Abstract
Drosophila Enhancer of Polycomb, E(Pc), is a suppressor of position-effect variegation and an enhancer of both Polycomb and trithorax mutations. A homologous yeast protein, Epl1, is a subunit of the NuA4 histone acetyltransferase complex. Epl1 depletion causes cells to accumulate in G2/M and global loss of acetylated histones H4 and H2A. In relation to the Drosophila protein, mutation of Epl1 suppresses gene silencing by telomere position effect. Epl1 protein is found in the NuA4 complex and a novel highly active smaller complex named Piccolo NuA4 (picNuA4). The picNuA4 complex contains Esa1, Epl1, and Yng2 as subunits and strongly prefers chromatin over free histones as substrate. Epl1 conserved N-terminal domain bridges Esa1 and Yng2 together, stimulating Esa1 catalytic activity and enabling acetylation of chromatin substrates. A recombinant picNuA4 complex shows characteristics similar to the native complex, including strong chromatin preference. Cells expressing only the N-terminal half of Epl1 lack NuA4 HAT activity, but possess picNuA4 complex and activity. These results indicate that the essential aspect of Esa1 and Epl1 resides in picNuA4 function. We propose that picNuA4 represents a nontargeted histone H4/H2A acetyltransferase activity responsible for global acetylation, whereas the NuA4 complex is recruited to specific genomic loci to perturb locally the dynamic acetylation/deacetylation equilibrium.
Keywords: Chromatin acetylation, histone H4, NuA4 complex, Epl1, Esa1, Yng2
Nucleosomes, the basic units of chromatin structure, suppress basal transcription initiation, an effect that is reinforced or alleviated by chromatin-modifying activities. Histone modifications are believed to regulate chromatin function by altering its structure through an epigenetic signature, the proposed histone code (Jenuwein and Allis 2001). Studies during recent years have identified several enzymes responsible for acetylation, methylation, ubiquitination, and phosphorylation of histones. These enzymes are often part of multiprotein complexes directly involved in gene regulation. Hyperacetylation of histone N-terminal tails is found in euchromatin and has been implicated in gene-specific activation, whereas hypoacetylation is found in heterochromatin and linked to gene repression. Histone acetyltransferase (HAT) and deacetylase (HDAC) complexes are recruited by activators and repressors to promoter regions to regulate transcription. This recruitment has been proposed to locally disturb the global highly dynamic acetylation/deacetylation equilibrium (for reviews, see Vogelauer et al. 2000; Katan-Khaykovich and Struhl 2002; Waterborg 2002). In budding yeast, it has been shown that HAT enzymes Gcn5 and Esa1 are present in complexes recruited to promoter regions by transcription activators (SAGA and NuA4, respectively) but are also responsible for global nontargeted acetylation (for review, see Utley and Côté 2002).
The mechanisms of epigenetic regulation of transcription have been studied intensively in Drosophila in part through the modulation of homeotic gene expression by the trithorax (activation) and Polycomb (repression) families of regulators. Position-effect variegation is also a hallmark of these mechanisms, as expression of genes located near heterochromatin is clonally, heritably, and reversibly turned off. Enhancer of Polycomb, E(Pc), was originally identified as a mutated gene that increased the homeotic phenotype of Polycomb group mutations without harboring one by itself (Stankunas et al. 1998). It was later also shown to enhance the phenotype of trithorax group mutants, supporting the argument for a role in positive and negative epigenetic regulation of homeotic gene expression (Gildea et al. 2000). Furthermore, E(Pc) mutation suppresses position-effect variegation (Sinclair et al. 1998). Enhancer of Polycomb is conserved throughout evolution (Stankunas et al. 1998). A mammalian homolog, EPC1, has been identified and implicated in gene regulation (Shimono et al. 2000). In Saccharomyces cerevisiae, Enhancer of Polycomb-like-1, Epl1, is essential for cell viability and copurifies with the NuA4 HAT complex (Galarneau et al. 2000).
NuA4 (nucleosome acetyltransferase of H4) is a 12-subunit histone acetyltransferase complex that modifies histone H4 and H2A N-terminal tails. Its catalytic subunit is Esa1, the only HAT essential for cell cycle progression in yeast and highly similar to human Tip60 (Allard et al. 1999; Clarke et al. 1999). It also contains Tra1, an essential ATM-family cofactor implicated in recruitment by transcription activators (Allard et al. 1999; Brown et al. 2001); Yng2, an ING1 tumor suppressor homolog implicated in transcription activation by p53 (Nourani et al. 2001); Arp4, an actin-related protein linked to epigenetic control of gene expression (Galarneau et al. 2000); and Eaf3, a chromodomain-containing protein related to dosage compensation in Drosophila and senescence in human cells (Eisen et al. 2001). NuA4 can be recruited by activators in vitro and in vivo to target a chromatin region for H4 hyperacetylation and stimulate transcription (Utley et al. 1998; Vignali et al. 2000; Nourani et al. 2001; A. Nourani, R.T. Utley, S. Allard, and J. Côté, in prep.). Esa1 has been found at the promoter of ribosomal protein genes in vivo (Reid et al. 2000). Mammalian homologs of NuA4 subunits have been implicated in transcription activation and cell transformation by c-Myc, E2F and E1A, p53 function, coactivation of steroid receptors and NFκB, DNA repair, apoptosis, and Alzheimer molecular onset (for review, see Utley and Côté 2002).
In the present study, we analyzed the function of Epl1, the yeast homolog of Enhancer of Polycomb. We confirm it as a stable stoichiometric subunit of the NuA4 complex and show that it, like its Drosophila counterpart, is implicated in positive and negative regulation of transcription as its mutation suppresses telomere position effect on gene expression. As for Esa1, Epl1 depletion provokes the accumulation of cells in G2/M and global loss of histone H4/H2A acetylation. Epl1 mutants are also sensitive to DNA-damaging agents and rapamycin, reflecting roles in DNA repair and ribosome biogenesis/translation.
We present evidence that Epl1 is strictly associated with Esa1 in NuA4 or a novel smaller HAT complex called Piccolo NuA4 (picNuA4). picNuA4 is highly active and prefers chromatin as a substrate. Epl1 bridges Esa1 and Yng2, and the trimeric complex is sufficient for the essential function of the Esa1 HAT. Our data suggest that picNuA4 is responsible for global nontargeted histone H4/H2A acetylation in vivo, whereas NuA4 is recruited to specific chromatin loci for gene regulation.
Results
Yeast Enhancer of Polycomb-like-1 is a stable stoichiometric subunit of NuA4
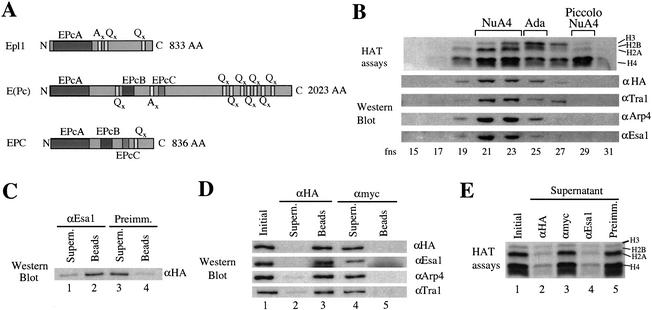
A homologous protein to Drosophila Enhancer of Polycomb is found in both yeast and human cells (see conserved domain regions in Fig. 1A). Epl1, the yeast protein, was identified by tandem mass spectrometry in the immunopurified NuA4 HAT complex, using the catalytic subunit Esa1 as epitope (Galarneau et al. 2000). Epl1 is essential for cell viability, and an epitope-tagged version overexpressed from a multicopy vector was shown to copurify with the NuA4 complex (Galarneau et al. 2000). To determine if Epl1 is a stable stoichiometric subunit of the NuA4 complex, we produced cells expressing physiological amounts of the HA-tagged Epl1 protein. Whole cell extracts were fractionated as before through three chromatographic steps (nickel-agarose, MonoQ, and Superose 6; Galarneau et al. 2000). Again, on gel filtration, Epl1 coeluted with known NuA4 subunits and HAT activity (Fig. 1B). Note that, in addition to the already identified NuA4 and Ada HAT complexes (Grant et al. 1997), a new smaller HAT complex was detected in fraction 29. Because it shares similar HAT specificity with NuA4 but is smaller in size, it was named Piccolo NuA4 (see below).
