Abstract
We have developed a new vector, named pDECAP, to express long double-strand RNA (ds-RNA) from an RNA polymerase II (Pol II) promoter. Because the transcripts from pDECAP lack both the 5′-cap structure and the 3′-poly(A) tail that facilitate ds-RNA export to the cytoplasm, long ds-RNA from pDECAP does not induce the interferon response. Transgenic mice embryos expressing long ds-RNA for the transcriptional corepressor Ski from this vector exhibited phenotypes that were remarkably similar to those of Ski-deficient embryos, including defects of neural tube closure and eye formation. Thus, this vector provides a new tool to efficiently generate tissue-specific knockdown mice for studying gene function in whole animal systems.
Keywords: RNA interference, double-strand RNA, transgenic mice, pol II promoter, Ski corepressor
Although mice mutants lacking a specific gene are very useful for analyzing gene function, the procedure used to generate mutant mice by homologous recombination in embryonic stem (ES) cells (Capecchi 1989) is time-consuming. RNA interference provides an alternative approach by inducing sequence-specific mRNA degradation with a 21- or 22-nucleotide (nt) small interfering RNA (siRNA) generated by ribonuclease III cleavage of longer double-strand RNA (ds-RNA; Hamilton and Baulcombe 1999; Hammond et al. 2000; Zamore et al. 2000; Bernstein et al. 2001; Elbashir et al. 2001a; Hannon 2002). Overexpression of 21-nt ds-RNA induces degradation of specific mRNA in mammalian cultured cells (Caplen et al. 2001; Elbashir et al. 2001b; Brummelkamp et al. 2002; Paddison et al. 2002a). The RNA polymerase III (Pol III) H1- and U6-RNA gene promoters were shown to efficiently express 21-nt ds-RNA in mammalian cultured cells (Brummelkamp et al. 2002; Paddison et al. 2002a). However, the Pol III promoter is active in all of the tissues and cannot be used to generate the tissue-specific knockout mice. The use of the RNA polymerase II (Pol II) promoter is essential for developing a method to efficiently generate tissue-specific knockout mice.
One of the difficulties in the use of long ds-RNA transcribed from the Pol II promoter is the interferon response. Pol II transcripts are transferred to the cytosol immediately after transcription, where they induce interferon synthesis (Stark et al. 1998). During this response, ds-RNA of >30 bp binds and activates the PKR protein kinase and 2′, 5′-oligoadenylate synthetase, which then leads to a block in translation and mRNA degradation (Minks et al. 1979; Manche et al. 1992). In fact, it was reported that long ds-RNA in the cytoplasm can induce the interferon response, which leads to a block in translation and sequence-nonspecific mRNA degradation (Elbashir et al. 2001b; Paddison et al. 2002b). Here, by overexpressing ds-RNA from the Pol II promoter in various tissues, we attempted to generate mice mutated in a specific gene. To prevent the interferon response, we have developed a new vector to express long ds-RNA from the Pol II promoter, which efficiently blocks the export of long ds-RNA to the cytosol. Because the ribonuclease(s) such as Dicer that cleave long ds-RNA are localized in both the nucleus and the cytoplasm (Lee et al. 2002), the long ds-RNA expressed from this vector is expected to be processed into siRNA in the nucleus, which then moves to the cytosol to induce the degradation of target mRNA. Transgenic mice embryos expressing long ds-RNA for the transcriptional corepressor Ski from this vector exhibited phenotypes that were remarkably similar to those of Ski-deficient embryos.
