Abstract
Generation of left–right asymmetry is an integral part of the establishment of the vertebrate body plan. Here we show that the Notch signaling pathway plays a primary role in the establishment of left–right asymmetry in mice by directly regulating expression of the Nodal gene. Embryos mutant for the Notch ligand Dll1 or doubly mutant for the Notch1 and Notch2 receptors exhibit multiple defects in left–right asymmetry. Analysis of the enhancer regulating node-specific Nodal expression revealed the presence of binding sites for the RBP-J protein, the primary transcriptional mediator of Notch signaling. Mutation of these sites destroyed the ability of this enhancer to direct node-specific gene expression in transgenic mice. Our results demonstrate that Dll1-mediated Notch signaling is essential for generation of left–right asymmetry, and that the Notch pathway acts upstream of Nodal expression during left–right asymmetry determination in mice.
Keywords: Notch signaling, Dll1, left–right determination, Nodal
Supplemental material is available at http://www.genesdev.org.
Generation of left–right asymmetry during development is an integral part of the establishment of the vertebrate body plan (Capdevila et al. 2000; Mercola and Levin 2001; Wright 2001; Yost 2001; Hamada et al. 2002). Specification of the left–right axis requires multiple steps: (1) generation of an initial asymmetric signal in or near the embryonic node, (2) transfer of asymmetric signals from the node to the lateral plate mesoderm (LPM), (3) induction of an evolutionarily conserved cascade of gene expression in the left LPM, and (4) transformation of these left–right asymmetric signals into morphological asymmetries of the visceral organs. In mice, generation of the initial asymmetric signal requires directional fluid flow on the ventral surface of the node (Nonaka et al. 2002). This fluid flow is generated by motile monocilia on cells of the node, and the presence of nodal cilia is conserved in other vertebrates (Essner et al. 2002). However, the mechanism by which directional fluid flow at the node specifies orientation of the left–right axis is controversial (Stern and Wolpert 2002; Tabin and Vogan 2003). In addition, the mechanism for transfer of the initial asymmetric signal from the node to the LPM is unknown.
The Notch signaling pathway is an evolutionarily conserved intercellular signaling mechanism. Mutations in Notch pathway components disrupt embryonic development in diverse multicellular organisms and cause inherited disease syndromes in humans (Artavanis-Tsakonas et al. 1999; Mumm and Kopan 2000; Allman et al. 2002; Gridley 2003). Genes of the Notch family encode large transmembrane receptors that interact with membrane-bound ligands encoded by Delta/Serrate/Jagged family genes. The signal induced by ligand binding is transmitted intracellularly by a process involving proteolytic cleavage of the receptor and nuclear translocation of the intracellular domain of the Notch family protein (Notch-IC). Once in the nucleus, Notch-IC forms a complex with the RBP-J protein, a sequence-specific DNA-binding protein that is the primary transcriptional mediator of Notch signaling. The Notch-IC/RBP-J complex then activates transcription of downstream target genes (Artavanis-Tsakonas et al. 1999; Mumm and Kopan 2000; Allman et al. 2002; Gridley 2003).
Here we demonstrate that the Notch signaling pathway plays a primary role in the establishment of left–right asymmetry in mice by directly regulating expression of the Nodal gene. Embryos mutant for the Notch ligand Dll1 or doubly mutant for the Notch1 and Notch2 receptors exhibit multiple defects in left–right asymmetry. Notably, Dll1−/− embryos do not express Nodal in the region around the node. Analysis of the enhancer regulating node-specific Nodal expression (termed the NDE) revealed the presence of binding sites for the RBP-J protein. Mutation of these sites destroyed the ability of the NDE to direct node-specific gene expression in transgenic mice. These results demonstrate that Dll1-mediated Notch signaling is essential for generation of left–right asymmetry, and indicate that perinodal expression of the Nodal gene is an essential component of left–right asymmetry determination in mice.
