Abstract
Hes7, a bHLH gene essential for somitogenesis, displays cyclic expression of mRNA in the presomitic mesoderm (PSM). Here, we show that Hes7 protein is also expressed in a dynamic manner, which depends on proteasome-mediated degradation. Spatial comparison revealed that Hes7 and Lunatic fringe (Lfng) transcription occurs in the Hes7 protein-negative domains. Furthermore, Hes7 and Lfng transcription is constitutively up-regulated in the absence of Hes7 protein and down-regulated by stabilization of Hes7 protein. Thus, periodic repression by Hes7 protein is critical for the cyclic transcription of Hes7 and Lfng, and this negative feedback represents a molecular basis for the segmentation clock.
Keywords: bHLH, Hes7, Lunatic fringe, negative feedback, segmentation clock, somite
Somites, the metameric units of vertebrate embryos, are aligned along both sides of the neural tube and give rise to repetitive structures including vertebrae, ribs, and skeletal muscles. A bilateral pair of somites buds off from the anteriormost end of the unsegmented presomitic mesoderm (PSM; Pourquié 2001; Saga and Takeda 2001). A new somite is formed every 120 min in the mouse, and this periodic event is believed to be governed by a molecular clock (Cooke 1998; Dale and Pourquié 2000). It is thus likely that, in somitogenesis, the temporal periodicity of the molecular clock is translated into the spatial periodicity of somites.
The first evidence of a molecular clock for somite segmentation was provided by the finding of the oscillatory expression of the basic helix–loop–helix (bHLH) gene c-hairy1 (Palmeirim et al. 1997). The expression of c-hairy1 mRNA sweeps across the PSM in a posterior-to-anterior direction repeatedly, and each cycle is synchronous with the somite formation. Interestingly, this wave-like propagation of gene expression is not caused by cell movement but is the result of synchronous oscillation of c-hairy1 expression in the PSM cells. Like c-hairy1, several other genes, all of which are involved in Notch signaling, also show cyclic expression in the PSM. They include bHLH genes such as Hes1, Hes7, and Hey2 in mouse and her1 and her7 in zebrafish (Holley et al. 2000; Jouve et al. 2000; Leimeister et al. 2000; Sawada et al. 2000; Bessho et al. 2001b; Dunwoodie et al. 2002; Oates and Ho 2002). In addition, the expression of zebrafish deltaC, which encodes a ligand for Notch, and mouse and chick Lfng, a gene for glycosyltransferase that modulates the Notch signaling, oscillates in the PSM (Forsberg et al. 1998; McGrew et al. 1998; Aulehla and Johnson 1999; Jiang et al. 2000). Genetic analyses revealed that at least some of these oscillating genes play a critical role in somitogenesis. Mutations for deltaC, her1, and her7 in zebrafish (Holley et al. 2000, 2002; Henry et al. 2002; Oates and Ho 2002) and Lfng and Hes7 in mouse (Evrard et al. 1998; Zhang and Gridley 1998; Bessho et al. 2001b) all exhibit defects of somite segmentation. Furthermore, persistent expression of Lfng also perturbs somite segmentation (Dale et al. 2003; Serth et al. 2003). Thus, periodic expression, but not the expression per se, of the cyclic genes is essential for coordinated somite segmentation. Despite these extensive studies, the precise mechanism for the segmentation clock is still obscure because, although Lfng establishes a negative feedback loop, which seems to underlie the segmentation clock in chick (Dale et al. 2003), it was recently shown that overexpression of Lfng does not inhibit the endogenous Lfng expression in mouse (Serth et al. 2003). Furthermore, the mechanism for c-hairy1-related gene oscillation as well as the causal link between Lfng and c-hairy1-related gene oscillations remained to be determined.