Figure 1.
Epl1 is a stable stoichiometric subunit of the NuA4 complex. (A) Diagram of homology regions of yeast Epl1 compared with Drosophila E(Pc) and human EPC. (B) Epl1 coelutes with NuA4 components and H4/H2A acetyltransferase activity. Whole cell extract from a HA-Epl1-expressing strain was fractionated over nickel-agarose, followed by a MonoQ column. Peak NuA4 fractions were pooled and loaded on a Superose 6 gel-filtration column. Fractions were tested for HAT activity and by Western blotting with the indicated antisera. (C) Epl1 is a stable Esa1-associated factor. Equal amounts of Superose 6 fraction 21 described in B were incubated with preimmune or anti-Esa1 beads. The majority of HA-Epl1 coimmunoprecipitates with Esa1. (D) HA-Epl1 coimmunoprecipitates Esa1, Arp4, Tra1, and NuA4 HAT activity, showing that Epl1 is a stable stoichiometric subunit of NuA4. The Superose 6 fraction was incubated with anti-HA or anti-Myc beads. After washes, equivalent amounts of initial samples, beads, and supernatant were analyzed by Western blotting with anti-HA, anti-Esa1, anti-Arp4, and anti-Tra1 as indicated. (E) Immunoprecipitation of HA-Epl1 fully depletes NuA4 HAT activity from the supernatant. Immunoprecipitations were performed as in C and D, and initial samples and supernatants were tested for NuA4 HAT activity.
Next, to confirm stable and stoichiometric association of Epl1 with NuA4, we performed immunoprecipitation experiments with a NuA4 peak gel filtration fraction. Antibodies against the Esa1 catalytic HAT subunit efficiently immunoprecipitate the majority of HA-Epl1 protein in this fraction (Fig. 1C, cf. the preimmune control). Reciprocally, known NuA4 subunits were fully immunoprecipitated by antibodies against the HA epitope (Fig. 1D, cf. the control antibody), and, accordingly, NuA4 HAT activity was efficiently depleted from the supernatant (Fig. 1E). Together these data let us conclude that Epl1 is a bone fide stoichiometric subunit of the NuA4 HAT complex.
Epl1 is required for cell cycle progression and global histone H4/H2A acetylation in chromatin
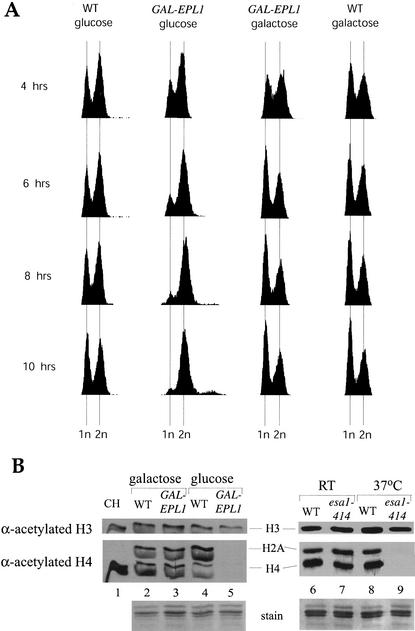
Both esa1 thermosensitive (ts) mutants and mutants for histone H4 lysines targeted by Esa1 provoke a RAD9-dependent G2/M delay (Megee et al. 1995; Clarke et al. 1999). The tumor-suppressor homolog Yng2, another NuA4 subunit, was also shown to be required for normal progression through mitosis (Choy et al. 2001). To determine if Epl1 essential function is related to the Esa1 role in cell cycle progression, we produced yeast cells in which the EPL1 promoter is replaced by the GAL1 promoter at the chromosomal loci. When these cells are placed in galactose media, Epl1 is expressed, whereas media containing glucose blocks its expression. Western blot analysis showed that efficient depletion of Epl1 protein (no detectable signal) is obtained within 8 h of incubation in glucose-containing media (A. Nourani, unpubl.). Time-course FACS analysis clearly shows that cells expressing Epl1 under GAL1 promoter control accumulate in G2/M (2n DNA content) upon Epl1 depletion (Fig. 2A). In comparison, cells grown in galactose-containing media or control cells do not change their cell distribution during the course of the experiment. These data clearly demonstrate that Epl1, like Esa1, is required for cell cycle progression through G2/M.
Figure 2.
Epl1 expression is essential for cell cycle progression and global histone H4 and H2A acetylation in chromatin. (A) Epl1 is required for progression through mitosis. FACS analysis of cells expressing Epl1 under control of the GAL1 promoter and the isogenic wild type for the indicated times in either galactose or glucose media. When Epl1 expression is repressed in glucose, cells accumulate in G2/M. (B) Histones were purified from wild-type or GAL1-EPL1-expressing cells after 12 h in galactose or glucose media and run on an 18% SDS-PAGE. Histones purified from wild-type and esa1 ts mutant cells grown at room temperature or at 37°C for 4 h were also examined for comparison. Gels were blotted and screened with the indicated antibodies.
Although Esa1 is required for accurate transcription of specific genes, its depletion has been shown to provoke a global loss of acetylated histone H4 isoforms in vivo (Clarke et al. 1999; Loewith et al. 2000; Reid et al. 2000; Vogelauer et al. 2000; Suka et al. 2001). We then tested if Epl1 depletion had a related impact on bulk chromatin acetylation status using the glucose-regulated EPL1 strain. We purified bulk histones from cells incubated in galactose or glucose as in Figure 2A and analyzed their acetylation status using antibodies for acetylated histone H3 and H4 (Fig. 2B). Note that the antiacetylated H4 serum also recognizes acetylated H2A in yeast histones but not in human cells (Fig. 2B, cf. lanes 1 and 2). Incubation of the GAL-EPL1 strain in glucose media did not significantly affect the levels of acetylated histone H3 (Fig. 2B, cf. lanes 3 and 5). In contrast, acetylated histone H4 and H2A signals are completely lost upon Epl1 depletion, identical to the effect in esa1 ts mutants at nonpermissive temperature (Fig. 2B, cf. lanes 7 and 9). In consequence, we conclude that Epl1 is strictly required for global histone H4 and H2A acetylation by Esa1 in vivo.
Mutations in EPL1 provoke sensitivity to DNA-damaging agents, create defects in gene-specific transcription, and suppress telomere-position effect
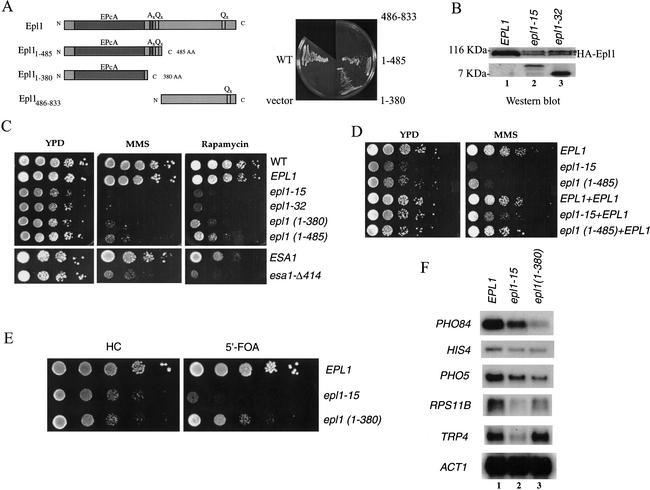
Because Epl1 is essential for cell viability, we determined which domains within the protein were required for growth. Truncated forms were produced to isolate the N-terminal conserved EPcA domain with or without the proximal stretches of alanine and glutamine residues (Epl11–380 and Epl11–485, respectively) and the nonconserved C-terminal domain (Epl1486–833; see Fig. 3A). The cell growth phenotype of cells expressing physiological levels of these mutant Epl1 proteins was analyzed after chasing the wild-type copy of EPL1 on 5′-FOA plates (Fig. 3A). Clearly, the conserved EPcA domain is by itself sufficient for cell viability. Although the C-terminal part of Epl1 is not required, its loss creates a significant slow-growth phenotype (Fig. 3C–E). Loss of the central stretches of alanine and glutamine residues does not seem to have a notable effect on growth.