Results and Discussion
Laterality defects in Dll1 mutant and Notch1/Notch2 double-mutant mouse embryos
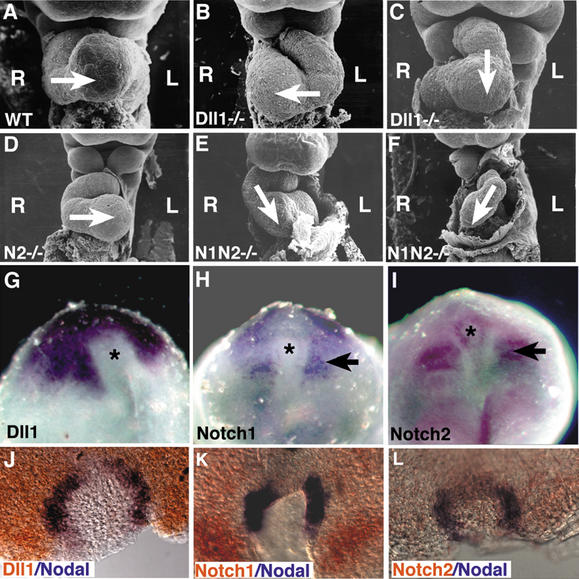
During studies on the role of the Dll1 gene during somitogenesis (Zhang et al. 2002), we observed that some Dll1−/− embryos (Hrabé de Angelis et al. 1997) exhibited reversed heart looping. We examined this phenotype more closely by performing scanning electron microscopy on Dll1−/− and control littermate embryos isolated at embryonic day 9.5 (E9.5). Normally, the linear heart tube loops from the left toward the right side (Fig. 1A). Thirty-two percent of Dll1−/− embryos exhibited normal rightward heart looping, and 42% exhibited reversed heart looping (Fig. 1B; Supplementary Table 1). We characterized the remaining 26% of the Dll1−/− embryos as having ventral heart looping, in which the developing right ventricle and conotruncal region of the heart was situated above the primitive left ventricle (Fig. 1C). Another morphological manifestation of left–right axis determination is the direction of embryonic turning. Wild-type embryos undergo a 180° axial rotation shortly after the initiation of heart looping. We observed defects in axial rotation in Dll1−/− embryos. Forty-three percent of the homozygous mutant embryos underwent reversed axial rotation, whereas the remaining 57% exhibited normal axial rotation (Supplementary Table 2). These morphological analyses suggested that laterality was randomized in Dll1−/− embryos.
Figure 1.
Heart looping defects in Notch pathway mutants. (A–F) Scanning electron micrographs of embryos at E9.5. The direction of heart looping is indicated by the arrow. The left (L) and right (R) sides of the embryo are indicated. (A) Wild-type embryo exhibiting normal right-to-left heart looping. (B) Dll1−/− mutant embryo exhibiting reversed heart looping. (C) Dll1−/− mutant embryo exhibiting ventral heart looping. (D) Embryo homozygous for the Notch2del1 hypomorphic allele (designated N2−/−) exhibiting normal right-to-left heart looping. (E) Notch1−/− Notch2−/− double-mutant embryo exhibiting mild ventral looping. (F) Severely affected Notch1−/− Notch2−/− double-mutant embryo exhibiting reversed looping and cardiac hypoplasia. (G–I) Expression of Notch pathway genes around the node (asterisk). (G) Dll1 expression. (H) Notch1 expression. (I) Notch2 expression. Both Notch1 and Notch2 are also expressed in the paraxial mesoderm (arrows). (J–L) Double-label whole-mount in situ hybridization with a digoxigenin-labeled riboprobe (blue) for Nodal and fluorescein-labeled riboprobes (orange) for Dll1 (J), Notch1 (K), or Notch2 (L).
We also examined embryos homozygous for a Notch1 null mutation (Swiatek et al. 1994) and a Notch2 hypomorphic mutation (McCright et al. 2001) for defects in heart looping and axial rotation. We did not observe any laterality defects in embryos singly homozygous for mutations in either of these genes. However, we did observe defects in both heart looping and axial rotation in Notch1/Notch2 double homozygous mutant embryos (Fig. 1E,F; Supplementary Tables 1, 2), suggesting functional redundancy between the Notch1 and Notch2 proteins during left–right asymmetry determination. Laterality defects in Notch1/Notch2 double-mutant embryos were less penetrant than in Dll1−/− embryos, suggesting either that the hypomorphic Notch2 allele retained partial function during left–right asymmetry determination or that other Notch family receptors also function in this process. Whole-mount in situ hybridization analyses confirmed previously reported (Bettenhausen et al. 1995; Williams et al. 1995) expression of the Dll1, Notch1, and Notch2 genes in the region around the node in gastrulating mouse embryos (Fig. 1G–I), and double-label in situ hybridization revealed overlap between expression of the Nodal gene and the Dll1, Notch1, and Notch2 genes in the region in and around the node (Fig. 1J–L).