Results and Discussion
Expression of long Ski ds-RNA using the pDECAP vector induces Ski mRNA degradation
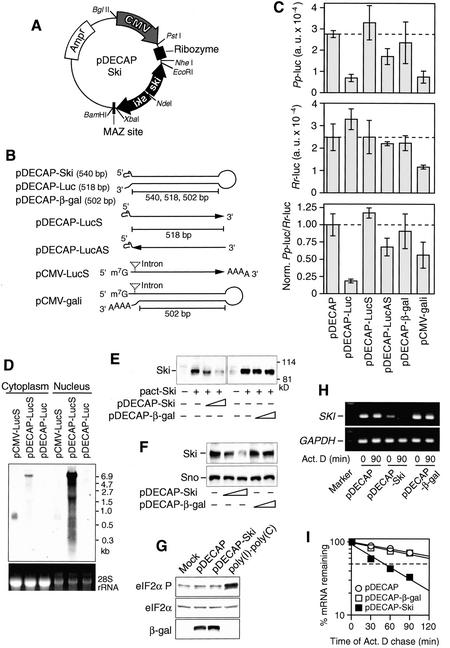
In order to prevent the interferon response when a long ds-RNA is expressed in mammalian cells, we generated a novel pDECAP vector [Deletion of Cap structure and poly(A)] that efficiently blocks the export of long ds-RNA to the cytosol. For efficient export of mRNA to the cytosol, all pre-mRNA processing, including 5′-capping, splicing, and 3′-polyadenylation, must be completed. The pDECAP vector was designed to express mRNA lacking 7-methylguanosine (m7G) cap structure at its 5′ end and a poly(A) tail at its 3′ end from the cytomegalovirus (CMV) promoter, which is active in various tissues of mouse embryos (Fig. 1A; Schmidt et al. 1990). Normally m7G cap is added to the nascent mRNA cotranscriptionally, after which the cap-binding complex accompanies the mRNA to the cytoplasm by interacting with nuclear eIF4F (McKendrick et al. 2001). The 3′ ends of Pol II transcripts are formed by cleavage and polyadenylation after transcription termination, and the polyadenylation is required for the nuclear-cytoplasmic transport of mRNA (Huang and Carmichael 1996). To remove the 5′-cap structure, we added the cis-acting hammerhead ribozyme (Huang and Carmichael 1996) at a site downstream of the RNA start site. In addition, the specific sequence that mediates Pol II transcriptional pausing and binding of the zinc finger protein MAZ was added (Yonaha and Proudfoot 1999, 2000). A MAZ site is present between the closely spaced human genes, complement C2 and factor B, g11-C4, and within an intron of the mouse IgM-D gene (Ashfield et al. 1994). No intron was added to the pDECAP vector because splicing sites recruit specific proteins to transcripts that enhance the rate of their export from the nucleus (Luo and Reed 1999).
Figure 1.
Overexpression of 540-nt Ski ds-RNA induces Ski mRNA degradation. (A) Structure of the pDECAP vector. The pDECAP vector contains the CMV promoter, a ribozyme cassette to cut off the m7G cap structure, and a MAZ site for Pol II pausing. The gene-specific targeting sequence was cloned between the ribozyme cassette and the MAZ site as an inverted repeat separated by a 12-nt spacer. (B) The predicted secondary structures of RNAs transcribed from pDECAP vectors. pDECAP-Ski, pDECAP-Luc, and pDECAP-β-gal encode a 540- to 502-bp hairpin ds-RNA for Ski, firefly luciferase, and β-galactosidase, respectively, which lack the 5′ cap structure and a poly(A) tail. Sense and antisense single-stranded RNA of the luciferase gene were transcribed from pDECAP-LucS and pDECAP-LucAS, respectively. pCMV-gali encodes the same hairpin ds-RNA as pDECAP-β-gal except that it contains the 5′ cap structure, a spliced intron, and a poly(A) tail. (C) Effect of long ds-RNA expressed from the pDECAP vector on the luciferase activity. The firefly luciferase (Pp-luc) expression plasmid was cotransfected with the plasmid shown below, which encodes various forms of RNA, and the internal control plasmid expressing sea pansy luciferase (Rr-luc) in mouse embryonic fibroblasts. (Top panel) Firefly luciferase (Pp-luc) activity plotted in arbitrary luminescence units (a.u.). (Middle panel) Sea pansy luciferase (Rr-luc) activity plotted in arbitrary luminescence unit. (Bottom panel) Ratios of normalized target to control luciferase. (D) RNA transcribed from pDECAP vector accumulated in the nucleus. 293T cells were transfected with indicated plasmids (0.8 μg) and cytoplasmic and nuclear RNA were extracted 48 h later. The subcellular distribution of RNAs was examined by Northern blot using a digoxigenin (DIG)-labeled LucS DNA probe. (E) Decrease in the Ski protein level induced by pDECAP-Ski vector. The Ski expression plasmid (pact-Ski, 0.4 μg) was transfected into 293T cells together with the indicated plasmid (0.2 or 0.4 μg) or the control empty vector (-). The luciferase gene expression plasmid (TK-Luc, 0.05 μg) was cotransfected as an internal control. Forty-two hours after transfection, cell extracts were prepared and immunoblotted detect Ski protein. The amounts of cell lysates were adjusted according to the transfection efficiency estimated by the luciferase assay. (F) Decrease in the endogenous Ski protein level induced by pDECAP-Ski vector. 293T cells were transfected with the indicated plasmids and endogenous Ski and Sno proteins were detected by immunoblotting. (G) Expression of long ds-RNA the nucleus did not affect the phosphorylation of eIF2α. 293T cells were cotransfected with pDECAP or pDECAP-Ski plasmid (0.6 μg) and pCMV-β-gal plasmid (0.3 μg) as a internal control. The cells were harvested 48 h later and subjected to immunoblot analysis using phospho-specific antibody against eIF2α phosphorylated on Ser 51 (eIF2α-P). The same membrane was reprobed with antibodies against total eIF2α and β-galactosidase. The lysate of 293T cells treated with poly(I)·poly(C) (100 μg/mL) for 24 h was used as a positive control. (H,I) Degradation of Ski mRNA by pDECAP-Ski. The indicated plasmid (1.5 μg) was transfected into 293T cells with the enhanced green fluorescent protein (EGFP) expression plasmid, pMSCV-ir-EGFP (0.5 μg). EGFP-positive cells were collected by sorting and treated with actinomycin D. Total RNA was isolated at various times and the amount of Ski mRNA was analyzed by RT–PCR (F) or quantitative real-time RT–PCR (G). The RT–PCR products were separated on 2% agarose gels and visualized by ethidium bromide staining. GAPDH mRNA was use as a control.
