Fig. 6H). GST-GPR-1 did not precipitate detectable amounts of GPB-1 Gβ, but did precipitate LIN-5 independent of guanine nucleotide (data not shown). In addition, the use of purified proteins expressed in bacteria demonstrated that GPR and Gαi/o proteins interact directly. GST-GPR-1 associated with purified GOA-1·GDP but not GOA-1·GTPγS (Fig. 6I). Further analysis showed that the GPR-1 C terminus containing the GoLoco domain was sufficient for this interaction (Fig. 6I). Together, these results demonstrate that GPR-1/GPR-2 act as canonical GPR/GoLoco proteins by binding GOA-1·GDP and that GDP-bound GOA-1 can interact either with Gβγ or with GPR-1/GPR-2.
Our in vivo and in vitro experiments support two different protein interactions for GPR-1/GPR-2: direct interaction with LIN-5, which determines its localization to the spindle and cortex, and direct interaction with GOA-1 Gαi/o, which depends on the nucleotide state of the G protein.
Discussion
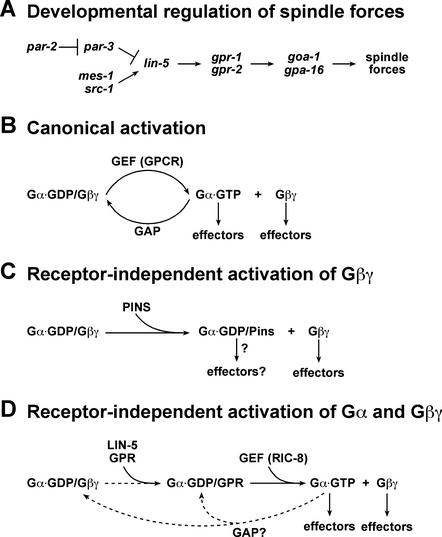
We have shown that the GoLoco/GPR motif proteins GPR-1/GPR-2 interact with the coiled-coil protein LIN-5 as well as a Gαi/o subunit of heterotrimeric G proteins. Both LIN-5 and GPR proteins are essential for chromosome and spindle movements, and LIN-5 is required for GPR-1/GPR-2 localization. Based on overlap in phenotype, presence of a GoLoco motif, and direct physical interaction with Gαi/o, GPR-1/GPR-2 act as regulators of heterotrimeric G protein signaling. LIN-5 and GPR-1/GPR-2 act genetically downstream of or parallel to the par and mes-1/src-1 spindle positioning genes. Our results suggest a pathway in which LIN-5/GPR/Gα act to regulate spindle forces (Fig. 7A). We will discuss this pathway, focusing on two important questions. What is the mechanism by which LIN-5/GPR affect G protein signaling? How do upstream components regulate LIN-5/GPR/Gα in positioning the spindle for asymmetric divisions?
Figure 7.
A model for GPR-mediated initiation of G protein signaling. (A) A pathway for spindle force regulation by LIN-5/GPR/Gα, which may be locally regulated by polarity genes. (B) Classical heterotrimeric G protein activation. Exchange of GDP for GTP associated with Gα is stimulated by the guanine nucleotide exchange activity of a GPCR. The heterotrimer is dissociated into two independent signaling modules, Gα·GTP and Gβγ, which are then competent to activate downstream effectors. (C) Activation of G protein signaling in the absence of a GPCR. A GoLoco/GPR motif protein, such as Pins, could compete with and dissociate Gβγ from the heterotrimer, thus initiating Gβγ signaling. It is not known if Gα·GDP/Pins is competent to signal. (D) Receptor-independent initiation of Gα·GTP signaling. The GPR-1/GPR-2 GoLoco proteins could dissociate Gβγ from the heterotrimer and allow the subsequent production of Gα·GTP through the activity of a GEF such as RIC-8. Additionally, free Gβγ could regulate its specific effectors. The signal could be attenuated by intrinsic or GAP-stimulated GTP hydrolysis, allowing for cycling activation of this pathway.