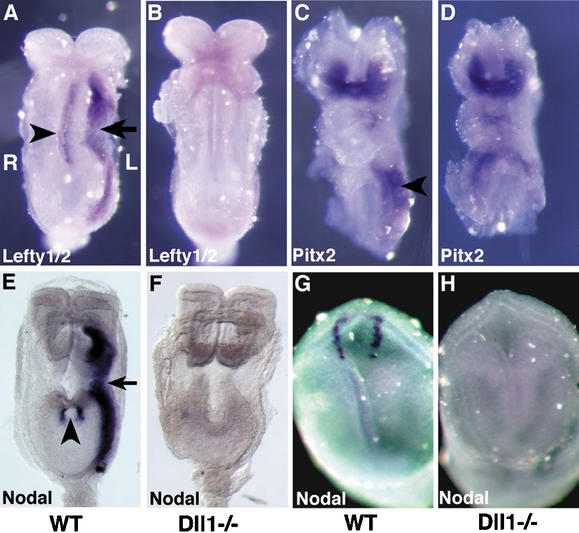
To understand the molecular basis for the laterality defects in the Dll1−/− mutant embryos, we examined expression of the Nodal, Lefty1, Lefty2, and Pitx2 genes. These genes are expressed on the left side of the embryo, and form part of an evolutionarily conserved signaling cascade responsible for left–right morphogenesis (Capdevila et al. 2000; Mercola and Levin 2001; Wright 2001; Yost 2001; Hamada et al. 2002). At early somite stages in mice, the Nodal, Lefty2, and Pitx2 genes all are expressed in the left LPM. In Dll1−/− embryos, no Nodal or Lefty2 expression was observed in the left LPM (Fig. 2B,F; Supplementary Tables 3, 4). Pitx2 expression, on the other hand, was randomized in Dll1−/− embryos. Twenty-five percent of Dll1−/− embryos exhibited normal expression of Pitx2 in the left LPM, 25% did not express Pitx2 in the LPM, and 50% expressed Pitx2 bilaterally in both the left and right LPM (Fig. 2D; Supplementary Table 3). In wild-type embryos, the Lefty1 gene is expressed along the midline in the prospective floorplate. Lefty1 expression is believed to serve a midline barrier function, preventing the spread of Nodal-induced patterning signals from the left to the right side of the embryo (Meno et al. 1998). No Lefty1 expression was observed in Dll1−/− embryos (n = 9; Fig. 2B). However, expression of the Shh and Foxa2 genes was observed in the embryonic midline of Dll1−/− embryos (Supplementary Fig. 1).
Figure 2.
Dll1−/− mutant embryos exhibit defects in the expression of left–right determinant genes. (A,B) Lefty1/2 expression. Embryos were hybridized with an antisense riboprobe that detects both the Lefty1 and Lefty2 genes. (A) In the wild-type embryo, Lefty1 is expressed along the midline in the prospective floorplate of the neural tube (arrowhead), and Lefty2 is expressed in the left LPM (arrow). (B) In the Dll1−/− embryo, no Lefty1 or Lefty2 expression is observed. Weak background staining is observed in the head of the mutant embryo. (C,D) Pitx2 expression. (C) In the wild-type embryo, Pitx2 is expressed in the head and in the left LPM (arrowhead). (D) In the Dll1−/− embryo shown, Pitx2 is expressed bilaterally in the LPM. Other Dll1−/− mutant embryos did not express Pitx2 in the LPM, or expressed it normally in the left LPM (see Supplementary Table 3). (E,F) Nodal expression at early somite stages. (E) In the wild-type embryo, Nodal is expressed in the node (arrowhead) and left LPM (arrow). (F) In the Dll1−/− embryo, Nodal is not expressed. (G,H) Nodal expression in headfold-stage embryos (E7.5). (G) In the wild-type embryo, Nodal is expressed in the perinodal region. (H) Nodal is not expressed in the Dll1−/− embryo. All images are ventral views, so that the left side of the embryo is toward the right of the panel (indicated in panel A).
Nodal expression in the left LPM is preceded by Nodal expression adjacent to the node (the perinodal region; Collignon et al. 1996). We observed that at early somite stages Nodal expression also was absent from the perinodal region of Dll1−/− embryos (Fig. 2F; Supplementary Table 4). In order to determine whether Dll1-mediated Notch signaling was required for the initiation or maintenance of node-specific Nodal expression, we examined Dll1−/− embryos and littermate controls isolated at E7.5, prior to the formation of somites and the expression of Nodal, Lefty2, and Pitx2 in the left LPM. Nodal was not expressed around the node in these headfold-stage mutant embryos (Fig. 2H; Supplementary Table 4), indicating that a Dll1-mediated Notch signal was essential for the initiation of Nodal expression around the node.