It was reported that ds-RNAs of >400 nt were quite effective for gene silencing, whereas ds-RNA of <100 nt had few effects (Hammond et al. 2000). Therefore, we first designed a gene-specific insert in the pDECAP vector so that it specified a 518-nt sequence derived from the firefly (Photinus pyralis) luciferase gene, separated by a short spacer from the reverse complement of the same 518-nt sequence (Fig. 1B). The nascent transcript from this vector was expected to form a long ds-RNA with a small loop during transcriptional pausing, and to be processed into siRNA in the nucleus because it lacked a 5′-cap structure, a poly(A) tail, and splicing sites as described earlier. This expression vector was cotransfected into mouse embryonic fibroblast cells with the firefly luciferase expression vector (TK-βREx2-Luc), which contains the thymidine kinase (TK) promoter and β-retinoic acid receptor-binding sites, and the internal control vector to express sea pansy (Renilla reniformis) luciferase from the CMV promoter. Firefly luciferase levels were reduced about fivefold on cotransfection with the pDECAP vector expressing firefly luciferase ds-RNA, whereas Renilla luciferase activity was not affected (Fig. 1C). Cotransfection of the pDECAP vector expressing sense RNA for the same region of the firefly luciferase gene did not affect the firefly luciferase activity, whereas cotransfection of the pDECAP vector expressing antisense RNA for the same region had a slight affect (about a 25% reduction), indicating that the structure of the ds-RNA is important for the efficient inhibition of firefly luciferase activity. Moreover, coexpression of a 502-bp ds-RNA for the β-galactosidase gene from the pDECAP vector did not affect firefly or Renilla luciferase activities, but coexpression of a β-galactosidase ds-RNA containing a 5′-cap structure, splicing sites, and a poly(A) tail from another vector (pCMV-gali), which had the CMV promoter but lacked both ribozyme and MAZ sites, decreased the activities of both luciferases, indicating that the removal of the cap structure and the poly(A) is required to suppress nonspecific inhibition of translation.
We analyzed the transcripts by Northern blotting using the cytoplasmic and nuclear RNAs prepared from 293T cells transfected with various constructs shown in Figure 1B. Most of the luciferase sense RNAs transcribed from pDECAP (pDECAP-LucS) were in the nucleus, whereas the luciferase sense RNA transcribed from the poly(A) site-containing control vector that lacked the ribozyme and MAZ sites (pCMV-LucS) were in the cytoplasm (Fig. 1D). The size of the luciferase transcripts from pDECAP-LucS corresponds to that of plasmid, suggesting that the long RNAs were transcribed by a read-through mechanism because of a lack of the poly(A) addition signal. The long ds-RNA transcribed from pDECAP-Luc was detected neither in the cytoplasm nor in the nucleus by Northern blotting, suggesting that it was immediately digested into small RNAs.
We next investigated the effect of a long ds-RNA for the Ski gene on Ski protein levels. Ski forms a complex with histone deacetylases and is required for the transcriptional repression by various repressors, including Mad, Rb, thyroid hormone receptor-β, MeCP2, and Gli3 (Nomura et al. 1999; Tokitou et al. 1999; Kokura et al. 2001; Dai et al. 2002). The pDECAP vector (pDECAP-Ski) expressing a 540-bp ds-RNA for the N-terminal portion of Ski was cotransfected into 293T cells together with the Ski expression vector, and Ski protein levels were examined by Western blotting (Fig. 1E). The levels of Ski protein were reduced by cotransfection of pDECAP-Ski in a dose-dependent manner. When the pDECAP-Ski vector alone was transfected into 293T cells, the levels of endogenous Ski protein were also similarly decreased (Fig. 1F). There was no apparent change in the endogenous levels of Sno protein, which has about 60% homology with the targeted sequence of pDECAP-Ski.
To investigate whether Ski ds-RNA expressed from pDECAP-Ski induces the interferon response, we examined the phosphorylation status of eIF2α, a substrate of the ds-RNA-activated PKR protein kinase. The levels of phosphorylated eIF2α were not enhanced by transfection of 293T cells with pDECAP-Ski, whereas the phosphorylation of eIF2α was stimulated in the control cells treated with poly(I) · poly(C) (Fig. 1G).