Heterotrimeric G proteins classically are known to transmit extracellular signals sensed by seven transmembrane receptors to targets within the cell (Gilman 1987). Upon ligand binding, the receptor stimulates release of GDP from the Gα subunit, which is then able to bind GTP. The Gα·GTP and Gβγ subunits then dissociate and can act on their respective downstream targets (Fig. 7B). Attenuation of the signal is achieved through hydrolysis of GTP by the intrinsic GTPase activity of Gα, which can be stimulated by GTPase-activating proteins (GAPs). Although G protein coupled receptors (GPCRs) mediating spindle positioning remain to be identified, our results and work by others point toward a receptor-independent mechanism for G protein activation mediated by GoLoco domain proteins. Biochemical evidence shows that the GoLoco motif acts as a guanine-nucleotide dissociation inhibitor (GDI), inhibiting the release of GDP from Gα (Bernard et al. 2001; Natochin et al. 2001). Moreover, the GoLoco domain competes with Gβγ for binding to Gα·GDP (Bernard et al. 2001; Natochin et al. 2001; Schaefer et al. 2001) and binds Gα in a region overlapping with Gβγ association (Kimple et al. 2002). Together, these results have led to a simple model in which binding of the GPR motif to Gα·GDP releases Gβγ, thus solely initiating Gβγ signaling (Fig. 7C; Takesono et al. 1999; Knust 2001; Schaefer et al. 2001). In contrast, our in vivo observations in C. elegans and some studies using Drosophila indicate that GoLoco proteins can also initiate Gα signaling.
In Drosophila, the GoLoco protein Pins activates G protein signaling to control spindle positioning in two model cell types for asymmetric division, embryonic neuroblasts and sensory organ precursor (SOP) cells (for review, see Knust 2001). It has been suggested that spindle orientation in neuroblasts is primarily mediated by Pins activation of Gβγ (Schaefer et al. 2001). These authors also suggest that spindle orientation in SOP cells requires Pins-mediated activation of both Gβγ and Gα. However, the mechanism by which Pins promotes Gα signaling and the specific contributions of Gα and Gβγ toward proper spindle orientation were not clear from these experiments.
Our results indicate that GPR-mediated spindle positioning occurs primarily through activation of Gα signaling in C. elegans. Loss of Gα, rather than Gβγ, mimicked the gpr-1/gpr-2 loss-of-function phenotype. Distinct phenotypes were observed following inactivation of Gα, Gβ, or both, indicating that they can be inactivated independently. Interestingly, another C. elegans gene, ric-8, is also required for embryonic spindle positioning and genetically interacts with goa-1 (Miller and Rand 2000). Recently, the mammalian RIC-8 homolog, RIC-8A/synembrin, has been shown to act as a guanine nucleotide exchange factor (GEF) for Gαo, Gαq, and Gαi1 subunits (Tall et al. 2003). The similar gpr-1/gpr-2 and ric-8 loss-of-function phenotypes indicate that both activate Gα signaling in C. elegans.
If GPR-1/GPR-2 act as GDIs and RIC-8 acts as a GEF, then how could these opposing activities have a similar effect on Gα signaling? An important clue might come from the observation that mammalian RIC-8A cannot bind the heterotrimeric GαGβγ complex (Tall et al. 2003). We propose a model in which GPR-1/GPR-2 competes with Gβγ for Gα·GDP association and subsequently allows RIC-8-mediated nucleotide exchange to produce active Gα·GTP independent of a GPCR (Fig. 7D). In this model, GPR and GEF proteins are both positive regulators of G protein signaling; GTP-bound Gα, rather than Gα·GDP, is the effector, consistent with all previous studies; and signaling could be attenuated by GTP hydrolysis. The previously described role in Gβγ activation is incorporated in this model, although our results suggest that Gβγ activation does not require GPR-1/GPR-2. This difference in Gβγ activation by GPR-1/GPR-2 and Pins may depend on cell type, expression levels, or Gα-binding affinity of the GPR/GoLoco protein. Receptor-independent activation of heterotrimeric G proteins allows signaling under cell intrinsic control and in close association with the spindle apparatus, which might be difficult to achieve through GPCRs.