Figure 3.
Epl1 mutations affect growth, sensitivity to DNA damage, target gene transcription, and telomeric silencing. (A) Schematic presentation of Epl1 truncations that were tested in growth complementation assay. Only the conserved EPc domain of Epl1 is required for cell viability. EPL1-deleted haploid strains expressing wild-type Epl1 from a URA3 vector and the indicated Epl1 truncations from a low-copy LEU2 vector were selected on 5′-FOA plates to evict the URA3 plasmid. (B) epl1 mutant clones obtained in a screen produce low amounts of full-length Epl1 protein. Western analysis of extracts from epl1-15 and epl1-32 mutants show that both strains produce a very short truncated protein, as expected based on the mutation, but also low amounts of full-length Epl1. The asterisk indicates a nonspecific band detected by anti-HA. (C) Cells expressing Epl1 C-terminal truncation and dosage mutants exhibit slow growth and are highly sensitive to MMS and rapamycin. Here, 10-fold serial dilutions of the EPL1, epl1-15, epl1-32, epl1 (1–380), and epl1 (1–485) strains were spotted on YPD, YPD + 0.03% MMS, or YPD + 25 nM rapamycin plates and incubated at 30°C for 2 d (YPD) and 4 d (MMS and rapamycin). esa1-Δ414 mutant cells were also tested and show the same sensitivity to the drugs. (D) The dominance/recessivity aspect of the epl1 mutants was tested by coexpression of the wild-type protein in the mutant cells. Spot dilution analyses as in C were performed and show that wild-type Epl1 rescues growth in the mutants, indicating that they are not dominant. (E) The conserved EPc domain of Epl1 is important for telomeric silencing. The 10-fold serial dilutions of the strains QY129 (EPL1), QY130 (epl1-15), and QY131 [epl1 (1–380)] containing a URA3 gene located next to the telomere region of Chromosome VII were spotted on Hartwell's complete (HC) or HC + 5′-FOA plates (to reflect expression of URA3) and incubated at 30°C for 2 and 3 d, respectively. (F) Epl1 is required for normal expression of specific genes in vivo. RNA was extracted from the indicated strains and analyzed by Northern blot using PHO84, HIS4, PHO5, RPS11B, TRP4, and ACT1 probes.
To further investigate Epl1 function, we performed a screen for thermosensitive mutants using standard procedures (see Materials and Methods). Unfortunately, out of 6000 clones tested, only two slow-growth mutants were obtained, epl1-15 and epl1-32. These clones are not cold or thermosensitive and are in fact base-substitution mutants that create early stop codons after amino acids 83 and 57, respectively. Whereas production of the small truncated proteins is seen by Western blot on whole cell extracts, a low amount of full-length protein is also detected, presumably by readthrough/tRNA suppression (Fig. 3B). Thus, we believe that these clones behave like Epl1 low-dosage mutants as the small peptides cannot support growth by themselves (data not shown). Along with the truncation mutants, we tested these strains for growth defects in varied environments. Striking sensitivities were found when the DNA-damaging agent methyl methanesulfonate (MMS) was included in the media (Fig. 3C, middle panel), suggesting that Epl1 is important for survival from DNA double-strand breaks. MMS sensitivity was also seen in an esa1 ts mutant strain, again linking Epl1 and Esa1 functions. Sensitivity to DNA-damaging agents has been observed in cells mutated for other NuA4 subunits, implicating the complex in DNA damage signaling (Nourani et al. 2001; N. Bouchard, D. Cronier, L. Galarneau and J. Côté, unpubl.). A recent study demonstrated that Esa1-dependent acetylation is required for efficient double-strand break repair and implicated an Arp4-containing complex at double-strand break sites (Bird et al. 2002). epl1 mutant cells are also very sensitive to rapamycin, a drug that targets the Tor signaling pathway, which controls cell growth in part through regulation of ribosome biogenesis and translation (Fig. 3C, right panel). Esa1 has been shown to be required for proper expression of ribosomal protein genes and nucleolar structure (Clarke et al. 1999; Reid et al. 2000). Accordingly, esa1 mutant cells are also sensitive to rapamycin. epl1 mutant cell sensitivity again establishes a close functional interaction with Esa1. To be sure that the phenotypes we see can be used to infer normal Epl1 function and are not caused by a dominant-negative artifact produced by the truncated proteins, we confirmed that growth was restored by coexpression of a wild-type copy of EPL1 (Fig. 3D).
Because Enhancer of Polycomb is a suppressor of position-effect variegation in Drosophila (Sinclair et al. 1998), we wanted to determine if the yeast homolog was implicated in a related epigenetic mechanism, that is, silencing by telomere-position effect (TPE). We introduced epl1 mutations in cells carrying a URA3 marker located next to the telomere region of the left arm of Chromosome VII (Gottschling et al. 1990). A mutation that decreases telomeric silencing would allow URA3 gene expression that is detectable by cell sensitivity to 5′-FOA. In these cells, Epl1 C-terminal truncation had no significant effect on telomeric silencing [Fig. 3E, epl1 (1–380) vs. wild type and growth on control plate]. On the other hand, depletion of Epl1 (epl1-15) made the cells very sensitive to 5′-FOA, portraying a defect in telomeric silencing. Thus, as for Drosophila E(Pc), yeast Epl1 is implicated in gene silencing by neighboring heterochromatin. Furthermore, the conserved N-terminal EPc domain is solely responsible for this role in silencing at telomeres.
Mutations in Esa1 and other NuA4 subunits have been shown to create gene-specific transcription defects and block transcription activation in vivo (Galarneau et al. 2000; Reid et al. 2000; Eisen et al. 2001; Nourani et al. 2001; Verdone et al. 2002). To obtain insight on the role of Epl1 in gene expression, we performed Northern analysis using RNA purified from epl1 mutant cells. Both dosage and truncation mutants show two-fold to fivefold decreased expression of specific genes also known to be affected by ESA1 mutation (Fig. 3F). Interestingly, Epl1 C-terminal truncation did not affect TRP4 gene expression, but the dosage mutant did (Fig. 3F, cf. lanes 2,3 and 1). Thus, like other NuA4 subunits, Epl1 is required for normal expression of specific genes in vivo.
Epl1 is also present with Esa1 in Piccolo NuA4, a small highly active and chromatin-preferring HAT complex
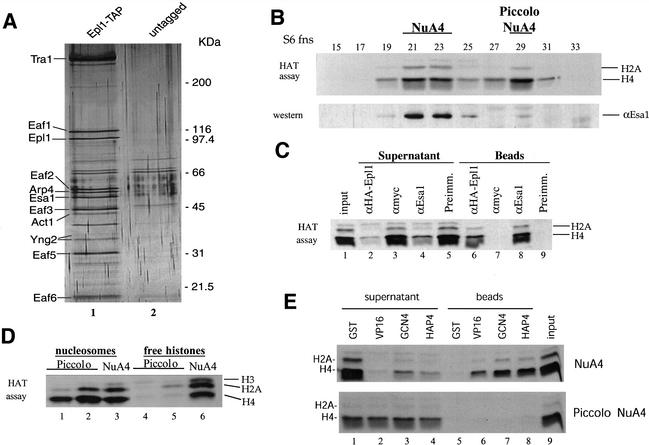
Because we established that Epl1 can have both positive and negative effect on gene transcription, we wanted to rule out the possibility of its presence in complexes other than NuA4 (NuA4 being historically linked to positive regulation). We inserted the TAP (tandem affinity purification; Puig et al. 2001) cassette at the C terminus of Epl1 in the endogenous gene. We accomplished two-step affinity purification of Epl1-associated proteins from whole cell extract and visualized them on SDS-PAGE (Fig. 4A). All identified protein bands turned out to be the complete set of NuA4 subunits (by a combination of Western analysis, apparent mobility, and reciprocal tagging; data not shown). Furthermore, mass spectrometric analysis of the material also identified only NuA4 subunits plus the usual set of nonspecific proteins contaminating TAP preps (protein chaperones, ribosomal proteins; N. Lacoste, unpubl.; see Pijnappel et al. 2001). Consequently, these data suggest that the large majority of soluble Epl1 is found associated with NuA4 components in the cell, supporting the argument that it strictly functions through its physical association with the Esa1 HAT, even in telomeric silencing (see Discussion).