The reverse transcriptase PCR (RT–PCR) analyses indicated that the levels of endogenous Ski mRNA were also reduced by transfection with pDECAP-Ski. To confirm that the decreased Ski mRNA levels were due to the degradation of Ski mRNA, we measured Ski mRNA levels at various times after treatment of the transfected cells with actinomycin D, an inhibitor of transcription (Fig. 1H,I). In the cells transfected with the pDECAP-Ski vector, the half-life of endogenous Ski mRNA was 0.9 h, whereas it was 2.7 and 3.1 h in cells transfected with the pDECAP vector expressing β-galactosidase ds-RNA and the empty vector, respectively. Thus, Ski ds-RNA expressed from the pDECAP vector stimulated the degradation of Ski mRNA.
Defects in neural tube and eye formation in pDECAP-Ski transgenic mice
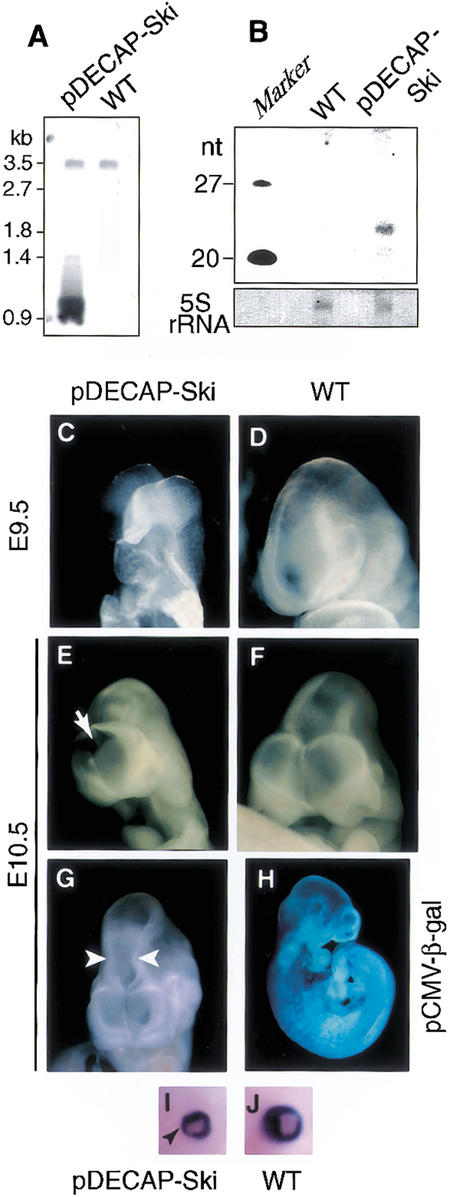
We then prepared a DNA fragment containing the expression unit of the 540-nt Ski ds-RNA from the pDECAP-Ski vector, injected it into the fertilized mouse oocytes, and generated transgenic mice. Integration of the DNA fragment was confirmed by Southern blotting (Fig. 2A). The copy number of the transgene was estimated to be 2–12 copies. To examine whether the small RNA was generated in the transgenic mouse embryo, we performed Northern blotting using the Ski sense-strand probe. The small RNA of about 21–22 nt was detected in the pDECAP-Ski transgenic mouse embryo, but not in the wild-type embryo (Fig. 2B).
Figure 2.
Defects in neural tube and eye formation in pDECAP-Ski transgenic mice. (A) Southern blot analysis of genomic DNA from animals carrying pDECAP-Ski sequence. (B) Detection of siRNA. Nuclear RNA (5 μg) extracted from the head of a transgenic mouse embryo was analyzed by Northern blotting. The membrane was probed with a DIG-labeled sense Ski probe corresponding to the targeting sequence. Shown here is 5S rRNA visualized by staining with methylene blue on the membrane as a loading control. (C–G) Neural tube closure defect and narrowing of the neural tube (second ventricle) were observed in pDECAP-Ski transgenic embryos. Wild-type (D,F) and pDECAP-Ski embryo (C,E,G) at E9.5 (C,D) and E10.5 (E–G) are shown. (H) Expression of β-galactosidase in the embryos of pCMV-β-gal transgenic mice. Staining for β-galactosidase activity at E10.5 is indicated. (I,J) Eye formation defects. Wild-type (I) and pDECAP-Ski (J) embryos at E14.5 are shown. Black arrowhead indicates hypoplasia of the distal end of retina.