Przemeck et al. (2003) also recently reported laterality defects in Dll1−/− embryos. Their results differ from ours and from those reported in the paper by Izpisúa Belmonte and colleagues (Raya et al. 2003) in that they observed Nodal expression (in either left LPM, right LPM, or both) in approximately half of the Dll1−/− embryos analyzed. They also observed Lefty2 expression in Dll1−/− embryos. We cannot explain the discrepancy between these results. We noted, however, that the whole-mount in situ hybridization conditions used by Przemeck et al. (2003) differed from ours. We therefore hybridized additional Dll1−/− embryos (n = 7) with a Nodal antisense riboprobe using the in situ hybridization conditions of Przemeck et al. (2003; kindly provided by Martin Hrabé de Angelis). We did not observe Nodal expression in any Dll1−/− embryos (n = 20) hybridized under either set of in situ hybridization conditions. It remains unclear, therefore, why the results of Przemeck et al. (2003) differ from the results reported here and in the paper by Izpisúa Belmonte and colleagues (Raya et al. 2003). We should note, however, that genetic background differences exist between the two populations of Dll1 mutant mice. The mice of Przemeck et al. (2003) were maintained on a mixed 129Sv × C57BL/6J background, whereas our mice had been extensively back-crossed to C57BL/6J.
Nodal cilia are present and functional in Dll1 mutant embryos
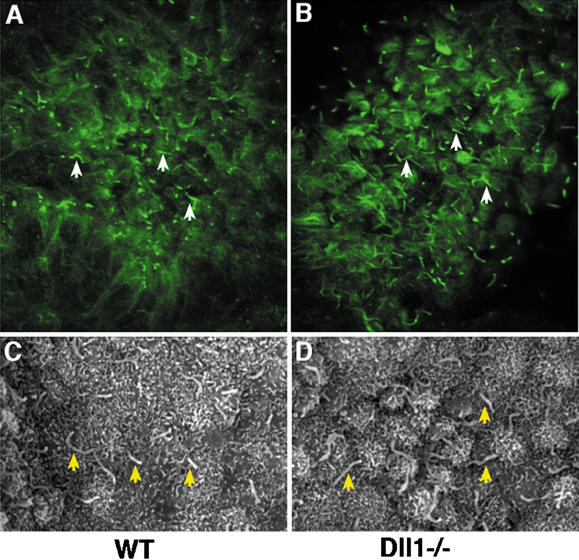
In mice, generation of the initial asymmetric signal involves a leftward fluid flow generated by the rotational movement of monocilia on the ventral surface of the node (Capdevila et al. 2000; Mercola and Levin 2001; Wright 2001; Yost 2001; Essner et al. 2002; Hamada et al. 2002; Nonaka et al. 2002; Tabin and Vogan 2003). We established by immunofluorescence and scanning electron microscopy that monocilia were present on node cells of Dll1−/− embryos (Fig. 3). To test whether these monocilia were functional, we examined nodal fluid flow by videomicroscopy. All Dll1−/− embryos tested (n = 7) exhibited normal leftward fluid flow at the node (Supplementary Video 1). Therefore, defects in nodal flow do not appear to contribute to the laterality defects observed in Dll1−/− embryos.
Figure 3.
Presence of monocilia on node cells of Dll1−/− mutant embryos. (A,B) Monocilia (some indicated with arrows) were detected by immunofluorescence with an antibody against acetylated tubulin. (C,D) Monocilia (arrows) were observed by scanning electron microscopy on both wild-type (C) and Dll1−/− mutant (D) node cells.
While performing videomicroscopy on these node explants, we noted that the nodes of some Dll1−/− embryos appeared distorted, although this distortion did not affect nodal fluid flow. Dll1−/− embryos with distorted nodes also appeared to be delayed in the formation of mesodermal somites, suggesting the possibility that the laterality defects observed in Dll1−/− embryos were secondary to defects in somitogenesis. We therefore examined other Notch pathway mutants exhibiting defects in somite formation (Zhang et al. 2002) for morphological laterality defects, such as reversed heart looping or axial rotation. No morphological laterality defects were observed in either Lfng-deficient (Zhang and Gridley 1998; n = 8) or Dll3-deficient (Kusumi et al. 1998; n = 12) embryos, indicating that the laterality defects observed in Dll1−/− embryos are not secondary to defects in somitogenesis.
