Abstract
MSL proteins and noncoding roX RNAs form complexes to up-regulate hundreds of genes on the Drosophila male X chromosome, and make X-linked gene expression equal in males and females. Altering the ratio of MSL proteins to roX RNA dramatically changes X-chromosome morphology. In protein excess, the MSL complex concentrates near sites of roX transcription and is depleted elsewhere. These results support a model for distribution of MSL complexes, in which local spreading in cis from roX genes is balanced with diffusion of soluble complexes in trans. When overexpressed, MSL proteins can recognize the X chromosome, modify histones, and partially restore male viability even in the absence of roX RNAs. Thus, the protein components can carry out all essential functions of dosage compensation, but roX RNAs facilitate the correct targeting of MSL complexes, in part by nucleation of spreading from their sites of synthesis.
Keywords: Drosophila dosage compensation, noncoding roX RNAs, MSL proteins
The establishment and maintenance of chromatin organization by histone modification and chromatin-remodeling complexes has been postulated to occur in some cases by initial recognition of a nucleation site, followed by spreading in cis into flanking sequences (Lee and Jaenisch 1997; Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001; Ho et al. 2002). In dosage compensation of the Drosophila male X chromosome, MSL (male-specific lethal) proteins and roX (RNA on X) RNAs form large complexes that modify histone tails and can spread long distances from initiation sites into flanking chromatin (Kelley et al. 1999; Meller et al. 2000; Park et al. 2002). The two known roX genes are located on the X chromosome (Amrein and Axel 1997; Meller et al. 1997) and are thought to have dual functions. First, roX RNAs are components of the MSL complex(Meller et al. 2000; Smith et al. 2000). The MSL proteins are unable to bind the X chromosome efficiently without roX RNA, resulting in male lethality (Franke and Baker 1999; Meller and Rattner 2002). Second, roX transgenes can function as nucleation sites for ectopic targeting and spreading of MSL complexes into flanking autosomes regardless of location (Kelley et al. 1999). We have inferred that similar spreading around the endogenous roX genes could contribute to targeting the MSL complex to the correct chromosome.
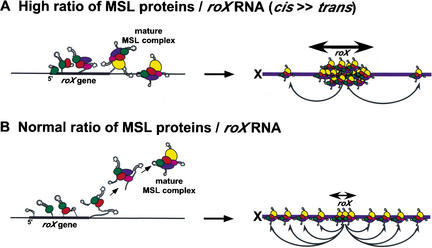
Ectopic spreading of MSL complexes on autosomes is profoundly influenced by the number of roX genes in the nucleus (Park et al. 2002). When both endogenous roX genes are deleted so that a single roX autosomal transgene is the sole source of roX RNA, consistent spreading of the MSL complex occurs for long distances (several megabase pairs) into flanking chromatin. However, when several roX genes compete for a limiting amount of MSL proteins, ectopic spreading from autosomal roX transgenes is very rare, and instead all MSL complexes diffuse to the X chromosome. A model based on these observations proposes that there are two pools of MSL complexes. If MSL proteins are abundant and rapidly assemble onto growing roX transcripts, functional complexes will be completed before release of the nascent roX transcript from the DNA template. These complexes are postulated to immediately bind the flanking chromosome regardless of sequence and begin spreading in cis (Fig. 1A). When multiple roX genes are present, they compete for a finite supply of MSL proteins, thus slowing the assembly of complete complexes. Under these conditions, nascent roX RNA will be released from the template with an incomplete set of MSL subunits. After maturation is completed in solution, these complexes are postulated to diffuse through the nucleus until encountering the X chromosome (Fig. 1B). How such soluble MSL complexes recognize and bind the X chromosome is not understood. We initially postulated that, besides the two roX genes, there were ∼35 additional “chromatin entry sites” that would also initiate MSL spreading (Kelley et al. 1999). However, the nature of these sites remains a mystery.
Figure 1.
Model for cis-versus-trans interaction of MSL complexes with the X chromosome. (A) If the local concentration of MSL proteins is high at the site of roX transcription, a complete set of MSL proteins will be assembled efficiently on the nascent roX RNA, leading to local MSL spreading in cis. If the rate of MSL protein assembly is accelerated by overexpressing MSL1 and MSL2, extensive cis spreading of MSL complexes will predominate and trans interactions will decrease. (B) Conversely, if the local concentration of MSL proteins is low, only partial MSL complexes will be assembled on the nascent roX RNA prior to release from the DNA template. Released complexes can finish assembly in the nucleoplasm, but are unlikely to return to the site of roX synthesis. In normal conditions, the rate of roX RNA transcription and the rate of MSL protein assembly on nascent roX RNA are optimized, resulting in balanced cis and trans interactions of MSL complexes with the X chromosome.