In both Drosophila and C. elegans, a conserved PAR protein complex establishes cell polarity and spindle position but is not required for chromosome movements. This PAR-determined polarity directs spindle positioning possibly through activation of G protein signaling mediated by Pins/Inscuteable (Insc) in Drosophila neuroblasts, Pins/Discs large (Dlg) in Drosophila SOP cells, and GPR/LIN-5 in C. elegans embryos (Bellaiche et al. 2001; Schaefer et al. 2001; this paper). Although Insc, Dlg, and LIN-5 all act to localize GoLoco proteins, their functions and localizations differ. LIN-5, GPR-1/GPR-2, and Gαi/o interactions appear to be required for cell division and chromosome segregation, whereas no such role has been shown for Drosophila Gαi or Pins. Consistent with a role in chromosome movements, GPR-1/GPR-2 proteins localize to the spindle apparatus, whereas Pins does not. This may indicate that an additional spindle-associated GoLoco protein exists, and/or possibly that in Drosophila multiple Gα subunits act redundantly in mitosis, as in C. elegans. Consistent with the former hypothesis, a mammalian homolog of Pins, LGN, is required for spindle assembly and localizes to spindle asters (Du et al. 2001).
In order to achieve asymmetric spindle movements, uneven spindle forces must be generated. This can be accomplished by asymmetric localization of spindle force regulators, as is observed for Drosophila Gαi and Pins (Schaefer et al. 2000, 2001; Yu et al. 2000). Alternatively, G protein signaling could be asymmetrically activated. In our studies, we did not observe asymmetric localization of GPR-1/GPR-2, GOA-1, or LIN-5 in the first two rounds of embryonic cell divisions. Thus, other asymmetrically localized positioning determinants may locally regulate GPR-mediated G protein signaling. Later, at the four-cell stage, LIN-5 and GPR-1/GPR-2 localize asymmetrically dependent on mes-1/src-1, which regulate spindle position in EMS redundantly with Wnt/Fz signaling (Bei et al. 2002). This may more closely resemble Drosophila SOP cells in which Pins and Gαi also act redundantly with Fz to orient the mitotic spindle (Bellaiche et al. 2001; Schaefer et al. 2001). Together, our results suggest that both mechanisms, asymmetric regulation and asymmetric localization of spindle force regulators, dictate spindle position in C. elegans.
The spindle and chromosome movement defects seen when lin-5, gpr-1/gpr-2, or goa-1 gpa-16 are inactivated are consistent with reduced spindle forces. The spindle behavior in these embryos resembles what is seen in par-2 mutants, where reduced cortical spindle forces have been directly demonstrated (Cheng et al. 1995; Grill et al. 2001). When par-3 or both par-2 and par-3 are inactivated, high cortical pulling forces are present on both sides of the spindle (Grill et al. 2001) and these forces appear to depend on LIN-5 and GPR-1/GPR-2. This suggests that LIN-5, GPR-1/GPR-2, and Gα act to promote spindle forces and that local regulation of LIN-5/GPR/Gα activity could create asymmetric forces that move the spindle. Because spindle forces involve a coordination of MT dynamics, motor proteins, and cortical attachments, GPR/LIN-5/Gα and other LIN-5 complex components may regulate the spindle at either one or multiple levels.
Materials and methods
C. elegans culture conditions and strains
All strains used in this study were derived from the wild-type Bristol strain N2 and cultured under standard conditions (Brenner 1974) and are listed in the Supplemental Material. Alleles used here are par-1(b274), par-2(it5), par-3(it71), par-4(it47), par-6(zu222), mom-5(zu193), mes-1(bn74), goa-1 (n1134), gpb-2(sa603), and rrf-3(pk1426).