Figure 4.
Epl1 is also present with Esa1 in Piccolo NuA4, a small highly active, chromatin-preferring, nontargeted histone H4/H2A acetyltransferase complex. (A) Soluble Epl1 is exclusively associated with NuA4 components in the cell. A yeast strain expressing Epl1 with a TAP (tandem affinity purification) cassette at its C terminus was produced by homologous recombination. Purified TAP-Epl1 and its associated proteins were run on SDS-PAGE and silver stained. Known NuA4 components are labeled on the left based on molecular weight and Western and mass spectrometric analysis. Fractionation from a control untagged strain is also presented to indicate nonspecific bands. (B) Identification of Piccolo NuA4 as a new histone acetyltransferase complex. Whole cell yeast extract purified over nickel-agarose followed by MonoQ was subsequently passed over a Superose 6 gel filtration column. Fractions were assayed for HAT activity (top panel) and Western blotted for the presence of Esa1 (bottom panel). An additional nucleosomal H4/H2A acetyltransferase activity is apparent in fraction 29. The 300-kD complex (Piccolo NuA4) has a similar level of HAT activity as NuA4 (fractions 21–23; ∼10,000 cpm/μL). Note that a low amount of Esa1 is also detected by Western blot in fraction 29. (C) Esa1 and Epl1 are stable components of Piccolo NuA4 as demonstrated by coimmunoprecipitation of picNuA4 HAT activity with Esa1 and HA-Epl1. Equal amounts of the Superose 6 fraction 29 obtained from an HA-Epl1-expressing strain were incubated with anti-HA, anti-Esa1, anti-Myc, and preimmune serum beads. After washes, equivalent amounts of input sample, supernatant, and beads were used in nucleosomal HAT assays. (D) NuA4 and picNuA4 complexes differ in their ability to acetylate histones. Complexes were immunopurified (with anti-Esa1 beads) from Superose 6 peak fractions of NuA4 and two different picNuA4 preps and assayed for HAT activity on oligonucleosomes or free histones. Although NuA4 acetylates free histones and nucleosomal histones at relatively similar levels (lanes 3,6), picNuA4 has a striking preference for chromatin (lanes 1,2 vs. 4,5). (E) Unlike NuA4, picNuA4 does not interact with the acidic transcription activation domains of VP16, Gcn4, and Hap4 in vitro. NuA4 or picNuA4 Superose fractions were incubated with GST-VP16, GST-Gcn4, GST-Hap4, or control GST beads. Supernatant was removed, beads were washed, and equivalent amounts were assayed for nucleosomal HAT activity.
During our multiple fractionations of yeast HAT complexes over the years, we often detected a novel activity with nucleosomal histone specificity similar to NuA4. Because of this shared specificity toward histone H4 and H2A N termini and the fact that it behaves as a relatively small complex by gel filtration (∼300 kD on a calibrated Superose 6 column; Figs. 1B, 4B), this activity was named Piccolo NuA4 (picNuA4). The relative amounts of total activities in this specific fractionation showed a ratio of 2:1 between NuA4 and picNuA4 (by liquid counts). Because Esa1 is required in vivo for global H4 and H2A acetylation in chromatin, it was surprising to find a new nucleosomal H4/H2A HAT that would be independent of Esa1. Upon closer examination, we noticed that there was a faint Western signal for Esa1 in the column fraction containing Piccolo NuA4 (Fig. 4B, fraction 29). Immunoprecipitations were performed with Esa1 antibodies on the Superose 6 peak fraction and allowed us to conclude that Esa1 was, indeed, the catalytic subunit of picNuA4 (Fig. 4C, lanes 8,9 vs. 4,5). These data indicate that picNuA4 has a much higher specific activity than the NuA4 complex (see ratio of Esa1 protein vs. HAT activity in Fig. 4B). Because Epl1 is also required for global H4/H2A acetylation in vivo (Fig. 2B), we tested for its presence in picNuA4. We used the Superose 6 peak fraction obtained from a strain expressing HA-Epl1 (Fig. 1B). Again, we found perfect coimmunoprecipitation of picNuA4 HAT activity with Epl1 (Fig. 4C, lanes 6,7 vs. 2,3). Preparative immunopurification of picNuA4 was accomplished following a procedure similar to the one used for NuA4 (Galarneau et al. 2000). Because of the low amount of picNuA4 in our extract, no clear specific protein band pattern was detected on gel (data not shown). On the other hand, tandem mass spectrometry allowed us to clearly identify by multiple peptide sequence hits Esa1, Epl1, and another protein also present in the NuA4 complex, Yng2. At least 15 hits were obtained for each protein covering 50%, 33%, and 59% of their respective peptide sequences (data not shown). Because these three proteins together fall short of accounting for the apparent molecular weight of picNuA4 by gel filtration, we cannot completely rule out the presence of other subunits in the complex. However, experiments presented here with recombinant proteins strongly suggest that picNuA4 contains only the three identified proteins (see below).
Whereas NuA4 and Piccolo NuA4 (picNuA4) have identical specificity toward chromatin substrates, a striking difference was seen when free histones were used in the HAT assay. It has been shown that NuA4 is able to acetylate free and nucleosomal histones at about the same rate, whereas its catalytic subunit Esa1 can only target free histones as a recombinant protein (Fig. 4D, cf. lanes 3 and 6; Allard et al. 1999). In contrast, Piccolo NuA4 strongly prefers chromatin over free histones as substrate (Fig. 4D, cf. lanes 1,2 and 4,5). This is reminiscent of a similar small chromatin preferring HAT specific for histone H4/H2A tails reported in Tetrahymena (Ohba et al. 1999).
To understand the possible specific function of picNuA4 in the cell, we continued to compare its biochemical characteristics with those of NuA4. NuA4 can be recruited to specific chromatin loci by DNA-bound transcription factors through interaction with their activation domains (Utley et al. 1998; Vignali et al. 2000). Gcn4, VP16, and HAP4 activation domains are known to interact with NuA4, an association easily tested by the GST pull-down assay. NuA4 and picNuA4 were incubated with GST, GST-Gcn4, GST-VP16, or GST-HAP4 beads, and HAT assays were performed on bound and unbound fractions (Fig. 4E). As expected, most of NuA4 activity was recovered on Gcn4, VP16, and HAP4 beads (Fig. 4E, upper panel, lanes 6–8). In contrast, Piccolo NuA4 activity fully remained in the supernatant (Fig. 4E, lower panel, lanes 2–4). This suggests that, unlike NuA4, picNuA4 cannot be recruited by transcription activators in vivo. This can be explained in part by the absence of Tra1, a known activator-interacting subunit in NuA4 (Brown et al. 2001). Along with the high specific activity and strong chromatin preference, these data indicate that picNuA4 could be the proposed general nontargeted HAT activity involved in the highly dynamic global acetylation/deacetylation equilibrium in the cell (Vogelauer et al. 2000; Katan-Khaykovich and Struhl 2002). The presence of Esa1 and Epl1 in picNuA4 would then account for their role in global acetylation of histone H4 and H2A, whereas their presence in NuA4 is implicated in targeted site-specific chromatin acetylation, for example, for transcription activation. Interestingly, deletion of YNG2, encoding for the third subunit of picNuA4, was also shown to provoke global loss of acetylated histone H4 in vivo (Loewith et al. 2000).