To date, all evidence for cis spreading comes from autosomal roX transgenes. Here, we demonstrate that MSL complexes do spread locally from the endogenous roX genes on the X chromosome, the natural target of dosage compensation. We find that wild-type males require a balance of MSL proteins and roX RNAs to evenly distribute MSL complexes both locally and at a distance along the X chromosome. When we artificially increase the amounts of MSL1 and MSL2, thought to be the limiting proteins (Kelley et al. 1997; Chang and Kuroda 1998; Park et al. 2002), MSL complexes spread predominantly over a local segment of the X chromosome surrounding a roX gene. More remote regions bind little MSL complex. This dramatically alters the morphology of polytene X chromosomes. Surprisingly, we found that overexpressing MSL1 and MSL2 partially restored viability to males lacking roX RNA. This indicates that the MSL proteins have intrinsic affinity for the X chromosome that is enhanced or stabilized in wild-type males by the roX RNAs.
Results and Discussion
Spreading of MSL complexes from roX genes on the X chromosome
There are two known roX genes located on the X chromosome; roX1 at 3F near the telomere and roX2 at 10C in the middle of the chromosome (Fig. 2A). The even distribution of MSL complexes in a banded, reproducible pattern in single roX mutants, with no preference for the regions surrounding 3F or 10C, does not suggest regional spreading (Meller et al. 1997; Kelley et al. 1999; Meller and Rattner 2002). Our recent finding, that the ratio of MSL proteins to roX RNAs affects whether or not MSL complexes spread in cis near autosomal roX transgenes (Park et al. 2002), prompted us to ask if increasing the levels of MSL proteins would alter balanced targeting of the X chromosome. We used the previously characterized Hsp83 promoter-driven transgenes, [H83M1] (previously known as M1-ECTOPIC) and [H83M2] (Chang and Kuroda 1998) to overexpress the msl1 and msl2 genes in roX1- or roX2- single mutants. Under these conditions, we found that the roX genotype greatly affected X-chromosome morphology. In roX1- larvae, MSL localization to the X chromosome was concentrated over the middle region of the X chromosome surrounding the roX2+ gene (Fig. 2B). In contrast, in roX2- larvae, MSL localization was extensive around the tip of the X chromosome around the roX1+ gene (Fig. 2C). More distant regions of the X chromosome were deficient for MSL complexes. The MSL-faint regions of the X chromosome displayed a collapsed, narrow morphology reminiscent of that seen in mle or roX1- roX2- mutants (Belote and Lucchesi 1980; Meller and Rattner 2002). MSL-bright regions surrounding either roX+ gene showed very diffuse DAPI staining, reminiscent of chromosome puffing. Although chromosome morphology varied from nucleus to nucleus within the same individual, ∼20%–30% of the nuclei showed local alteration of morphology around roX genes. These results strongly suggest that spreading really occurs from the roX1 and roX2 genes in their native locations, and can be exaggerated by overexpression of MSL proteins. The width of polytene chromosomes is often correlated with transcriptional activity (Offermann 1936; Dobzhansky 1957). The gross alteration in X-chromosome morphology seen in these males suggests that affected regions might be overexpressed and depleted regions under expressed. This could lead to reduced male viability (Table 1).
Figure 2.
(A–G) Local cis spreading of MSL complexes on the X chromosome. Male polytene chromosomes were stained with anti-MSL1 antibodies and visualized with Texas Red-conjugated secondary antibody (red). DNA was stained with DAPI (blue). Arrowheads and arrows indicate the locations of the roX1 and roX2 genes, respectively. The genotypes are wild type (A), roX1ex6; [M1][M2] (B), Df(1)roX252; [M1][M2] (C), wild type; [M1][M2] (D), roX1ex6 Df(1)roX252 [w+ GMroX1-13A]; [M1][M2] (E), roX1ex6 Df(1)roX252 [w+ GMroX2–14F]; [M1][M2] (F), roX1ex6 Df(1)roX252 [w+ GMroX1–18C] without extra MSL1 and MSL2 (G). [M1][M2] stands for [w+ H83M1][w+ H83M2]/+ on the third chromosome. Full genotypes of crosses are listed in Materials and Methods. (H) Northern blot showing that the steady-state level of roX RNA is increased with overexpression of MSL1 and MSL2. The same membrane was hybridized to an rp49 probe as a loading control.
Table 1.