We analyzed for abnormalities in the embryos of pDECAP-Ski transgenic mice at stages of E9–E12 and E14–E16. Among 13 embryos containing the exogenous pDECAP-Ski DNA fragment, 5 embryos (38.5%, 5/13) exhibited defects in neural tube closure or narrowing of the second ventricle at E9–E12 (Figs. 2C–G, 3A; Table 1). In addition, eye formation defects, including hypoplasia of the distal end of retina, which forms the iris, were observed in two embryos analyzed at E14–E16 (100%, 2/2; Figs. 2I,J, 3E; Table 1). These defects could not be correlated with the copy number of the transgene, suggesting that the expression levels of Ski ds-RNA may depend on the integration site of the transgene, and not on the copy number. The defects were similar to those seen in Ski-deficient embryos with mixed genetic backgrounds generated by homologous recombination using ES cells (Berk et al. 1997; Colmenares et al. 2002). The abnormalities seen in the Ski-deficient mice, including the cranial neural tube closure defect and the eye abnormalities, were reported to be affected by genetic background (Colmenares et al. 2002), and we confirmed that most of the Ski-deficient mice generated by using ES cells still had these abnormalities after 10 generations of back-crossing with C57BL/6N mice. Based on this observation, the fertilized mouse oocytes derived from matings between C57BL/6N male and C57BL/6N female mice were used to generate transgenic mice in this study.
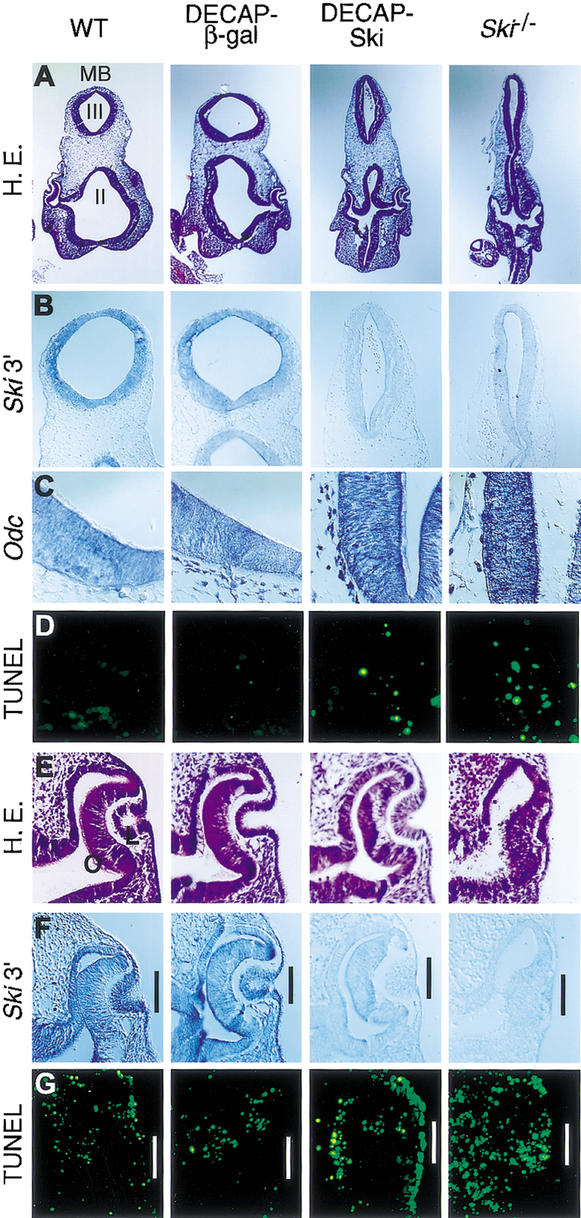
Figure 3.
Defects in pDECAP-Ski transgenic mice is caused by a loss of Ski mRNA. Developmental stage-matched wild-type, pDECAP-β-gal transgenic, pDECAP-Ski transgenic, and Ski-deficient embryos were analyzed. (A) Hematoxylin-eosin staining (H&E) of head sections. (B,C) Ski and Odc mRNA expression in midbrain and mesenchyme as detected by RNA in situ hybridization. For detection of Ski mRNA, the 3′-half of the Ski gene was used as the probe. (D) Apoptotic cells were assessed by the TUNEL assay on the neuroepithelium of midbrain and mesenchyme. (E) H&E staining on eyes. (F) Ski mRNA expression in eyes as detected by in situ hybridization. (G) TUNEL assay on eyes. MB, midbrain; II, second ventricle; III, third ventricle; O, optic cup; L, lens vesicle.
Table 1.
Generation and analysis of transgenic mice
| Number of embryos exhibiting phenotypes/number of embryos analyzed
|
|||||
|---|---|---|---|---|---|
| DNA injected | Stage of analyzed embryos | Number of transgenic embryos obtained | neural tube defect | eye defect | β-gal expression |
| pDECAP-Ski | E9-E12 | 13 | 5/13 (38.5%) | 2/2 (100%)a | ND |
| E14-E16 | 2 | 0/2 (0%) | 2/2 (100%) | ND | |
| pDECAP-β-gal | E9-E11 | 9 | 0/9 (0%) | ND | ND |
| pCMV-β-gal | E9-E12 | 6 | 0/6 (0%) | ND | 2/6 (33.3%) |
| E15 | 1 | 0/1 (0%) | 0/1 (0%) | 1/1 (100%) | |
(ND) Not determined.