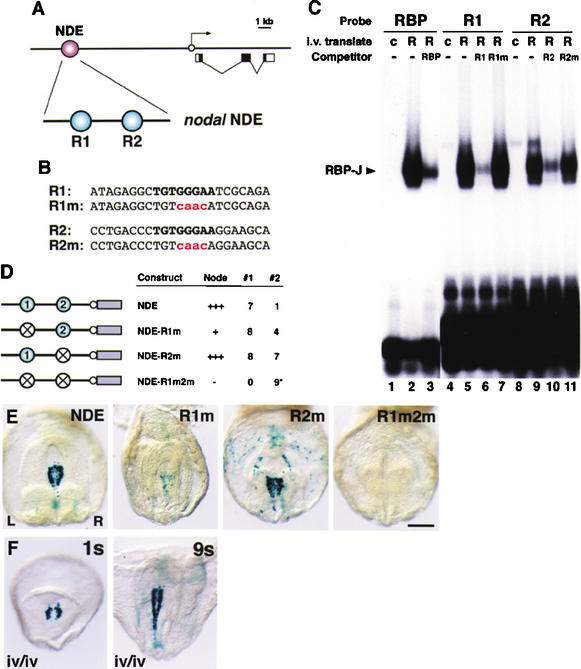
The NDE enhancer contains RBP-J-binding sites essential for activity
Expression of the Nodal gene in the mouse node is regulated by a specific enhancer element termed the node-specific enhancer (NDE; Adachi et al. 1999; Norris and Robertson 1999). The NDE has been localized to a 0.8 kb-fragment located between −9.5 and −8.7 kb upstream of the Nodal gene (Fig. 4A; Adachi et al. 1999). Analysis of the nucleotide sequence of the NDE revealed the presence of two consensus binding sites (T/CGTGGGAA) for the RBP-J protein (Tun et al. 1994; Honjo 1996; Kato et al. 1997), the primary transcriptional mediator of the Notch signal (Fig. 4A,B). Electrophoretic mobility shift assays confirmed that these consensus RBP-J-binding sites bound recombinant RBP-J protein (Fig. 4C). Analysis of RBP-J RNA expression during mouse embryogenesis revealed that RBP-J is expressed ubiquitously, including in node cells that express the Nodal gene (Supplementary Fig. 2).
Figure 4.
The Nodal node-specific enhancer (NDE) contains two RBP-J-binding sites essential for directing lacZ reporter gene expression. (A) The NDE enhancer resides in a 0.8-kb region upstream of Nodal coding sequences (−9.5 to −8.7 kb). Within the NDE, two motifs identical to the consensus binding sequence of RBP-J were present. The relative positions of the 5′ motif (site R1) and 3′ motif (site R2) are indicated. (B) The nucleotide sequences of sites R1 and R2 and of R1m and R2m containing four base pair substitutions (shown in red) in the RBP-J-binding consensus (shown in bold). (C) Electrophoretic mobility shift assay was performed with in vitro translated RBP-J protein (R; lanes 2,3,5–7,9–11) or control translates without RBP-J (c; lanes 1,4,8). Three labeled oligonucleotides were used: a consensus RBP-J sequence (RBP; lanes 1–3), site R1 from NDE (lanes 4–7), and site R2 from NDE (lanes 8–11). Binding reactions were performed in the absence (−) or presence of a 50-fold excess of unlabeled homologous oligonucleotides (lanes 3,6,10), or unlabeled mutated oligonucleotides R1m and R2m (lanes 7,11). (D) Summary of transient transgenic analysis of NDE constructs bearing single or double mutations in the two RBP-J-binding sites. The wild-type construct (NDE) contains the 0.8-kb NDE region linked to a hsp68–lacZ promoter (white circle) lacZ gene (gray bar) reporter cassette. The 5′ RBP-J-binding site, the 3′ RBP-J-binding site, and both RBP-J-binding sites were mutated in NDE-R1m, NDE-R2m, and NDE-R1m2m, respectively. For each construct, the level of NDE activity in the node, the number of embryos that showed NDE activity (column #1), and the number of embryos that carried the transgene but showed either only ectopic expression or no expression at all (column #2) are indicated. Four of the nine R1m2m embryos (indicated by an asterisk in column #2 of the table) showed weak, patchy expression in the center of the node, where the endogenous Nodal gene is not expressed. (E) Ventral views of X-gal-stained E8.2 embryos transgenic for the indicated constructs. The left (L) and right (R) sides of the axis are shown. Bar, 250 μm. (F) X-gal-stained iv/iv embryos transgenic for the NDE construct. One-somite (1s) and nine-somite (9s) iv/iv embryos exhibiting NDE activity are shown. The nine-somite embryo shown had reversed axial rotation.
To assess the function of these RBP-J-binding sites in directing node-specific Nodal expression, we tested mutations of these binding sites in transgenic mouse embryos. Control experiments demonstrated that oligonucleotides containing these binding site mutations were unable to compete binding of wild-type oligonucleotides in electrophoretic mobility shift assays (Fig. 4C). As demonstrated previously, pronuclear injection of a transgene consisting of the 0.8-kb Nodal NDE linked to the hsp68 promoter and β-galactosidase (lacZ) coding sequence (Kothary et al. 1989) gave rise to X-gal staining around the node in embryos isolated at early somite stages (Fig. 4D,E). The role of the two RBP-J-binding sites was investigated by mutating them separately or in combination. Mutation of the 5′ RBP-J-binding site (R1m) had a more severe effect on lacZ expression than mutation of the 3′ RBP-J-binding site (R2m). However, node-specific lacZ expression was lost completely when both RBP-J-binding sites were mutated (R1m2m; Fig. 4D,E).