Effect of overexpression of MSL proteins on male viability of roX mutants
| % viability of progeny overexpressing MSL1 and MSL2
| |||||
|---|---|---|---|---|---|
| Cross | roX genotypea | Female | Female; [M1][M2]/+ | Male | Male; [M1][M2]/+ |
| 1 | WT | 100b (1181)c | 0.9 (10) | 56.5 (667) | 31.3 (370) |
| 2 | roX1- | 100 (1364) | 0.4 (6) | 63.4 (865) | 34.2 (466) |
| 3 | roX2- | 100 (646) | 0.5 (3) | 86.5 (559) | 75.5 (488) |
| 4 | roX1-roX2- | 100 (1286) | 11.1 (143)d | 0.1 (1) | 7.2 (93) |
| 5 | roX1-roX2- | 100 (1185) | 9.3 (110) d | 0.9 (11) | 25.4 (301) |
| 6 | roX1-roX2- | 100 (2239) | 16.0 (358) d | 6.6 (147) | 44.3 (992) |
Full genotypes of crosses:
- y w × w/Y; msl2 cn; [M1][M2}/+
- y w rox1ex6 × w/Y; msl2 cn; [M1][M2]/+
- w Df(1)roX252; [w+ 4Δ4.3] × w/Y; msl2 cn; [M1][M2]/+
- y w rox1ex6 Df(1)roX252 [w+ 4Δ4.3] × w/Y; msl2 cn; [M1][M2]/+
- y w rox1ex6 Df(1)roX252; [w+ 4Δ4.3] × w/Y; msl2 cn; [M1][M2]/+
- rox1ex6 Df(1)roX252; [w+ 4Δ4.3] × w/Y; msl2 cn/+; [M1][M2]/TM3 Sb
The number of XO males and XXY females is not included.
This indicates the male X genotype; females are heterozygous, as they also carry one wild-type X chromosome.
Numbers represent the percent of expected when compared with control females that do not overexpress MSL1 and MSL2.
Numbers in parentheses show actual number of progeny.
Compared with the survival of wild-type females overexpressing MSL1 and MSL2, these females show enhanced viability, possibly because they have one wild-type and one roX mutant X chromosome.
To determine whether increased spreading would be evident when both roX genes were functional, we overexpressed MSL1 and MSL2 in otherwise wild-type males. These males displayed short, wide X chromosomes that stained strongly with MSL antibodies but showed faint, diffuse staining with DAPI (Fig. 2D). This morphology suggests overall enhanced MSL binding and function on the X chromosome that could be caused by extensive spreading from both roX loci. This is somewhat similar to the phenotype of iswi or nurf301 mutants (Deuring et al. 2000; Badenhorst et al. 2002), raising the strong possibility that it reflects aberrant chromatin organization. Although male viability was decreased by overexpression of MSL1 and MSL2 (Table 1), both male and female flies can tolerate surprisingly large perturbations in MSL localization patterns on the X chromosome (Kelley et al. 1997, 1999).
To test our assumption that the roX genes are the precise sites of initiation of MSL spreading, we moved the roX1+ or roX2+ genes to new locations along the roX- X chromosome. In each of 10 sites tested, additional MSL1 and MSL2 protein produced bright MSL staining and diffuse chromosome morphology surrounding the relocated roX+ gene (Fig. 2E,F; data not shown). Again, more distant regions of the X chromosome had much less MSL staining. Because a strong bias for local MSL spreading was seen at each segment of the X chromosome tested, the sequences surrounding the endogenous roX genes are unlikely to contain special spreading elements. Moreover, some roX transgenes displayed a strong preference for regional spreading from their site of insertion with only wild-type levels of MSL1 and MSL2 (Fig. 2G). This indicates that our conditions of MSL1 and MSL2 overexpression were not extreme, and that changes in the MSL protein:roX RNA ratio or chromatin environment can produce large shifts in the pattern of local MSL spreading.
To determine whether overexpression of MSL1 and MSL2 affected the steady-state levels of roX RNAs, we performed Northern analysis (Fig. 2H). Compared with normal males (Fig. 2H, lanes 1,3,5), the levels of roX RNAs were increased when MSL1 and MSL2 were overexpressed (Fig. 2H, lanes 2,4,6). Under these conditions, the local assembly of MSL complexes might be amplified through a positive feedback loop. For example, the transcription of roX genes might be increased by the high concentration of MSL complexes, or the accelerated rate of MSL protein assembly with nascent roX RNA could lead to increased stabilization of the RNA that might otherwise be degraded. Thus, the higher density of MSL complexes around the roX genes (Fig. 2B–G) probably results from two factors. There is an overall increase in MSL complexes, and complexes normally destined for remote parts of the X chromosome instead remain local. This shows that the wild-type male X chromosome is not saturated with MSL complex, but rather that the distribution must be under active control.