The EPc domain of Epl1 interacts independently with Esa1 and Yng2 to potentiate chromatin acetylation
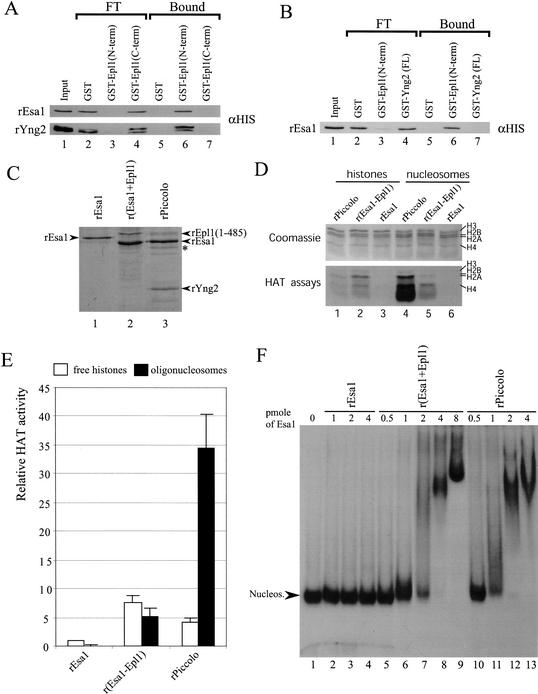
Because of the striking enzymatic characteristics of picNuA4, we were interested in deciphering the physical interactions occurring in picNuA4 and their relation to the highly active nucleosomal HAT activity. Using GST pull-down assays with recombinant proteins, we determined that both Esa1 and Yng2 interact very strongly with the Epl1 N-terminal (amino acids 1–485) but not the C-terminal region (amino acids 486–833; Fig. 5A, lanes 6,7 vs. 3,4). Interestingly, this N-terminal region is mostly the conserved EPc domain, which is sufficient for cell viability. We then examined if Esa1 was able to interact directly with Yng2 (Fig. 5B). Clearly this is not the case, as Esa1 fully remains in the supernatant of GST-Yng2 beads, but it is efficiently brought down by GST-Epl1 beads (Fig. 5B, cf. lanes 3,4 and 6,7). The physical interactions detected here are most likely specific because near equimolar ratios of recombinant proteins were used in the binding studies. These data suggest that the Epl1 N-terminal region independently binds Esa1 and Yng2, physically linking them together in Piccolo NuA4. We then wanted to analyze if these interactions modulate the properties of Esa1 HAT activity toward free histones and chromatin substrate. As previously shown, recombinant Esa1 can only acetylate free histones (rEsa1; Allard et al. 1999). The addition of Yng2 did not affect Esa1 activity, as expected because no direct interaction was detected between the two proteins (data not shown). On the other hand, the addition of Epl1 to Esa1 had a striking positive effect on its activity toward free histones but did not enable activity on chromatin. This indicates that Epl1 association with Esa1 increases its specific enzymatic activity. Finally, mixture of all three proteins (rEsa1, rEpl1, and rYng2) did increase Esa1 specific activity toward free histones to about the same level as Epl1 binding did by itself. This supports the argument that Yng2 does not have much effect on Esa1 specific activity on free histones. On the other hand, addition of Yng2 clearly enabled Esa1 activity on chromatin. These data suggest that the Epl1 N-terminal region physically links Esa1 to Yng2, creating a structure that allows Esa1 to acetylate nucleosomal histone H4/H2A. The mechanism by which Epl1–Yng2 addition enables Esa1 action on chromatin could be related to an affinity for chromatin because both Yng2 and Epl1 (1–485) can interact with nucleosomes in vitro (data not shown).
Figure 5.
The “Enhancer of Polycomb” domain of Epl1 bridges Esa1 and Yng2 to enable nucleosomal acetyltransferase activity. (A,B) GST pull-downs using a near stoichiometric ratio of the indicated recombinant proteins were carried out and assayed by Western blotting. As shown using αHIS Western blots, both rEsa1p and rYng2p are pulled down using GST-Epl1 (N-term) but not using the C terminus of the same protein, and GST-Yng2 (FL) does not associate with rEsa1. (C) Coomassie staining of the purified recombinant proteins and bacterially coexpressed complexes used for HAT and mobility shift assays. The slight size difference of rEsa1 in lane 1 versus lanes 2 and 3 is caused by different cloning strategies. The band marked by an asterisk is a degradation product of rEpl1. The loaded amount of proteins as standardized to rEsa1 is 4, 8, and 8 pmole (lanes 1,2,3, respectively). (D) Relative HAT activity of rEsa1 alone versus polycistronic recombinant complexes. HAT assays with the recombinant proteins shown in C were carried out with free histones or nucleosomes. rEsa1 was kept constant in all assays. (E) Quantification of HAT assays done on free (open box) or nucleosomal (black box) histones. Data are representative of two different experiments done in duplicate. (F) Mobility shift assay with mononucleosomes shows the formation of ternary complexes: Nucleosome binding is detected with r(Esa1–Epl1) (lanes 5–9) and rPiccolo (lanes 10–13), whereas rEsa1 shows no interaction (lanes 2–4).
Even though we could recreate chromatin acetylation by just mixing the three proteins identified as subunits of Piccolo NuA4, we could not recreate the strong chromatin preference of the native complex. To produce a fully recombinant picNuA4 complex, we used a polycistronic bacterial vector for coexpression, increasing the chance of proper folding and association (Tan 2001). Recombinant monomeric Esa1, dimeric Esa1–Epl1, and trimeric Esa1–Epl1–Yng2 (rPiccolo) were expressed and purified over cobalt metal affinity resin, ion exchange, and gel filtration chromatography. Only the N-terminal (amino acids 1–485) region of Epl1 was used for convenience. Both Epl1–Esa1 and rPiccolo complexes could be purified, whereas coexpressed rEsa1 and rYng2 did not copurify (W. Selleck and S. Tan, unpubl.), confirming their lack of direct interaction. The purified recombinant protein complexes used in the assays are presented in Figure 5C. Some degradation of Epl1 has occurred, accounting for the apparent substoichiometry versus Esa1. These complexes were assayed for acetyltransferase activity on free histones and nucleosomes with a constant amount of Esa1 protein (Fig. 5D,E). Again, Epl1 association with Esa1 increased the specific enzymatic activity of Esa1 on free histones (Fig. 5D, lane 2 vs. 3). Furthermore, the purified dimer is able to acetylate chromatin substrates at about the same level as free histones (Fig. 5D, lane 2 vs. 5). This indicates that the Epl1 N terminus allows Esa1 access to histone H4/H2A tails in the nucleosome. When rPiccolo is tested on free histones, the level of detected HAT activity is similar to the one obtained with r(Esa1–Epl1) (Fig. 5D, lane 1 vs. 2). Strikingly, rPiccolo was sevenfold more active on oligonucleosomes versus free histones (Fig. 5D, lane 4 vs. 1), a unique characteristic similar to that observed with endogenous yeast Piccolo NuA4 (Fig. 4D). To further understand the chromatin preference of rPiccolo versus the Epl1–Esa1 dimer, we performed mononucleosome mobility shift assays using the different recombinant proteins and a radiolabeled 5S rDNA nucleosome (Fig. 5F). Although rEsa1 showed no interaction by itself, both dimeric and trimeric complexes showed good affinity for nucleosome particles, even creating a distinct ternary complex (Fig. 5F, lanes 1–4, 5–9, 10–13). rPiccolo may have a slightly higher affinity, which does not fully account for its chromatin preference in HAT assays. Nevertheless, Yng2 is the key to the preference of chromatin as substrate. The Yng2 PHD finger is not required for this activity, as a purified rPiccolo complex lacking this domain possess the same enzymatic characteristics (W. Selleck and S. Tan, unpubl.). Because Epl1 bridges Yng2 to Esa1 in Piccolo NuA4, the combined structural functions of Epl1 and Yng2 could allow Esa1 to preferentially acetylate chromatin. In conclusion, purified rPiccolo has enzymatic properties very similar to endogenous yeast picNuA4, including high specific enzymatic activity and strong preference for chromatin substrates, both certainly an asset for a low-abundance putative nontargeted enzyme required for global H4/H2A acetylation in vivo.