We also examined activity of the Nodal NDE in embryos homozygous for the iv mutation, in which laterality is randomized. Previous work had demonstrated that an enhancer in the Nodal gene that directs expression in the left LPM (termed the asymmetric enhancer, or ASE) was responsive to the iv mutation (Adachi et al. 1999; Norris and Robertson 1999). However, iv/iv embryos transgenic for the NDE construct exhibited the same perinodal lacZ expression as wild-type embryos (Fig. 4F), indicating that the NDE enhancer functions independently of the iv mutation.
Our results demonstrate that Dll1-mediated Notch signaling is essential for expression of Nodal in the perinodal region, and that Notch signaling acts upstream of Nodal in left–right asymmetry determination in mice. Prior work supports the model that Nodal expression in the LPM is a positive regulator of subsequent Pitx2 expression (Capdevila et al. 2000; Mercola and Levin 2001; Wright 2001; Yost 2001; Hamada et al. 2002). However, uncoupling of a strict correlation between expression of Nodal and Pitx2 in the LPM has been observed previously (Meyers and Martin 1999; Constam and Robertson 2000a,b; Pennekamp et al. 2002). For example, some mouse embryos mutant for the Pkd2 gene do not express Nodal in the LPM but exhibit bilateral expression of Pitx2 in the LPM (Pennekamp et al. 2002). We observe these same patterns of expression in Dll1−/− embryos, confirming that Nodal and Pitx2 expression in the LPM need not be strictly correlated.
Our work also indicates that expression of the Nodal gene around the node is an essential component of left–right axis determination in mice. During the establishment of left–right asymmetry, positional information must be transferred from the node to the lateral plate mesoderm. Our results indicate that perinodal expression of the Nodal gene is essential for activation of asymmetric gene expression in the left LPM. A similar conclusion was reached in recent studies that used genetic strategies to selectively remove Nodal activity from the node (Brennan et al. 2002; Saijoh et al. 2003). These results are consistent with a model for asymmetric signal transfer in which Nodal protein produced in the perinodal region induces its own expression in the LPM, although a model in which perinodal expression of Nodal regulates production of another molecule that induces asymmetric gene expression in the left LPM cannot be excluded. The loss of perinodal expression of the Nodal gene and the laterality defects observed in Dll1 mutant and Notch1/Notch2 double-mutant mice demonstrate that Notch signaling plays a significant and early role in regulating development of the left–right axis.
Materials and methods
Mutant mouse strains
Mutant mice used in these studies were Dll1tm1Go (Hrabé de Angelis et al. 1997), Notch1in32 (Swiatek et al. 1994), Notch2del1 (McCright et al. 2001), LfnglacZ (Zhang and Gridley 1998), and Dll3pu (Kusumi et al. 1998). Mice heterozygous for these mutant alleles were maintained by back-crossing to C57BL/6J (B6) mice. Notch1in32, Notch2del1, LfnglacZ, and Dll3pu mutant embryos were genotyped by allele-specific PCR. Dll1tm1Go mutant embryos were genotyped using a closely linked marker (D17Mit156) that is polymorphic between strains 129 and B6, permitting identification of the 129-derived Dll1tm1Go allele and the B6-derived wild-type Dll1 allele. PCR primer sequences are available on request.
Whole-mount in situ hybridization and immunofluorescence
Single- and double-label whole-mount in situ hybridization was performed as described previously (Krebs et al. 2001). Presence of node monocilia was detected using a monoclonal antibody against acetylated tubulin (Sigma), as described (Essner et al. 2002).
Visualization of nodal flow
Embryos were dissected and cultured as described previously (Nonaka et al. 1998, 2002). Fluid flow was visualized by addition of polystyrene beads to the culture medium. Images were collected with a monochrome CCD camera. Embryos were genotyped by PCR after observation.