Overexpressed MSL1 and MSL2 can partially overcome a lack of roX RNAs
Males lacking both roX1 and roX2 have only small amounts of MSL proteins bound to a subset of sites on the X chromosome (Meller and Rattner 2002). Depending on poorly understood differences in genetic background, ∼95%–99.9% of such males die before adulthood. Surprisingly, a small number of adult males somehow survive without any detectable roX RNA (Meller and Rattner 2002; Stuckenholz 2002). Larval polytene chromosomes from such animals display very poor morphology, with ectopic MSL binding to the chromocenter and some autosomal sites (Fig. 3A; Meller and Rattner 2002). For comparison, mutants for each of the five known msl genes show a complete male lethal phenotype with no escapers. Because wild-type and single roX mutant males each had altered X-chromosome morphology when MSL1 and MSL2 were overexpressed, we asked whether roX1- roX2- double-mutant X chromosomes would also be affected by these conditions. We found that overexpression of MSL1 and MSL2 improved the morphology of all polytene chromosomes in roX- males. In addition, the X chromosome displayed much more robust binding of MSL1 (Fig. 3B) and MSL2 (data not shown) than brothers with only wild-type levels of MSL proteins (Fig. 3A). This indicates that the MSL proteins have an intrinsic ability to recognize the X chromosome. At normal MSL levels, this binding is weak but greatly enhanced by roX RNA. When MSL proteins are artificially overproduced, however, MSL binding to the X chromosome is at least partially RNA-independent.
Figure 3.
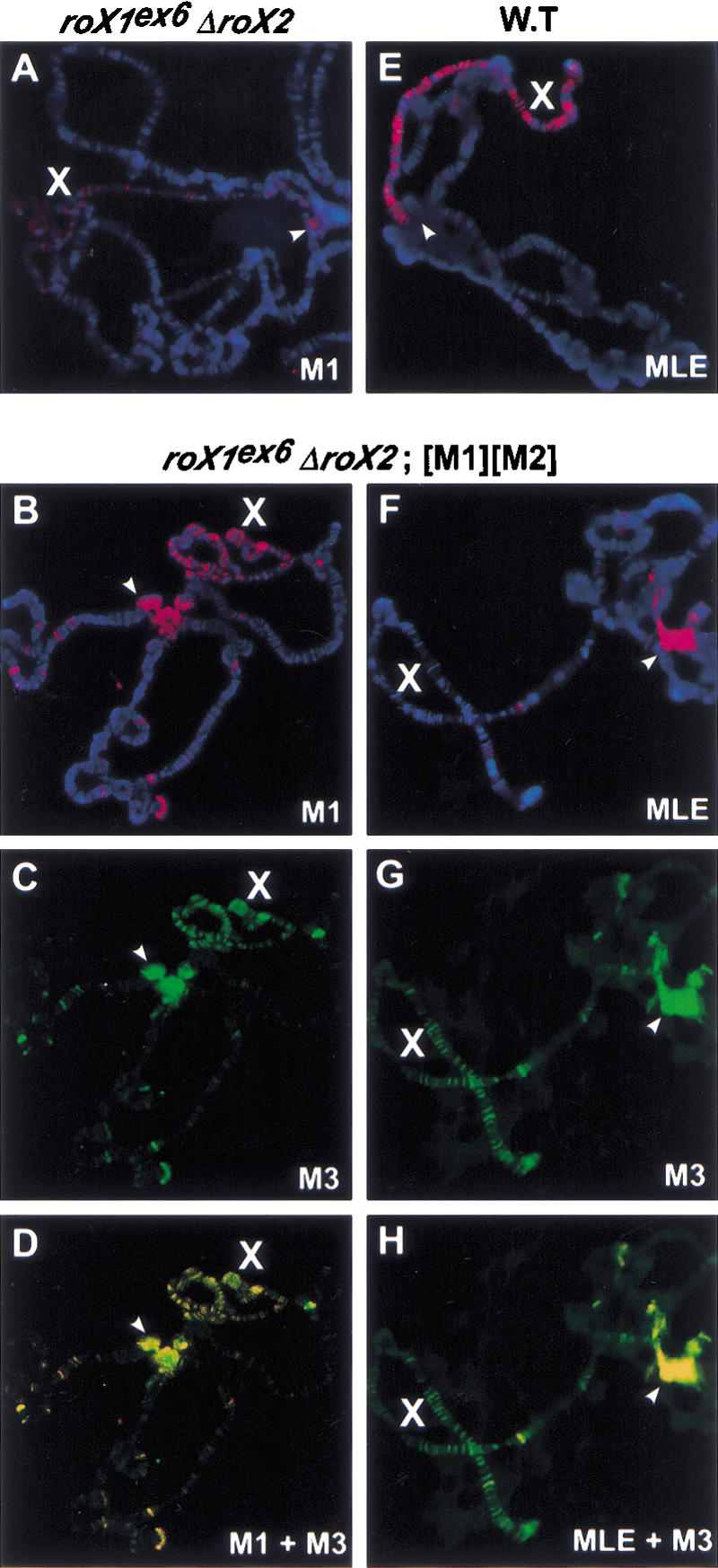
Colocalization of MSL proteins to the X chromosome without roX RNA. Male polytene chromosomes were immunostained and visualized by FITC-conjugated secondary antibodies for anti-MSL3 (green) or Texas Red-conjugated secondary antibodies for anti-MSL1 or anti-MLE (red). DNA was stained with DAPI (blue). Arrowheads indicate the heterochromatic chromocenter, which shows robust MSL binding in roX- mutants. The genotypes of male larvae are roX1ex6 Df(1)roX252; [w+ 4Δ4.3] (A), y w roX1ex6 Df(1)roX252 [w+ 4Δ4.3]; msl2 cn/+; [M1][M2]/+ (B–D,F–H), and y w (wild type; E). roX- mutants with different genetic backgrounds and escaper frequencies showed similar chromosomal staining (data not shown).