RNA-mediated interference
Double-stranded RNA produced from full-length cDNAs was introduced by either injection or feeding, and animals were harvested after 2 or 3 d, respectively, for phenotypic analysis (Fire et al. 1998; Timmons et al. 2001). Injection was used to inactivate more than one gene by RNAi. RNAi of ags-3 yielded no apparent phenotype.
Embryo lysates and gel filtration chromatography
For biochemical experiments, embryo protein lysates were created from large liquid cultures of N2 animals. For gel filtration chromatography, lysates made without detergent were loaded onto a preequilibrated Sephacryl S300-16/40 gel filtration column and 1.5-mL fractions were collected (see Supplemental Material for details).
Immunoprecipitations and identification of LIN-5 associated proteins
Mouse ascites from two anti-LIN-5 hybridomas, hel-1 and hel-2 (Lorson et al. 2000), was used in IP reactions, as well as SD15 ascites (negative control, anti-human p107, a kind gift from E. Harlow), anti-GPR-1 affinity-purified serum, anti-GOA-1 serum (a kind gift from M. Koelle), anti-GPB-1 serum (a kind gift from R. Plasterk), normal rabbit serum, and normal mouse serum. LIN-5 complexes were purified using hel-1 and hel-2 cross-linked, respectively, to Protein G or A Sepharose beads, which were then incubated with precleared embryo lysate. Specific coprecipitating proteins were identified by liquid chromatography followed by tandem mass spectrometric measurements of trypsinized peptides (Taplin Spectrometry Core, Harvard Medical School; see Supplemental Material for details).
Recombinant protein production and antiserum generation
A full-length cDNA clone of gpr-1 (yk103.a4, obtained from Y. Kohara) was used to express full-length or fragments of GPR-1 fused to either 10X Histidine, GST or GSTFlag epitopes in bacteria. Purified denatured full-length His-GPR-1 was used to generate and purify anti-GPR-1 serum.
GOA-1 guanine nucleotide loading and GST pulldowns
GOA-1 was loaded with either GDP or GTPγS (Sigma) as described previously (Dong et al. 2000): 100 ng GOA-1 (a generous gift from M. Koelle) was incubated at room temperature for 3 h in 100 μL 2× guanine nucleotide (GNP) loading buffer (20 mM HEPES at pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol) with either 10 μM GDP or GTPγS. This mixture was incubated with ∼500 ng GST fusion protein bound to glutathione agarose beads, incubated for 1.5 h at 4°C, washed extensively in 1× GNP loading buffer plus appropriate guanine nucleotide, boiled in sample buffer, and subjected to SDS-PAGE and Western blotting.
Antibody and propidium iodide staining
Immunostaining of C. elegans embryos was performed as previously described (Lorson et al. 2000). When staining for PAR proteins, embryos were fixed only in methanol. Washes were extended from 10 to 20 min for GPR-1/GPR-2 immunostainings. We used anti-LIN-5 hel-1 (diluted 1:2), and rabbit polyclonal sera of the following dilutions: anti-GPR-1/GPR-2 (1:100), anti-GOA-1 (1:100), anti-PAR-1 (1:50), anti-PAR-2 (1:10), anti-PAR-3 (1:30), and anti-PAR-6 (1:20; all PAR antisera kindly provided by K. Kemphues). Secondary FITC- or Texas red-conjugated antibodies were used at 1:100 dilution (Jackson ImmunoResearch Laboratories). To visualize DNA, we stained embryos with 1 μg/mL DAPI. For PAR-2 detection, the PAR-2∷GFP strain was used in combination with anti-PAR-2 antibody staining. To examine DNA content, we treated methanol/acetone-fixed embryos and Carnoy's-fixed larvae with 200 μg/mL RNase A overnight at room temperature and stained with 1 μg/mL propidium iodide. Animals were mounted on slides using glycerol with 10% phoshpate-buffered saline and 2.3% Dabco (Sigma).