Ski expression in lens vesicle of E10.5 embryos.
The CMV promoter was reported to be active in various regions of mouse embryos (Schmidt et al. 1990). We confirmed this by generating the transgenic mice embryos containing the β-galactosidase expression vector driven by the CMV promoter (pCMV-β-gal). In two of six transgenic mice embryos containing the β-galactosidase expression vector driven by the CMV promoter (pCMV-β-gal), the β-galactosidase was expressed in almost all of the tissues, including the neural epithelial cells, at E9–E12 (Fig. 2H). This frequency (33.3%, 2/6) is similar to that of the pDECAP-Ski transgenic mice embryos exhibiting abnormalities and is within the average frequency (10%–40%) of transgenic mice showing specific phenotype caused by the injected DNA. Consistent with the high frequency of pDECAP-Ski transgenic embryos exhibiting eye defect at E14–E16, β-galactosidase expression was frequently observed in various tissues, including the eye, of the pCMV-β-gal transgenic mice embryo at E15. As control experiments, we also generated transgenic mice by injecting a fragment containing the expression unit of 502-nt β-galactosidase ds-RNA prepared from the pDECAP-β-gal. No abnormality was observed at all in the nine embryos analyzed (Table 1), indicating that the abnormalities observed in the pDECAP-Ski transgenic mice were specifically caused by Ski ds-RNA, and not nonspecifically by long ds-RNA. Although the typical abnormalities of Ski-deficient mice such as neural tube defects and eye defect were seen in pDECAP-Ski transgenic mice, other defects such as postaxial polydactyly of Ski-deficient mice (Colmenares et al. 2002) were not observed in the pDECAP-Ski transgenic mice. This may be because the CMV promoter was not sufficiently active in some tissues.
Defects in pDECAP-Ski transgenic mice is caused by a loss of Ski mRNA
To confirm that the levels of endogenous Ski mRNA were lower in the tissues of the transgenic mice showing abnormalities, in situ hybridization experiments were performed using a probe corresponding to the 3′-half of the Ski gene (Fig. 3B). In the neural epithelial cells of the pDECAP-Ski transgenic mice and the Ski-deficient mice, which were generated by homologous recombination in ES cells, Ski mRNA was not detected. In contrast, Ski mRNA was expressed in the neural epithelial cells of pDECAP-β-gal transgenic mice and wild-type mice. These results were confirmed by RT–PCR (data not shown). The neural tube closure defects of Ski-deficient mice were caused by the ectopic expression of the ornithine decarboxylase (Odc) gene followed by apoptosis in neural epithelial cells, because Ski is required for Mad-mediated transcriptional repression of the Odc gene (Nomura et al. 1999). Similar ectopic expression of the Odc gene was observed in neural epithelial cells of pDECAP-Ski transgenic mice embryos (Fig. 3C). Furthermore, in regions where the levels of Odc expression were high, ectopic apoptosis was observed (Fig. 3D). These results indicate that the defects in neural tube closure observed in pDECAP-Ski transgenic mice are caused by the same mechanism that operates in Ski-deficient mice generated by homologous recombination in ES cells.
During eye development, different retinal cell types, including retinal ganglion cells, cone photoreceptors, amacrine cells, and rod photoreceptors, are generated from the retinal progenitor cells in a fixed chronological sequence (Young 1985). In wild-type embryo, Ski mRNA was detected in the lens vesicle, optic cup, and surface epithelium at E10.5 (Fig. 3F). In embryos of pDECAP-Ski transgenic mice and Ski-deficient mice, the levels of Ski mRNA in these regions were dramatically reduced. Consistent with these observations, we observed ectopic apoptosis in the lens vesicle and the surface epithelium of pDECAP-Ski transgenic mice and Ski-deficient mice, but not in wild-type embryos (Fig. 3G). However, ectopic expression of the Odc gene was not seen in theses regions (data not shown), suggesting that some other uncharacterized target gene, which is negatively regulated by Ski, may be up-regulated. Ectopic apoptosis of the optic cup was observed in the Ski-deficient embryos but was not apparent in pDECAP-Ski transgenic embryos, suggesting that low levels of Ski mRNA may remain at this region.