Given their greatly improved X-chromosome morphology and MSL localization, we examined roX1- roX2-; [H83M1][H83M2]/+ males for viability (Table 1). We crossed roX mutant females from three genetically different backgrounds to male flies overexpressing MSL1 and MSL2. Progeny from all three lines showed enhanced male viability (up to 90-fold improvement) with overexpression of MSL1 and MSL2. This suggests that the protein components of the MSL complex mediate the chromatin changes during dosage compensation. Normally the roX RNAs play a crucial role in assembly or targeting of complexes, which can be partially recapitulated by MSL overexpression.
Screening for additional noncoding RNA component(s) of MSL complexes
One qualification of our results is that the roX1- roX2- double-mutant stocks might not be truly null for all roX RNAs. Although the roX2 allele is a full deletion, the roX1ex6 allele removes only the 5′ half of the gene, but retains the last ∼2.3 kb of the locus (Kelley et al. 1999). If a truncated roX1 transcript were incorporated into MSL complexes, we would expect it to accumulate to detectable levels. However, using Northern analysis (Fig. 4A) or reverse transcriptase PCR (RT–PCR; Fig. 4B), we were unable to detect any roX1 RNA in roX- escaper males carrying the roX1ex6 allele. Coupled with the observation that the roX1ex6 allele showed no evidence of MSL attraction and spreading in our polytene chromosome assay (Fig. 2B,E–G), these results suggest that the roX1ex6 allele is nonfunctional.
Figure 4.
MSL proteins are partially functional without roX RNA. To determine whether an incompletely deleted roX1 RNA retains residual expression, Northern blotting (A) and RT–PCR (B) were performed. (A) A full-length roX1 genomic fragment was used as a probe to detect any aberrant RNA from the roX1ex6 gene in males. The full genotype of the roX mutant is roX1ex6 Df(1)roX252; [w+ 4Δ4.3], which showed the highest escaper frequency. Hybridization to an rp49 probe is a loading control. (B) cDNA from RNA of adult males from each genotype was amplified by PCR (30 cycles). Primers from sequences retained in roX1ex6 were used for roX1 (261 bp) amplification. rps17 (210 bp) was amplified in the same tube as a positive control. No-RT controls produced no PCR products (data not shown). (C) Western analysis of roX- mutants. The total cell extract from one adult fly was loaded in each lane; 80 μg of total SL-2 cell extract was loaded as a positive control. The antibodies used for blotting are shown at left. The full genotype of the roX mutant is y w roX1ex6 Df(1)roX252 [w+ 4Δ4.3]; msl2 cn/+, which showed the lowest escaper frequency. roX- mutants with different genetic backgrounds and escaper frequencies showed similar results (data not shown). A similar level of MSL3 was also detected with or without roX RNA (data not shown). A Western blot for α-tubulin was a loading control.