Here, we found that, like Hes7 mRNA, the expression of Hes7 protein oscillates in the PSM. Spatial comparison revealed that Hes7 and Lfng transcription occurs in the Hes7 protein-negative domains, suggesting that in these domains both Hes7 and Lfng transcription is repressed by Hes7 protein. Agreeing with this theory, Hes7 overrides the Notch-induced transcription from the Hes7 promoter in transfection assays. Furthermore, Hes7 and Lfng transcription is constitutively up-regulated in the absence of Hes7 protein and constitutively down-regulated by stabilization of Hes7 protein. Thus, periodic repression by Hes7 protein is critical for the cyclic transcription of both Hes7 and Lfng, indicating that this negative feedback represents a molecular basis for the segmentation clock.
Results and Discussion
Expression of Hes7 protein is dynamic in the PSM
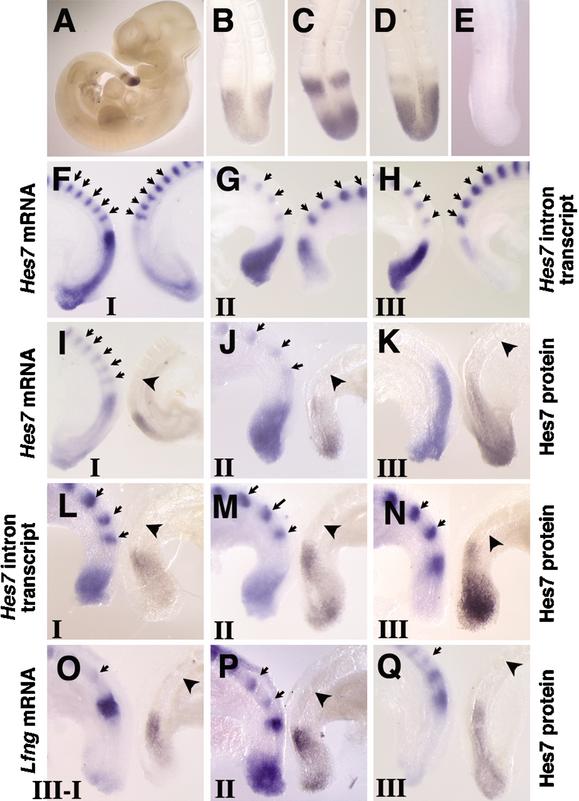
To examine the expression profiles of Hes7 protein, we generated anti-Hes7 antibody and carried out whole-mount immunostaining of mouse embryos (Fig. 1A–E). The Hes7 immunoreactivity is specifically observed in the PSM but not in other regions, including formed somites (Fig. 1A), whereas it is not detected at all in Hes7-null mice (Fig. 1E). This result is highly consistent with the expression patterns of Hes7 mRNA (Bessho et al. 2001a), indicating that the antibody reacts specifically with Hes7 protein.
Figure 1.
Spatial relationship of Hes7 protein, Hes7 mRNA, Hes7 nascent transcript, and Lfng mRNA in the PSM. (A–E) The expression pattern of Hes7 protein was examined by whole-mount immunochemistry. Hes7 immunoreactivity is specifically observed in the PSM of wild-type embryos (A). At E10.5, wild-type embryos display various expression patterns (B, n = 8; C, n = 5; D, n = 8). No signal is detected in Hes7-null mice (E, n = 3). (F–H) Comparison of the expression of Hes7 mRNA (left) and Hes7 nascent transcript (right) in the bisected caudal portions of E9.5 embryos. The latter is detected by the Hes7 intron probe. (I–Q) The expression of Hes7 protein (right) was compared with the expression of Hes7 mRNA (I–K, left), Hes7 nascent transcript (L–N, left), and Lfng mRNA (O–Q, left). The expression patterns are categorized into three phases: phase I (I,L,O), phase II (J,M,P), and phase III (K,N,Q). Hes7 protein-positive domains and the regions for Hes7 nascent transcript and Lfng mRNA are mutually exclusive. The established somites are stained with the Uncx4.1 probe for spatial alignment (arrows). The boundary between S0 and S-I is indicated by an arrowhead.