Piccolo NuA4 encompasses the essential in vivo functions of Esa1 and Epl1
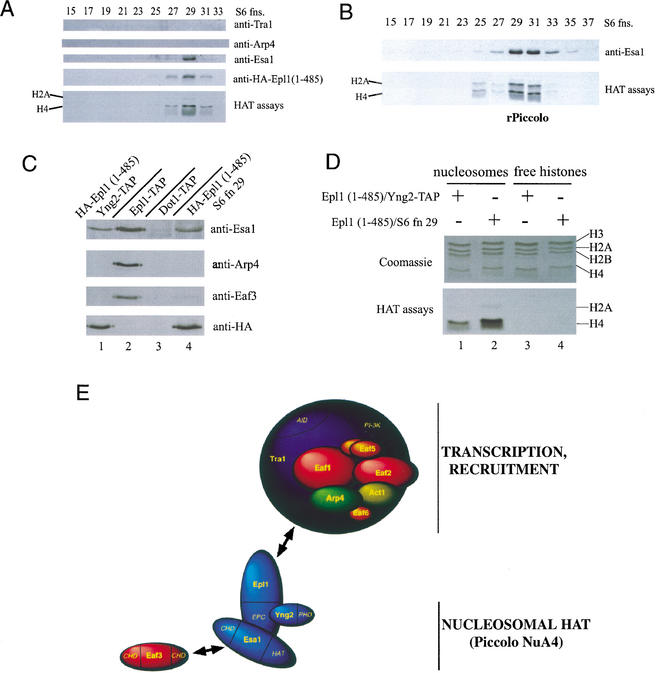
Because the conserved EPc domain of Epl1 is sufficient for cell viability (Fig. 3A) and directly interacts with Esa1 and Yng2 (Fig. 5), we were interested in looking at the status of the NuA4 complex in cells expressing only this domain of Epl1. We fractionated whole cell extracts as before and performed Western analysis and HAT assays on fractions from the gel filtration column (Fig. 6A). Strikingly, the usual nucleosomal H4 HAT activity associated with NuA4 is now absent (no activity in fractions 19–23). Instead, this activity now elutes exclusively around fraction 29, where Piccolo NuA4 is usually found (Fig. 4B). Furthermore, only known picNuA4 subunits are detected by Western analysis in these fractions (Fig. 6A). The elution profile is also similar to the one obtained when recombinant Piccolo NuA4 is applied to the same gel filtration column (Fig. 6B). NuA4 subunits Tra1 and Arp4 are not detected throughout the column elution (Fig. 6A). Signals for both of these proteins were detected on the previous MonoQ column eluting away from the single peak of Esa1, Epl1, and nucleosomal H4 HAT activity (data not shown). Thus, these cells do not seem to contain NuA4 complexes, but, instead, solely possess picNuA4 as a source of nucleosomal histone H4 acetyltransferase activity. To confirm this, we TAP-tagged the Yng2 endogenous gene in cells expressing only Epl1 (1–485). Western analysis of the affinity-purified fraction confirmed that Yng2 is part of a trimeric complex with Esa1 and Epl1 in vivo (Fig. 6C). No other NuA4 subunit was detected (Fig. 6C, cf. lane 1 to NuA4 complex in lane 2). Furthermore, the TAP- and conventionally purified complexes greatly prefer chromatin as substrate, a characteristic of endogenous and recombinant picNuA4 complexes (Fig. 6D). Thus, we conclude that the epl1 (1–485) cells do not contain NuA4 complexes, but possess instead a picNuA4-like trimeric complex. Additionally, the data indicate that the Epl1 non-essential C-terminal region is responsible for anchoring the Esa1–Epl1–Yng2 complex (picNuA4) to the rest of NuA4 (model in Fig. 6E). Altogether these results demonstrate that, during normal growth in rich media, all the essential functions of Esa1 and Epl1 are accomplished through their presence in Piccolo NuA4. In these growth conditions, this would support the argument that nontargeted histone H4 acetylation (by picNuA4) is essential, whereas site-specific recruitment (through NuA4) is not (but is required for normal growth; Fig. 3). Of course, in specific growth conditions inducing various signaling pathways such as genome integrity checkpoint, the specific function of NuA4 becomes essential to survival (Fig. 3C).
Figure 6.
Essential functions of Esa1 and Epl1 through their presence in the Piccolo NuA4 complex. (A) The C-terminal portion of Epl1 is important to maintain integrity of the NuA4 complex. Extract from an epl1 deletion strain covered by a HA-Epl1 (1–485)-expressing plasmid, was purified over nickel-agarose, followed by MonoQ column fractionation. MonoQ peak fractions were pooled and loaded on a Superose 6 gel filtration column. Fractions were tested for HAT activity and analyzed by Western blotting with the indicated antisera. (B) Recombinant Piccolo NuA4 from the Source S peak fraction was loaded on a Superose 6 filtration column; fractions were analyzed as in A. (C) Tandem affinity purification of Yng2 from epl1 (1–485) cells brings only picNuA4 components. Western analysis of TAP-purified material obtained with YNG2–TAP/HA–EPL1 (1–485), EPL1–TAP (NuA4 control), and DOT1–TAP (control) cells. NuA4-specific components are only detected in EPL1–TAP cells. Superose 6 fraction 29 from A is also shown for comparison. (D) Affinity- and conventionally purified picNuA4-like complexes from A and C were tested in a HAT assay on oligonucleosomes and free histones. Both complexes show strong preference for chromatin substrates (cf. lanes 1,2 and 3,4). (E) Model of Piccolo NuA4 anchoring itself to the rest of the NuA4 complex through the Epl1 C-terminal region. Eaf3 is depicted separately because of a previously reported direct interaction with Esa1 (Eisen et al. 2001). Subunit names are indicated, and protein domains are italicized. EPC, conserved “Enhancer of Polycomb” domain; PHD, PHD finger domain; HAT, histone acetyltransferase domain; CHD, chromo domain; AID, activator interaction domain; PI-3K, phospho-inositol-3 kinase family domain.
Discussion
Esa1 has been shown to be responsible for global histone H4 acetylation in yeast chromatin (Clarke et al. 1999; Loewith et al. 2000; Reid et al. 2000; Vogelauer et al. 2000; Suka et al. 2001). In consequence, Esa1 is responsible for the global nontargeted histone H4/H2A acetylation implicated in the very dynamic acetylation–deacetylation equilibrium. Here we showed that the yeast homolog of Drosophila Enhancer of Polycomb, Epl1, seems always directly associated with Esa1 in the cell and is required for its function in global nontargeted acetylation of chromatin and cell cycle progression. Epl1 and Esa1 are stable stoichiometric subunits of the NuA4 histone acetyltransferase complex. The NuA4 complex is recruited by transcription activators to promoter regions to stimulate gene expression at least in part through targeted histone acetylation (Utley et al. 1998; Reid et al. 2000; Brown et al. 2001; Nourani et al. 2001; A. Nourani, R. Utley, S. Allard, and J. Côté, in prep.). Because NuA4 is a HAT complex that can be targeted to specific loci to locally perturb the highly dynamic equilibrium of acetylation/deacetylation, it was puzzling to think that the same large complex would also be responsible for global histone H4/H2A acetylation.
In this report, we present the identification and characterization of a new Esa1-containing nucleosomal H4/H2A acetyltransferase complex called Piccolo NuA4. We propose that this small trimeric complex is responsible for nontargeted global chromatin acetylation in vivo. This is supported by several lines of evidence. First, this complex also contains Epl1 and Yng2, two proteins also required for global acetylation of histone H4 in chromatin (Fig. 2; Loewith et al. 2000). Second, its high specific activity and strong preference for chromatin substrates are characteristics that would be an important asset for such a general nontargeted HAT. Third, the essential functions of Esa1 and Epl1 in normal growth conditions reside in their presence in Piccolo NuA4 because cells lacking the NuA4 complex are viable. Fourth, picNuA4 does not interact with transcription activators in vitro (Fig. 4E). On the other hand, because we failed to identify subunits in picNuA4 that are not present in the larger NuA4 complex, one could raise the possibility that picNuA4 is a breakdown product of the larger complex during the purification procedure. We think this is unlikely because (1) a similar small chromatin-preferring histone H4 HAT complex has been found in Tetrahymena (Ohba et al. 1999); (2) picNuA4 enzymatic characteristics are strikingly different from those of NuA4 (specific activity and strong preference for chromatin); (3) picNuA4 is preferentially recovered in extracts from cells grown in specific conditions, that is, at high density/near stationary phase (G0) and from cells blocked with nocodazole (G2/M; data not shown); (4) microarray data of cells in G0 show that, although most genes for NuA4 subunits are repressed (six) or not greatly affected (two), gene expression of picNuA4 subunits (ESA1, EPL1, and YNG2) is stimulated between fourfold and sixfold (Gasch et al. 2000); and finally, (5) both a large and a smaller nucleosomal H4 HAT complex were obtained from human cells when the Esa1-human homolog Tip60 was immunopurified and fractionated by gel filtration (Y. Doyon and J. Côté, unpubl.). Although the low abundance of picNuA4 in our extracts is apparently detrimental to a role in global chromatin acetylation, this can be corrected in part by the complex high specific activity and avidity for chromatin. Furthermore, global acetylases/deacetylases could be tightly associated to chromatin in vivo. For example, the nucleosomal H4 HAT activity from Tetrahymena was extracted by DNAse I digestion of the nuclei (Ohba et al. 1999). We found that using DNase I and harsher cell disruption techniques also increased picNuA4 recovery (data not shown). In any case, the work presented here clearly identified and characterized the Esa1–Epl1–Yng2 trimer as the catalytic nucleosomal acetyltransferase core of NuA4, a subcomplex essential for cell viability.