The present study indicates that ectopic expression of a 540-bp Ski ds-RNA, which lacks both a 5′-cap structure and a 3′-poly(A) tail, induces abnormalities that are nearly identical to those seen in Ski-deficient mice. Our results are consistent with reports that siRNA functions in both mouse embryo and adult mice (Wianny and Zernicka-Goetz 2000; Lewis et al. 2002; McCaffrey et al. 2002). Expression of long ds-RNA has been shown to induce gene silencing only in a limited number of cell types. In most cell types, it causes nonspecific mRNA degradation and translation blockage. The present results indicate that the removal of both the 5′-cap structure and 3′-poly(A) from long ds-RNA is effective in suppressing the interferon response. It was reported that adenosine in ds-RNA is deaminated to inosine by adenosine deaminases that act on RNA (ADARs) and that the editing of ds-RNA by ADARs blocks the processing of ds-RNA to siRNA (Knight and Bass 2002). Although ADAR1 is expressed at high levels in brain (Paul and Bass 1998), the endogenous Ski gene was effectively silenced in the neuroepithelium of pDECAP-Ski transgenic mice. These results suggest that the activity of ADARs is not high enough to block the formation of siRNA in vertebrates. Although pDECAP-Ski did not down-regulate Sno, it should be noted that one potential disadvantage of using long ds-RNA as compared with small hairpin RNAs is that if there are regions containing homology with other related genes, nonspecific silencing might occur. The use of the pDECAP vector containing various promoters for tissue-specific ds-RNA expression is expected to accelerate the study of gene function in the whole animal body system by allowing the efficient generation of tissue-specific knockdown mice with specific defects in certain genes.
Materials and methods
Construction of Ski ds-RNA expression vector
The plasmid to express Ski ds-RNA (pDECAP-Ski) was constructed by cloning the first 540 bp of the mouse Ski coding region into the pDECAP vector as an inverted repeat with a 12-bp spacer (GGTGCGCATATG). The pDECAP vector contains CMV promoter, ribozyme cassette (Huang and Carmichael 1996) to cut off the m7G cap structure at the 5′-end of the transcribed RNA (CTGCAGCTCGAGATGCATGTCGCGGCCGC CTCCGCGGCCGCCTGATGAGTCCGTGAGGACGAAACATGCAT AGGCTAGC), and MAZ site (Yonaha and Proudfoot 2000) for Pol II pausing (GGCCCTTATCAGGGCCTCTGGCCTTGGGGGAGGGGGA GGCCAGAATG). There is no poly(A) addition signal in the construct. In pDECAP-Luc and pDECAP-β-gal, the first 518 bp of the firefly luciferase gene-coding region and spanning nucleotides 443–944 of the β-galactosidase gene are cloned as inverted repeats with a 12-bp spacer (TTTACC GGGCCC and TATCGGCATATG), respectively. The pDECAP plasmid containing the inverted repeat was amplified in Escherichia coli Sure 2 strain (Stratagene), which allows the accurate replication of DNA containing inverted repeats, and purified using the EndoFree Plasmid Maxi Kit (Qiagen).
Luciferase assay
Mouse embryonic fibroblast cells were cultured in Dulbeco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) and antibiotics. Cells (0.5 × 105 cells) were transferred to 12-well plates (1 mL per well) and transfected with mixtures of the firefly luciferase expression plasmids (TK-βREx2-Luc, 0.1 μg), plasmids indicated in Figure 1C (0.4 μg), and the Renilla luciferase expression plasmid (pRL-CMV, 0.02 μg) by using Lipofectamine (Invitrogen) according to the manufacture's instructions. Twelve hours later, 1 μM of all-trans retinoic acid was added to induce expression of the firefly luciferase gene and cells were cultured for a further 36 h. Luciferase expression was subsequently monitored with the dual luciferase assay (Promega).
Generation of transgenic mice
The 2.2-kb BglII-BamHI fragment of pDECAP-Ski or pDECAP-β-gal was freed from background sequences, purified, and injected into fertilized mouse oocytes. The fertilized mouse oocytes were derived from matings between C57BL/6N male mice and C57BL/6N or (C57BL/6J× DBA) F1 female mice. Embryos were analyzed for the presence and copy number of the transgene by Southern blot analysis with a probe for the first 540 bp of the Ski coding region. Genomic DNA used for Southern blot analysis was extracted from extraembryonic tissue.
Histological analysis
In situ hybridization was performed essentially as described (Butler et al. 2001). To prepare the c-Ski 3′ probe used, a plasmid spanning nucleotides 754–2223 of the mouse c-Ski cDNA was linearized with BamHI and transcribed with T7 RNA polymerase. The ODC probe has been described previously (Nomura et al. 1999). The TUNEL procedure was used to study apoptosis, using a commercially available apoptosis kit (in situ cell death detection kit, Roche) according to the supplier's instructions.
Other methods are described in Supplemental Material.
Acknowledgments
We are grateful to T. Maekawa for technical assistance for DNA injection, C. Colmenares for the Ski-deficient mice, and K. Umesono for the TK-βREx2-Luc plasmid. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science and Technology, by the Industrial Technology Research Grant Program in 2002 from NEDO of Japan, and by a grant from the Human Frontier Science Program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1073003.
Footnotes
Supplemental material is available at http://www.genesdev.org.