We next asked whether additional species of roX RNA from unidentified genes might have gone undetected. We immunoprecipitated MSL complexes from wild-type embryos, recovered RNA, prepared cDNA, performed subtractive hybridization using excess female cDNA to deplete nonspecific contaminants, and cloned the enriched products. Out of 15 randomly picked colonies, we found that the resulting small cDNA library contained ∼70% roX1 and roX2 subclones (Supplementary Table 1A). To increase the chance of discovering new roX RNA(s), colonies containing roX1 or roX2 were excluded by colony hybridization with roX1 and roX2 probes. Another 31 clones were sequenced, but all represented nonspecific abundant RNAs (Supplementary Table 1B). Thus, we found no candidates for additional roX species. We conclude that if the MSL complex contains other RNA species, they have very different characteristics from roX1 and roX2. The fact that the MSLs spread from only the roX2+ locus in a roX1- mutant (Fig. 2B), or the roX1+ locus in a roX2- background (Fig. 2C), also strongly suggests that there are no additional RNAs functioning like roX1 and roX2.
Colocalization of MSL proteins to the X chromosome without roX RNA
We further investigated how loss of roX RNA affected MSL complexes in males overexpressing MSL1 and MSL2. Polytene chromosomes were stained with antibodies to each of the five MSL proteins and histone H4 acetylated at Lys 16. We found that MSL1, MSL2, MSL3, MOF, and H4Ac16 were colocalized to the X chromosome under these conditions (Fig. 3B–D; data not shown), showing substantial rescue of MSL localization to the X chromosome in the roX- mutant. The restoration of the precise MSL pattern was incomplete, as MSL proteins were still present at an increased number of autosomal sites and the heterochromatic chromocenter, as is seen in roX- mutants with wild-type levels of MSL proteins (Fig. 3A). Surprisingly, localization of the MLE helicase differed significantly from the other MSL proteins. Although MLE binds the X chromosome robustly in wild-type nuclei (Fig. 3E), it shows only very faint staining on the X chromosome in the absence of roX RNAs. However, it is still colocalized to some autosomal sites and the chromocenter with the other MSL proteins (Fig. 3F–H). These results demonstrate that stable localization of MLE to the male X chromosome is strongly dependent on roX RNAs, and is consistent with previous results showing that association of MLE with the X chromosome is particularly RNase-sensitive (Richter et al. 1996). Earlier work demonstrated the reciprocal result: roX RNA cannot bind the X chromosome without MLE helicase (Meller et al. 2000). We do not understand what attracts MLE to the ectopic autosomal sites and chromocenter, but note that MLE has an RNase-sensitive affinity for all chromosomes when overexpressed (Richter et al. 1996).
Given that males cannot live without mle function, why do some of these males survive without MLE enrichment on the X chromosome? We tested the possibility that the mle requirement also could be overcome by overexpression of MSL1 and MSL2. However, we recovered no mle escaper males in crosses in which excess MSL1 and MSL2 were supplied (data not shown). Therefore, although MLE is no longer concentrated on the X chromosome in roX mutant males, it still performs an essential function in dosage compensation. Perhaps it acts catalytically, associating only transiently with the other MSL proteins (Copps et al. 1998), or is sufficient when present at substoichiometric levels relative to the other MSL subunits.
If roX RNAs are not absolutely essential for MSL targeting, could they play some other role? One possibility is that assembly with the RNAs might stabilize the MSL proteins. To test this possibility, we checked the level of each MSL protein in roX- adult male escapers by Western blot (Fig. 4C). The amount of each MSL protein in these males was similar to that of wild-type males. This does not exclude a transient role for roX RNAs in MSL protein integrity but suggests that they instead play a specific role in efficient assembly or X localization of the MSL complex.
Model for MSL targeting to the X chromosome
Earlier observations that the MSL complex could spread in cis from an autosomal roX transgene lead to speculation that complexes normally spread from the endogenous roX loci on the X chromosome to paint the entire chromosome (Kelley et al. 1999). However, the initial characterization of roX1 clearly demonstrated that soluble MSL complexes could diffuse between chromosomes (Meller et al. 1997; Kelley et al. 1999). More recent work revealed that the ability of the MSL complex to spread from a site of roX transcription, or diffuse away, is highly sensitive to the balance between MSL proteins and roX transcripts in the nucleus (Park et al. 2002). Here, we demonstrate that the wild-type pattern of MSL complexes along the male X chromosome is the result of a delicate interplay between two targeting strategies. Local spreading from roX loci operates in parallel with a second route where soluble MSL complexes diffuse and reattach to distant segments of the X chromosome. We can alter the proportion of MSL complexes entering each pathway by manipulating the amount of MSL proteins or roX RNAs present. We speculate that the underlying mechanism controlling these two outcomes rests on how efficiently MSL subunits can assemble into functional complexes (Fig. 1).