Fluorescence intensity measurements
To determine relative fluorescence intensity (RFI), regions of interest of approximately the same area were drawn to encompass the cortices between AB.a and EMS (1), AB.p and P2 (2), and P2 and EMS (3), and the mean fluorescence intensity for each was recorded. Mean fluorescence intensities of equivalent areas adjacent to each cortex were subtracted to account for background fluorescence. The relative fluorescence intensity for the AB.p/P2 and P2/EMS cortex was determined as follows:
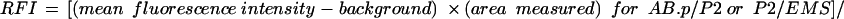 |
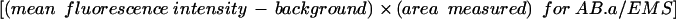 |
Acknowledgments
We are indebted to Michael Koelle and Ken Kemphues for reagents and helpful discussions, and to Steve Gygi for tandem mass spectrometry. We thank John White, Chris Malone, Yanxia Bei, and Craig Mello for strains; Anders Näär for GST expression vectors; Chidi Ezuma-Ngwu and Michaela Monaghan for help with hybridomas; Yuji Kohara and Marc Vidal for cDNA clones; and Ronald Plasterk for reagents. We are grateful to Jeff Settleman for helpful discussions and Mike Boxem, Abha Chandra, Anne Hart, Mako Saito, and Cindy Voisine for critically reviewing the manuscript. The Caenorhabditis Genetics Center, supported by the National Institutes of Health National Center for Research Resources, provided several strains for this work. This work was funded by a Howard Hughes Medical Institute Predoctoral Fellowship to R.M.F., a Massachusetts General Hospital Fund for Medical Discovery Predoctoral Fellowship to D.G.S., and grants to S.v.d.H. from the March of Dimes Birth Defects Foundation and the NIH (GM57990-05).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL heuvel@helix.mgh.harvard.edu; FAX (617) 724-9648.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1081203.
References
- Albertson DG, Thomson JN. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Albertson DG, Sulston JE, White JG. Cell cycling and DNA replication in a mutant blocked in cell division in the nematode Caenorhabditis elegans. Dev Biol. 1978;63:165–178. doi: 10.1016/0012-1606(78)90122-7. [DOI] [PubMed] [Google Scholar]
- Bei Y, Hogan J, Berkowitz LA, Soto M, Rocheleau CE, Pang KM, Collins J, Mello CC. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev Cell. 2002;3:113–125. doi: 10.1016/s1534-5807(02)00185-5. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Radovic A, Woods DF, Hough CD, Parmentier ML, O'Kane CJ, Bryant PJ, Schweisguth F. The Partner of Inscuteable/Discs-large complex is required to establish planar polarity during asymmetric cell division in Drosophila. Cell. 2001;106:355–366. doi: 10.1016/s0092-8674(01)00444-5. [DOI] [PubMed] [Google Scholar]
- Bernard ML, Peterson YK, Chung P, Jourdan J, Lanier SM. Selective interaction of AGS3 with G-proteins and the influence of AGS3 on the activation state of G-proteins. J Biol Chem. 2001;276:1585–1593. doi: 10.1074/jbc.M005291200. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NN, Kirby CM, Kemphues KJ. Control of cleavage spindle orientation in Caenorhabditis elegans: The role of the genes par-2 and par-3. Genetics. 1995;139:549–559. doi: 10.1093/genetics/139.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong MQ, Chase D, Patikoglou GA, Koelle MR. Multiple RGS proteins alter neural G protein signaling to allow C. elegans to rapidly change behavior when fed. Genes & Dev. 2000;14:2003–2014. [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- Du Q, Stukenberg PT, Macara IG. A mammalian Partner of Inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol. 2001;3:1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gilman AG. G proteins: Transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Goldstein B. Cell contacts orient some cell division axes in the Caenorhabditis elegans embryo. J Cell Biol. 1995;129:1071–1080. doi: 10.1083/jcb.129.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of spindle positioning: Focus on flies and worms. Trends Cell Biol. 2002;12:332–339. doi: 10.1016/s0962-8924(02)02306-1. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Echeverri G, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Gotta M, Ahringer J. Distinct roles for Gα and Gβγ in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat Cell Biol. 2001;3:297–300. doi: 10.1038/35060092. [DOI] [PubMed] [Google Scholar]
- Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple RJ, Kimple ME, Betts L, Sondek J, Siderovski DP. Structural determinants for GoLoco-induced inhibition of nucleotide release by Galpha subunits. Nature. 2002;416:878–881. doi: 10.1038/416878a. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Asymmetric cell division during animal development. Nat Rev Mol Cell Biol. 2001;2:11–20. doi: 10.1038/35048085. [DOI] [PubMed] [Google Scholar]
- Knust E. G protein signaling and asymmetric cell division. Cell. 2001;107:125–128. doi: 10.1016/s0092-8674(01)00534-7. [DOI] [PubMed] [Google Scholar]
- Lorson MA, Horvitz HR, van den Heuvel S. LIN-5 is a novel component of the spindle apparatus required for chromosome segregation and cleavage plane specification in Caenorhabditis elegans. J Cell Biol. 2000;148:73–86. doi: 10.1083/jcb.148.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Rand JB. A role for RIC-8 (Synembryn) and GOA-1 (G(o)alpha) in regulating a subset of centrosome movements during early embryogenesis in Caenorhabditis elegans. Genetics. 2000;156:1649–1660. doi: 10.1093/genetics/156.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natochin M, Gasimov KG, Artemyev NO. Inhibition of GDP/GTP exchange on G alpha subunits by proteins containing G-protein regulatory motifs. Biochemistry. 2001;40:5322–5328. doi: 10.1021/bi015505w. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Knoblich JA. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Diverse-Pierluissi M, De Vries L. The GoLoco motif: A Gαi/o binding motif and potential guanine-nucleotide exchange factor. Trends Biochem Sci. 1999;24:340–341. doi: 10.1016/s0968-0004(99)01441-3. [DOI] [PubMed] [Google Scholar]
- Straight AF, Field CM. Microtubules, membranes and cytokinesis. Curr Biol. 2000;10:R760–R770. doi: 10.1016/s0960-9822(00)00746-6. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 1998;125:3607–3614. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- Takesono A, Cismowski MJ, Ribas C, Bernard M, Chung P, Hazard S, III, Duzic E, Lanier SM. Receptor-independent activators of heterotrimeric G-protein signaling pathways. J Biol Chem. 1999;274:33202–33205. doi: 10.1074/jbc.274.47.33202. [DOI] [PubMed] [Google Scholar]
- Tall GG, Krumins AM, Gilman AG. Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. J Biol Chem. 2003;278:8356–8362. doi: 10.1074/jbc.M211862200. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Wallenfang MR, Seydoux G. Polarization of the anterior-posterior axis of C. elegans is a microtubule-directed process. Nature. 2000;408:89–92. doi: 10.1038/35040562. [DOI] [PubMed] [Google Scholar]
- White J, Strome S. Cleavage plane specification in C. elegans: How to divide the spoils. Cell. 1996;84:195–198. doi: 10.1016/s0092-8674(00)80974-5. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Hyman A, Desai A. The spindle: A dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:E28–E34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- Zwaal RR, Ahringer J, van Luenen HG, Rushforth A, Anderson P, Plasterk RH. G proteins are required for spatial orientation of early cell cleavages in C. elegans embryos. Cell. 1996;86:619–629. doi: 10.1016/s0092-8674(00)80135-x. [DOI] [PubMed] [Google Scholar]