References
- Ashfield R., Patel, A.J., Bossone, S.A., Brown, H., Campbell, R.D., Marcu, K.B., and Proudfoot, N.J. 1994. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 13: 5656–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk M., Desai, S.Y., Heyman, H.C., and Colmenares, C. 1997. Mice lacking the ski proto-oncogene have defects in neurulation, cranio-facial, patterning, and skeletal muscle development. Genes & Dev. 11: 2029–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards, R., and Agami, R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. [DOI] [PubMed] [Google Scholar]
- Butler K., Zorn, A.M., and Gurdon, J.B. 2001. Nonradioactive in situ hybridization to Xenopus tissue sections. Methods 23: 303–312. [DOI] [PubMed] [Google Scholar]
- Capecchi M.R. 1989. Altering the genome by homologous recombination. Science 244: 1288–1292. [DOI] [PubMed] [Google Scholar]
- Caplen N.J., Parrish, S., Imani, F., Fire, A., and Morgan, R.A. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. 98: 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares C., Heilstedt, H.A., Shaffer, L.G., Schwartz, S., Berk, M., Murray, J.C., and Stavnezer, E. 2002. Loss of the SKI proto-oncogene in individuals affected with 1p36 deletion syndrome is predicted by strain-dependent defects in Ski-/- mice. Nat. Genet. 30: 106–109. [DOI] [PubMed] [Google Scholar]
- Dai P., Shinagawa, T., Nomura, T., Harada, J., Kaul, S.C., Wadhwa, R., Khan, M.M., Akimaru, H., Sasaki, H., Colmenares, C., et al. 2002. Ski is involved in transcriptional regulation by the repressor and full-length forms of Gli3. Genes & Dev. 16: 2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel, W., and Tuschl, T. 2001a. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl T. 2001b. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Huang Y. and Carmichael, G.C. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.W. and Bass, B.L. 2002. The role of RNA editing by ADARs in RNAi. Mol. Cell 10: 809–817. [DOI] [PubMed] [Google Scholar]
- Kokura K., Kaul, S.C., Wadhwa, R., Nomura, T., Khan, M.M., Shinagawa, T., Yasukawa, T., Colmenares, C., and Ishii, S. 2001. The Ski family is required for MeCP2-mediated transcriptional repression. J. Biol. Chem. 276: 34115–34121. [DOI] [PubMed] [Google Scholar]
- Lee Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21: 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D.L., Hagstrom, J.E., Loomis, A.G., Wolff, J.A., and Herweijer, H. 2002. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat. Genet. 32: 107–108. [DOI] [PubMed] [Google Scholar]
- Luo M.J. and Reed, R. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. 96: 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche L., Green, S.R., Schmedt, C., and Mathews, M.B. 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12: 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey A.P., Meuse, L., Pham, T.T., Conklin, D.S., Hannon, G.J., and Kay, M.A. 2002. RNA interference in adult mice. Nature 418: 38–39. [DOI] [PubMed] [Google Scholar]
- McKendrick L., Thompson, E., Ferreira, J., Morley, S.J., and Lewis, J.D. 2001. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol. 21: 3632–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minks M.A., West, D.K., Benvin, S., and Baglioni, C. 1979. Structural requirements of double-stranded RNA for the activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J. Biol. Chem. 254: 10180–10183. [PubMed] [Google Scholar]
- Nomura T., Khan, M.M., Kaul, S.C., Dong, H.-D., Wadhwa, R., Colmanares, C., Kohno, I., and Ishii, S. 1999. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes & Dev. 13: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P.J., Caudy, A.A., Bernstein, E., Hannon, G.J., and Conklin, D.S. 2002a. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 16: 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P.J., Caudy, A.A., and Hannon, G.J. 2002b. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. 99: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M.S. and Bass, B.L. 1998. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 17: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E.V., Christoph, G., Zeller, R., and Leder, P. 1990. The cytomegalovirus enhancer: A pan-active control element in transgenic mice. Mol. Cell. Biol. 10: 4406–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Kerr, I.M., Williams, B.R., Silverman, R.H., and Schreiber, R.D. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67: 227–264. [DOI] [PubMed] [Google Scholar]
- Tokitou F., Nomura, T., Khan, M.M., Kaul, S.C., Wadhwa, R., Yasukawa, T., Kohno, I., and Ishii, S. 1999. Viral-Ski inhibits retinoblastoma protein (Rb)-mediated transcriptional repression in a dominant negative fashion. J. Biol. Chem. 274: 4485–4488. [DOI] [PubMed] [Google Scholar]
- Wianny F. and Zernicka-Goetz, M. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2: 70–75. [DOI] [PubMed] [Google Scholar]
- Yonaha M. and Proudfoot, N.J. 1999. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell 3: 593–600. [DOI] [PubMed] [Google Scholar]
- ____. 2000. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 19: 3770–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.W. 1985. Cell differentiation in the retina of the mouse. Anat. Rec. 212: 199–205. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. [DOI] [PubMed] [Google Scholar]