Materials and methods
Fly stocks and genetic crosses
Flies were raised on standard cornmeal–yeast–agar–molasses medium containing propionic acid. To obtain X-chromosome insertions of roX transgenes, [w+ GMroX1-69C] and [w+ GMroX2-40A] were remobilized in a roX- mutant background {y w roX1ex6 Df(1)roX252 [w+ 4Δ4.3]; Park et al. 2002} using genomic P transposase (Robertson et al. 1988). The new transgenic insertion sites were mapped by inverse PCR and sequencing. To overexpress MSL1 and MSL2, y w (wild type), y w roX1ex6 (Kelley et al. 1999), w Df(1)roX252; [w+ 4Δ4.3] (Meller and Rattner 2002), y w roX1ex6 Df(1)roX252 [GMroX1-13A][w+ 4Δ4.3] or y w roX1ex6 Df(1)roX252 [GMroX2–14F][w+ 4Δ4.3] females were crossed with w/Y; msl2, cn/+; [w+ H83M1][w+ H83M2]/TM6B, Tb males, and non-Tb male third-instar larvae were collected for anti-MSL1 immunostaining of polytene chromosomes. For Northern and Western blots, y w (wild type), y w roX1ex6, w Df(1)roX252; [w+ 4Δ4.3] or yw roX1ex6 Df(1)roX252 [w+ 4Δ4.3] females were crossed with w/Y; msl2 cn; [w+ H83M1][w+ H83M2]/+ males. The resulting adult male progeny carrying the MSL1 and MSL2 transgenes were selected by eye color. [w+ 4Δ4.3] supplies essential genes lost in Df(1)roX252 (Meller and Rattner 2002).
Immunostaining, Northern, and Western analyses
Immunostaining of polytene chromosomes, Northerns, and Westerns were as described previously (Kelley et al. 1999).
RT–PCR
Five micrograms of total RNA from adult males was reverse-transcribed using SuperScriptII reverse transcriptase (Invitrogen) and oligo(dT) primers, as described previously (Meller et al. 2000). Ten percent (2 μL) of the RT reaction was used in a 50-μL PCR amplification by using 0.2 μM rps17 primers (F: 5′-CGAACCAAGACGGTGAAGAAG-3′ and R: 5′-CC TGCAACTTGATGGAGATACC-3′) and roX1 primers (5A: 5′-CCCAGA AGAAACTGCCACTGC-3′ and 7B′: 5′-AATGTCCCTTTTCGAGCG-3′). PCR was performed as follows: at 94°C for 4 min, 30 cycles (at 94°C for 30 sec, at 55°C for 30 sec, at 72°C for 1 min), and at 72°C for 10 min. RT–PCR products were electrophoresed and stained with EtBr.
Immunoprecipitation and subtractive hybridization of RNA from MSL complexes
Total protein extracts of wild-type embryos (0∼24 h old) were prepared by homogenization and sonication as described previously (Smith et al. 2000) in extract buffer [20 mM HEPES at pH 7.6, 70 mM KCl, 2 mM DTT, 0.1% NP-40, 8% glycerol, 1 mM PMSF, 1 U/μL RNAsin (Promega), protease inhibitor cocktail; Lanz et al. 1999]. Extracts containing 10 mg of total protein were incubated with 10 μL of rabbit polyclonal α-MLE antibody, and the RNA from that immunoprecipitate was isolated as described previously (Meller et al. 2000). This RNA was used to make tester cDNA, whereas 10 μg of total RNA from adult female flies was used to make driver cDNA for subtraction (Clontech PCR-select cDNA subtraction kit). The resulting MSL-complex-specific cDNA fragments that survived the subtraction were amplified and subcloned for sequencing.
Acknowledgments
We are grateful to R.L. Kelley for help with Figure 1, for critical reading of the manuscript, and for many helpful discussions. We thank A. Alekseyenko, X. Bai, and C. Stuckenholz for critical reading of the manuscript and C. Stuckenholz for fly stocks and sharing unpublished data. We thank H. Kennedy and R. Richman for excellent technical support. This work was supported by the Welch Foundation (Q-1359), the National Institutes of Health (GM45744), and the Howard Hughes Medical Institute. M.I.K. is an HHMI Investigator.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1082003.
Footnotes
Supplemental material is available at http://www.genesdev.org.