Electrophoretic mobility shift assay
RBP-J protein was synthesized with an in vitro transcription–translation coupled system (Promega). An electrophoretic mobility shift assay was performed with the in vitro translated RBP-J protein (Okamoto et al. 1990). Three oligonucleotides were used for the binding assay: a consensus binding sequence for RBP-J (AAACACGCCGTGGGAAAAAA TTTGG) derived from the Epstein-Barr virus C promoter region (Lu and Lux 1996); the R1 site from NDE (ATAGAGGCTGTGGGAATCGCA GA); and the R2 site from NDE (CCTGACCCTGTGGGAAGGAAGCA). The mutant oligonucleotides R1m (ATAGAGGCTGTCAACATCG CAGA) and R2m (CCTGACCCTGTCAACAGGAAGCA) were used as competitors to assess binding specificity.
lacZ constructs and transient transgenic assay
For the NDE–lacZ construct, the 0.8-kb NDE region was subcloned into the hsp68–lacZ vector (Kothary et al. 1989). To generate site-directed mutant constructs, we subcloned the 0.8-kb NDE region into pKF18 (Takara), and the RBP-J-binding sites (TGTGGGAA) were mutated with oligonucleotides harboring four base substitutions (TGTCAACA). The mutated NDE fragments were similarly subcloned into the hsp68–lacZ vector. The resulting lacZ plasmids were digested with Not I to remove vector sequences. The lacZ DNA fragments were purified by gel electrophoresis and were microinjected into fertilized mouse eggs, as described (Saijoh et al. 1999). Embryos were recovered at E8.2 and examined for lacZ expression by X-gal staining according to standard protocols. Transgene integration was determined by PCR amplification of the lacZ gene.
Acknowledgments
We thank Juan Carlos Izpisúa Belmonte and Martin Hrabé de Angelis for helpful discussions and for communicating unpublished results; Tasuku Honjo for RBP-J plasmids; Christine Norton, Hiromi Hashiguchi, Yayoi Ikawa, and Kyoko Mochida for technical assistance; and John Eppig and Sue Ackerman for helpful discussions and comments on the manuscript. This work was supported by grants to T.G. from the NIH (NS36437) and the March of Dimes Birth Defects Foundation; to T.O. from the NIH (HD36434 and HD41066); and to H.H. from CREST (Core Research for Evolutional Science and Technology) of the Japan Science and Technology Corporation. This work was also supported by a grant from the National Cancer Institute (CA34196) to the Jackson Laboratory.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
School of Medicine and Dentistry, Rochester, NY 14642, USA.
Corresponding authors.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1084703.
References
- Adachi H, Saijoh Y, Mochida K, Ohishi S, Hashiguchi H, Hirao A, Hamada H. Determination of left/right asymmetric expression of nodal by a left side-specific enhancer with sequence similarity to a lefty-2 enhancer. Genes & Dev. 1999;13:1589–1600. doi: 10.1101/gad.13.12.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman D, Punt JA, Izon DJ, Aster JC, Pear WS. An invitation to T and more: Notch signaling in lymphopoiesis. Cell. 2002;109:S1–S11. doi: 10.1016/s0092-8674(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet J-L, Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left–right asymmetry. Genes & Dev. 2002;16:2339–2344. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Vogan KJ, Tabin CJ, Izpisúa Belmonte JC. Mechanisms of left–right determination in vertebrates. Cell. 2000;101:9–21. doi: 10.1016/S0092-8674(00)80619-4. [DOI] [PubMed] [Google Scholar]
- Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- Constam DB, Robertson EJ. SPC4/PACE4 regulates a TGFbeta signaling network during axis formation. Genes & Dev. 2000a;14:1146–1155. [PMC free article] [PubMed] [Google Scholar]
- ————— Tissue-specific requirements for the proprotein convertase furin/SPC1 during embryonic turning and heart looping. Development. 2000b;127:245–254. doi: 10.1242/dev.127.2.245. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Vogan KJ, Wagner MK, Tabin CJ, Yost HJ, Brueckner M. Conserved function for embryonic nodal cilia. Nature. 2002;418:37–38. doi: 10.1038/418037a. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet Suppl 1. 2003;12:R9–R13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left–right asymmetry. Nat Rev Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- Honjo T. The shortest path from the surface to the nucleus: RBP-J kappa/Su(H) transcription factor. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- Hrabé de Angelis M, McIntyre J, II, Gossler A. Maintenance of somite borders in mice requires the Delta homologue Dll1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68–lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Krebs LT, Deftos ML, Bevan MJ, Gridley T. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the Notch signaling pathway. Dev Biol. 2001;238:110–119. doi: 10.1006/dbio.2001.0408. [DOI] [PubMed] [Google Scholar]
- Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi D-C, Bulotsky MS, Spencer J, Birren BW, Frankel WN, Lander ES. Delta3 is disrupted in the pudgy somite patterning mutant. Nat Genet. 1998;19:274–278. doi: 10.1038/961. [DOI] [PubMed] [Google Scholar]
- Lu FM, Lux SE. Constitutively active human Notch1 binds to the transcription factor CBF1 and stimulates transcription through a promoter containing a CBF1-responsive element. Proc Natl Acad Sci. 1996;93:5663–5667. doi: 10.1073/pnas.93.11.5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development. 2001;128:491–502. doi: 10.1242/dev.128.4.491. [DOI] [PubMed] [Google Scholar]
- Meno C, Shimono A, Saijoh Y, Yashiro K, Mochida K, Ohishi S, Noji S, Kondoh H, Hamada H. lefty-1 is required for left–right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–297. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- Mercola M, Levin M. Left–right asymmetry determination in vertebrates. Annu Rev Cell Dev Biol. 2001;17:779–805. doi: 10.1146/annurev.cellbio.17.1.779. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Martin GR. Differences in left–right axis pathways in mouse and chick: Functions of FGF8 and SHH. Science. 1999;285:403–406. doi: 10.1126/science.285.5426.403. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Kopan R. Notch signaling: From the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left–right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Shiratori H, Saijoh Y, Hamada H. Determination of left–right patterning of the mouse embryo by artificial nodal flow. Nature. 2002;418:96–99. doi: 10.1038/nature00849. [DOI] [PubMed] [Google Scholar]
- Norris DP, Robertson EJ. Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes & Dev. 1999;13:1575–1588. doi: 10.1101/gad.13.12.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell. 1990;60:461–472. doi: 10.1016/0092-8674(90)90597-8. [DOI] [PubMed] [Google Scholar]
- Pennekamp P, Karcher C, Fischer A, Schweickert A, Skryabin B, Horst J, Blum M, Dworniczak B. The ion channel polycystin-2 is required for left–right axis determination in mice. Curr Biol. 2002;12:938–943. doi: 10.1016/s0960-9822(02)00869-2. [DOI] [PubMed] [Google Scholar]
- Przemeck GK, Heinzmann U, Beckers J, Hrabe De Angelis M. Node and midline defects are associated with left–right development in Delta1 mutant embryos. Development. 2003;130:3–13. doi: 10.1242/dev.00176. [DOI] [PubMed] [Google Scholar]
- Raya, Á., Kawakami, Y., Rodriguez-Esteban, C., Büscher, D., Koth, C.M., Itoh, T., Morita, M., Raya, R.M., Dubova, I., Bessa, J.G., et al. 2003. Notch activity induces Nodal expression and mediates the establishment of left–right asymmetry in vertebrate embryos. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Saijoh Y, Adachi H, Mochida K, Ohishi S, Hirao A, Hamada H. Distinct transcriptional regulatory mechanisms underlie left–right asymmetric expression of lefty-1 and lefty-2. Genes & Dev. 1999;13:259–269. doi: 10.1101/gad.13.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijoh Y, Oki S, Ohishi S, Hamada H. Left–right patterning of the mouse lateral plate requires Nodal produced in the node. Dev Biol. 2003;256:161–173. doi: 10.1016/s0012-1606(02)00121-5. [DOI] [PubMed] [Google Scholar]
- Stern CD, Wolpert L. Left–right asymmetry: All hands to the pump. Curr Biol. 2002;12:R802–R803. doi: 10.1016/s0960-9822(02)01312-x. [DOI] [PubMed] [Google Scholar]
- Swiatek PJ, Lindsell CE, Franco del Amo F, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes & Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- Tabin CJ, Vogan KJ. A two cilia model for vertebrate left–right axis specification. Genes & Dev. 2003;17:1–6. doi: 10.1101/gad.1053803. [DOI] [PubMed] [Google Scholar]
- Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R, Lendahl U, Lardelli M. Complementary and combinatorial patterns of Notch gene family expression during early mouse development. Mech Dev. 1995;53:357–368. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- Wright CV. Mechanisms of left–right asymmetry: What's right and what's left? Dev Cell. 2001;1:179–186. doi: 10.1016/s1534-5807(01)00036-3. [DOI] [PubMed] [Google Scholar]
- Yost HJ. Establishment of left–right asymmetry. Int Rev Cytol. 2001;203:357–381. doi: 10.1016/s0074-7696(01)03011-x. [DOI] [PubMed] [Google Scholar]
- Zhang N, Gridley T. Defects in somite formation in Lunatic fringe deficient mice. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]
- Zhang N, Norton CR, Gridley T. Segmentation defects of Notch pathway mutants and absence of a synergistic phenotype in lunatic fringe/radical fringe double-mutant mice. Genesis. 2002;33:21–28. doi: 10.1002/gene.10081. [DOI] [PubMed] [Google Scholar]