Our deletion of the nonessential C-terminal region of Epl1 allowed us to artificially create a cell containing only the Piccolo NuA4 complex as the source of nucleosomal histone H4/H2A acetylation. It also allowed us to conclude that the Esa1–Epl1–Yng2 catalytic nucleosmal HAT is anchored to the rest of NuA4 through the Epl1 C-terminal region. In consequence, phenotypic analysis of this epl1 mutant strain will be a useful tool to ask specific questions about NuA4 complex function versus nonrecruitable picNuA4. We already know that the DNA-damaging-agent and rapamycin sensitivities are related to NuA4 function because truncated Epl1-expressing strains are sensitive as well as mutations in NuA4-specific subunits (Fig. 3C; data not shown). On the other hand, the role of Epl1 in telomeric silencing seems to be dependent solely on Piccolo NuA4 function because only the dosage mutant strain shows a defect (Fig. 3E). The same can be said for the normal expression of the TRP4 gene (Fig. 3F). Another important finding presented here is the primary role of Yng2 in the catalytic acetylation of chromatin substrates by Esa1 and chromatin preference within picNuA4. Yng2 is part of a family of proteins homologous to the human ING1 tumor suppressor. Yeast paralogs of Yng2 are found in the Sin3–Rpd3 deacetylase complex and in the NuA3 HAT complex and are important for their activity (Loewith et al. 2001; Howe et al. 2002). A common role for this family of proteins in chromatin recognition/binding seems an attractive concept.
A role for a smaller HAT complex in global acetylation of chromatin versus larger recruitable complexes containing the same catalytic subunit may not be unique to Esa1 and picNuA4. The well characterized histone H3-specific HAT Gcn5 is also found in a smaller trimeric complex with Ada2 and Ada3 proteins, also present with Gcn5 in at least two other large complexes (Grant et al. 1997; Saleh et al. 1997; Sendra et al. 2000; Balasubramanian et al. 2002). Because Gcn5 has also been implicated in global nontargeted acetylation of H3 (Vogelauer et al. 2000) and the small trimeric complex does not interact with activators in vitro (Brown et al. 2001), it is tempting to speculate that this could be a partner of picNuA4 for global background acetylation of all histone tails in vivo.
Global acetylation by Esa1 is not required per se for gene-specific transcription (Reid et al. 2000). The state of chromatin acetylation is in constant flux. Globally acting HAT complexes like Piccolo NuA4 would allow rapid return to the steady-state level of histone acetylation upon removal of a transcription regulator that locally perturbed the acetylation/deacetylation equilibrium.
Materials and methods
Yeast strains and plasmids
Due to space limitation, details on strains, plasmids, and oligonucleotides used in this study are available upon request. Yeast culture, transformation, mating, sporulation, and dissection were done according to standard protocols. The EPL1 open reading frame and promoter sequence were amplified by PCR from yeast genomic DNA and cloned in yeast low-copy (ARS, CEN) plasmids after addition of three HA epitopes at the N terminus. The truncation-mutant-expressing plasmids were produced by PCR. The strains used carry the different low-copy plasmids in an EPL1-deleted background. For the telomere-silencing assay, the EPL1 gene was deleted in the UCC1001 strain (Gottschling et al. 1990) transformed with a wild-type Epl1-expressing vector. The vector was then replaced by the different epl1 mutant plasmids following standard procedures. Screening for epl1 mutants was done as described previously using hydroxylamine (Adams et al. 1997), and 6000 leu+/ura+ transformants were tested for growth and thermosensitivity. Two slow-growth mutants were isolated and named epl1-15 and epl1-32. The protocol used to insert the TAP (tandem-affinity purification) cassette at the C terminus of Epl1 and Yng2 in the endogenous gene is described by Puig et al. (2001). Details on bacterial expression vectors for different constructs of Esa1, Epl1, and Yng2 are available upon request.
HAT complexes purification and immunoprecipitations
Partial purification of the NuA4 and picNuA4 complexes by fractionation over Ni2+-nitrilotriacetic acid (NTA) agarose (QIAGEN), MonoQ HR5/5, and Superose-6 HR 10/30 columns (Pharmacia) and HAT assay on oligonucleosomes or HeLa core histones and Western blotting were previously described (Grant et al. 1997; Allard et al. 1999). The immunoprecipitation of NuA4 and picNuA4 complexes was performed on the peak Superose 6 fraction as described (Allard et al. 1999). Ion trap mass spectrometry and peptide sequencing of proteins present in the immunopurified picNuA4 complex were performed as described previously (Eisen et al. 2001). At least 15 unique peptide sequences were obtained for each of the Esa1, Epl1, and Yng2 proteins. Other major peptide hits were for Ty1A/B, Rsp5, and Snf4, none of which were shown to coimmunoprecipitate picNuA4 activity (data not shown). NuA4 purification using the TAP system was performed as described (Puig et al. 2001) from 250 mL of yeast cells grown to an OD600 of ∼2.5. The cell extracts were precleared before purification with 50 μL of IgG and calmodulin affinity resins. The final elution of the purified complex from the calmodulin resin was performed with 50 μL of 10 mM EGTA buffer. Recombinant Epl1 (1–485)/Esa1 and Epl1 (1–485)/Yng2/Esa1 complexes containing an N-terminal hexahistidine tag on Esa1 were reconstituted in vivo by coexpressing the two or three components in Escherichia coli using the T7-promoter-driven pST39 polycistronic expression plasmid (Tan 2001). The complexes were purified by Talon metal affinity, SourceQ anion exchange, Source S cation exchange, and Superose 6 gel filtration chromatography. HAT assays with different Esa1-containing complexes were standardized by the amount of Esa1 protein as evaluated by Western blotting.
MMS- and rapamycin-sensitivity assays, telomere silencing assay, and Northern blot analysis
Yeast strains were grown overnight at 30°C in nonselective medium, diluted to an OD600 of 0.5, and grown for 3 h in nonselective medium. Then 10-fold serial dilutions of cultures were plated onto medium containing 0.03% methyl methanesulfonate (MMS) or 25 nM rapamycin. For telomere-silencing assays, overnight precultures were diluted in YPD to an OD600 of 0.25 and incubated at 30°C for 5 h. Cultures were then diluted to an OD600 of 0.5, and 10-fold dilutions were spotted onto HC and HC/5′-FOA media and incubated at 30°C for 2 and 3 d (respectively). Total yeast RNA was isolated by the hot-phenol method; 15 μg of RNA was analyzed by Northern blotting as described (Galarneau et al. 2000). The probes used were ORFs from HIS4, PHO5, TRP4, PHO84, RPS11B, and ACT1, obtained by PCR and radiolabeled by using the Multiprime Labeling System (Amersham Bioscience).