References
- Amrein H. and Axel, R. 1997. Genes expressed in neurons of adult male Drosophila. Cell 88: 459–469. [DOI] [PubMed] [Google Scholar]
- Badenhorst P., Voas, M., Rebay, I., and Wu, C. 2002. Biological function of the ISWI chromatin remodeling complex NURF. Genes & Dev. 16: 3186–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. [DOI] [PubMed] [Google Scholar]
- Belote J.M. and Lucchesi, J.C. 1980. Control of X chromosome transcription by the maleless gene in Drosophila. Nature 285: 573–575. [DOI] [PubMed] [Google Scholar]
- Chang K.A. and Kuroda, M.I. 1998. Modulation of MSL1 abundance in female Drosophila contributes to the sex specificity of dosage compensation. Genetics 150: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copps K., Richman, R., Lyman, L.M., Chang, K.A., RampersadAmmons, J., and Kuroda, M.I. 1998. Complex formation by the Drosophila MSL proteins: Role of the MSL2 RING finger in protein complex assembly. EMBO J. 17: 5409–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R., Fanti, L., Armstrong, J.A., Sarte, M., Papoulas, O., Prestel, M., Daubresse, G., Verardo, M., Moseley, S.L., Berloco, M., et al. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5: 355–365. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. 1957. The X-chromosome in the larval salivary glands of hybrids Drosophila insularis and Drosophila tropicalis. Chromosoma 8: 691–698. [DOI] [PubMed] [Google Scholar]
- Franke A. and Baker, B.S. 1999. The roX1 and roX2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol. Cell 4: 117–122. [DOI] [PubMed] [Google Scholar]
- Ho Y., Elefant, F., Cooke, N., and Liebhaber, S. 2002. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9: 291–302. [DOI] [PubMed] [Google Scholar]
- Kelley R.L., Wang, J., Bell, L., and Kuroda, M.I. 1997. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387: 195–199. [DOI] [PubMed] [Google Scholar]
- Kelley R.L., Meller, V.H., Gordadze, P.R., Roman, G., Davis, R.L., and Kuroda, M.I. 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98: 513–522. [DOI] [PubMed] [Google Scholar]
- Lachner M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120. [DOI] [PubMed] [Google Scholar]
- Lanz R.B., McKenna, N.J., Onate, S.A., Albrecht, U., Wong, J., Tsai, S.Y., Tsai, M.-J., and O'Malley, B.W. 1999. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97: 17–27. [DOI] [PubMed] [Google Scholar]
- Lee J.T. and Jaenisch, R. 1997. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature 386: 275–279. [DOI] [PubMed] [Google Scholar]
- Meller V.H. and Rattner, B.P. 2002. The roX RNAs encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 21: 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller V.H., Wu, K.H., Roman, G., Kuroda, M.I., and Davis, R.L. 1997. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88: 445–457. [DOI] [PubMed] [Google Scholar]
- Meller V.H., Gordadze, P.R., Park, Y., Chu, X., Stuckenholz, C., Kelley, R.L., and Kuroda, M.I. 2000. Ordered assembly of roX RNAs into MSL complexes on the dosage compensated X chromosome in Drosophila. Curr. Biol. 10: 136–143. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Rice, J.C., Strahl, B.D., Allis, D.C., and Grewal, S.I. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. [DOI] [PubMed] [Google Scholar]
- Offermann C.A. 1936. Branched chromosomes as symmetrical duplications. J. Genet. 32: 103–116. [Google Scholar]
- Palmer M.J., Richman, R., Richter, L., and Kuroda, M.I. 1994. Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes & Dev. 8: 698–706. [DOI] [PubMed] [Google Scholar]
- Park Y., Kelley, R.L., Oh, H., Kuroda, M.I., and Meller, V.H. 2002. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298: 1620–1623. [DOI] [PubMed] [Google Scholar]
- Richter L., Bone, J.R., and Kuroda, M.I. 1996. RNA-dependent association of the Drosophila maleless protein with the male X chromosome. Genes Cells 1: 325–336. [DOI] [PubMed] [Google Scholar]
- Robertson A.M., Preston, C.R., Phillis, R.W., Johnson-Schlitz, D., Benz, W.K., and Engels, W.R. 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118: 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.R., Pannuti, A., Gu, W., Steurnagel, A., Cook, R.G., Allis, C.D., and Lucchesi, J.C. 2000. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20: 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckenholz C. 2002. “Functional redundancy within roX1, a noncoding RNA involved in dosage compensation in Drosophila melanogaster.” Ph.D. thesis, Baylor College of Medicine, Houston, TX. [DOI] [PMC free article] [PubMed]