GST pull-down and electromobility shift assays
GST- and HIS-recombinant proteins were expressed and purified on glutathione Sepharose or nickel-agarose beads following standard procedures. HAT/GST pull-downs were performed as described (Utley et al. 1998). HIS-tagged recombinant proteins were precleared on GST-Sepharose at 4°C for 1 h prior to pull-down. Then 300 ng of rEsa1 and 200 ng of rYng2 were incubated (at 4°C for 3 h) with 300 ng of appropriate GST fusion (or 1 μg of GST control). Equivalent amounts of input, bound, and free fractions were analyzed by Western blot using αHIS (BabCo) or HAT assays. For nucleosome-binding experiments, labeled 5S RNA gene was reconstituted in mononucleosomes as described (Côté et al. 1998). rEsa1, the rEsa1–Epl1 duplex, or the rPiccolo triplex were incubated with 12.5 ng of total nucleosomes at 30°C for 30 min. Binding reactions were resolved by 4% PAGE in 0.5× TBE.
Acknowledgments
We are grateful to A. Nourani for the Gal-Epl1 yeast strain; Y. Doyon for recombinant Yng2 expression vectors; and B. Séraphin, L. Pillus, D. Stillman, and J. Workman for strains and reagents. We thank A. Gagné-Henley and N. Roberge for technical help. We also thank M. Vignali for the name “Piccolo NuA4.” This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to J.C. and NIH to S.T. A.B. was a Natural Sciences and Engineering Research Council (NSERC) graduate student and R.T.U. a CIHR postdoctoral fellow. N.L. acknowledges support from the Canadian Office of Foreign Affairs. S.T. is a Pew Scholar in the Biomedical Sciences. J.C. is a CIHR New Investigator.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1056603.
References
- Adams A., Gottschling, D.E., Kaiser, C.A., and Stearns, T. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Allard S., Utley, R.T., Savard, J., Clarke, A., Grant, P., Brandl, C.J., Pillus, L., Workman, J.L., and Côté, J. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18: 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R., Pray-Grant, M.G., Selleck, W., Grant, P.A., and Tan, S. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277: 7989–7995. [DOI] [PubMed] [Google Scholar]
- Bird A.W., Yu, D.Y., Pray-Grant, M.G., Qiu, Q., Harmon, K.E., Megee, P.C., Grant, P.A., Smith, M.M., and Christman, M.F. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415. [DOI] [PubMed] [Google Scholar]
- Brown C.E., Howe, L., Sousa, K., Alley, S.C., Carrozza, M.J., Tan, S., and Workman, J.L. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333–2337. [DOI] [PubMed] [Google Scholar]
- Choy J.S., Tobe, B.T., Huh, J.H., and Kron, S.J. 2001. Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 276: 43653–43662. [DOI] [PubMed] [Google Scholar]
- Clarke A.S., Lowell, J.E., Jacobson, S.J., and Pillus, L. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19: 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J., Peterson, C.L., and Workman, J.L. 1998. Perturbation of nucleosome core structure by the SWI/SNF complex persists following its detachment, enhancing subsequent transcription factor binding. Proc. Natl. Acad. Sci. 95: 4947–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A., Utley, R.T., Nourani, A., Allard, S., Schmidt, P., Lane, W.S., Lucchesi, J.C., and Cote, J. 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem. 276: 3484–3491. [DOI] [PubMed] [Google Scholar]
- Galarneau L., Nourani, A., Boudreault, A.A., Zhang, Y., Héliot, L., Allard, S., Savard, J., Lane, W.S., Stillman, D.J., and Côté, J. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5: 927–937. [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea J.J., Lopez, R., and Shearn, A. 2000. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156: 645–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio, O.M., Billington, B.L., and Zakian, V.A. 1990. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Duggan, L., Côté, J., Roberts, S.M., Brownell, J.E., Candau, R., Ohba, R., Owen-Hughes, T., Allis, C.D., Winston, F., et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 11: 1640–1650. [DOI] [PubMed] [Google Scholar]
- Howe L., Kusch, T., Muster, N., Chaterji, R., Yates III, J.R., and Workman, J.L. 2002. Yng1p modulates the activity of Sas3p as a component of the yeast NuA3 histone acetyltransferase complex. Mol. Cell. Biol. 22: 5047–5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. and Allis, C.D. 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Katan-Khaykovich Y. and Struhl, K. 2002. Dynamics of global histone acetylation and deacetylation in vivo: Rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes & Dev. 16: 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Meijer, M., Lees-Miller, S.P., Riabowol, K., and Young, D. 2000. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20: 3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Smith, J.S., Meijer, M., Williams, T.J., Bachman, N., Boeke, J.D., and Young, D. 2001. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem. 276: 24068–24074. [DOI] [PubMed] [Google Scholar]
- Megee P.C., Morgan, B.A., and Smith, M.M. 1995. Histone H4 and the maintenance of genome integrity. Genes & Dev. 9: 1716–1727. [DOI] [PubMed] [Google Scholar]
- Nourani A., Doyon, Y., Utley, R.T., Allard, S., Lane, W.S., and Côté, J. 2001. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 21: 7629–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba R., Steger, D.J., Brownell, J.E., Mizzen, C.A., Cook, R.G., Côté, J., Workman, J.L., and Allis, C.D. 1999. A novel H2A/H4 nucleosomal histone acetyltransferase in Tetrahymena thermophila. Mol. Cell. Biol. 19: 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel W.W., Schaft, D., Roguev, A., Shevchenko, A., Tekotte, H., Wilm, M., Rigaut, G., Seraphin, B., Aasland, R., and Stewart, A.F. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes & Dev. 15: 2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Seraphin, B. 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Reid J.L., Iyer, V.R., Brown, P.O., and Struhl, K. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6: 1297–1307. [DOI] [PubMed] [Google Scholar]
- Saleh A., Lang, V., Cook, R., and Brandl, C.J. 1997. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J. Biol. Chem. 272: 5571–5578. [DOI] [PubMed] [Google Scholar]
- Sendra R., Tse, C., and Hansen, J.C. 2000. The yeast histone acetyltransferase A2 complex, but not free Gcn5p, binds stably to nucleosomal arrays. J. Biol. Chem. 275: 24928–24934. [DOI] [PubMed] [Google Scholar]
- Shimono Y., Murakami, H., Hasegawa, Y., and Takahashi, M. 2000. RET finger protein is a transcriptional repressor and interacts with Enhancer of Polycomb that has dual transcriptional functions. J. Biol. Chem. 275: 39411–39419. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A., Clegg, N.J., Antonchuk, J., Milne, T.A., Stankunas, K., Ruse, C., Grigliatti, T.A., Kassis, J.A., and Brock, H.W. 1998. Enhancer of Polycomb is a suppressor of position-effect variegation in Drosophila melanogaster. Genetics 148: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankunas K., Berger, J., Ruse, C., Sinclair, D.A., Randazzo, F., and Brock, H.W. 1998. The Enhancer of Polycomb gene of Drosophila encodes a chromatin protein conserved in yeast and mammals. Development 125: 4055–4066. [DOI] [PubMed] [Google Scholar]
- Suka N., Suka, Y., Carmen, A.A., Wu, J., and Grunstein, M. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8: 473–479. [DOI] [PubMed] [Google Scholar]
- Tan S. 2001. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21: 224–234. [DOI] [PubMed] [Google Scholar]
- Utley R.T. and Côté, J. 2002. The MYST family of histone acetyltransferases. Curr. Topics Microbiol. Immunol. 274: 203–236. [DOI] [PubMed] [Google Scholar]
- Utley R.T., Ikeda, K., Grant, P.A., Côté, J., Steger, D.J., Eberharter, A., John, S., and Workman, J.L. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394: 498–502. [DOI] [PubMed] [Google Scholar]
- Verdone L., Wu, J., van Riper, K., Kacherovsky, N., Vogelauer, M., Young, E.T., Grunstein, M., Di Mauro, E., and Caserta, M. 2002. Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J. 21: 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M., Steger, D.J., Neely, K.E., and Workman, J.L. 2000. Distribution of acetylated histones resulting from Gal4–VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 19: 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Wu, J., Suka, N., and Grunstein, M. 2000. Global histone acetylation and deacetylation in yeast. Nature 408: 495–498. [DOI] [PubMed] [Google Scholar]
- Waterborg J.H. 2002. Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochem. Cell Biol. 80: 363–378. [DOI] [PubMed] [Google Scholar]