The expression of Hes7 protein is variable in the PSM even at the same developmental stages and could be categorized into three patterns (Fig. 1B–D), which are similar to the patterns of transcripts of cyclic genes (Pourquié and Tam 2001). In the first pattern, embryos display Hes7 immunoreactivity mainly in the middle PSM (Fig. 1B). In the second pattern, embryos show strong Hes7 immunoreactivity in the posterior PSM as well as in the anterior PSM (Fig. 1C). In the third pattern, the posterior signal extends to the middle and the anterior signal becomes weaker and soon disappears (Fig. 1D). This anterior signal reaches the prospective somite termed S-I (Pourquié and Tam 2001). Hes7 protein is not expressed in the forming somite termed S0 in any embryos.
To determine the spatial relationship between Hes7 protein and mRNA, we bisected the PSM of embryos and carried out immunochemistry for Hes7 protein and in situ hybridization for Hes7 mRNA, separately. The established somites are also stained by in situ hybridization with the Uncx4.1 probe for spatial alignment (Fig. 1F–Q, arrows). At phase I (n = 7), when a weak Hes7 mRNA expression occurs in the posterior PSM, in addition to a strong signal in the anterior PSM (Fig. 1I, left), Hes7 protein is found mainly in the middle PSM (Fig. 1I, right). At phase II (n = 9), when Hes7 mRNA-positive domain extends to the middle from the posterior PSM (Fig. 1J, left), Hes7 protein disappears from the middle PSM but appears in the anterior and posteriormost PSM (Fig. 1J, right). At phase III (n = 11), when Hes7 mRNA expression extends to the anterior PSM (Fig. 1K, left), Hes7 protein is expressed in the posterior to the middle PSM but disappears from the anterior region (Fig. 1K, right). These results indicate that the expression domains of Hes7 protein and mRNA are overlapped but different from each other.
Transcription of Hes7 and Lfng occurs in the Hes7 protein-negative domains in the PSM
Because the Hes7 mRNA levels could be posttranscriptionally regulated, the mRNA-positive domains do not necessarily reflect the regions in which the gene is actively transcribed. Recently, Morales et al. (2002) carried out in situ hybridization with Lfng intron probes and successfully detected the regions in which Lfng is actively transcribed, because intron probes recognize only nascent transcripts in the nucleus (Shermoen and O'Farrell 1991). We thus examined the regions in which Hes7 is actively transcribed, by using the first intron of Hes7 as a probe. The PSM of embryonic day 9.5 (E9.5) embryos was bisected, and in situ hybridization was performed with the Hes7 intron and exon probes, separately. During phase I (n = 5), when Hes7 mRNA expression occurs in the posterior PSM in addition to a strong signal in the anterior PSM (Fig. 1F, left), the Hes7 intron signal is present mainly in the posterior PSM (Fig. 1F, right). At phase II (n = 7), when the Hes7 mRNA domain extends to the middle from the posterior PSM (Fig. 1G, left), the Hes7 intron signal is also present in the middle PSM but disappears from the posterior PSM (Fig. 1G, right). At phase III (n = 7), when the Hes7 mRNA expression domain extends to the anterior PSM (Fig. 1H, left), the Hes7 intron signal is found only in the anterior PSM (Fig. 1H, right). During phases II to III, Hes7 mRNA still remains in the posterior PSM, whereas Hes7 intron signal does not (Fig. 1G,H), suggesting that Hes7 mRNA persists for a while after Hes7 transcription is turned off. These results indicate that Hes7 transcription also occurs in a dynamic manner.
To determine the spatial relationship between the Hes7 protein expression and Hes7 transcription, we bisected the PSM of embryos and carried out immunochemistry for Hes7 protein and in situ hybridization for the Hes7 intron, separately. At phase I (n = 10), Hes7 transcription occurs in the posterior PSM (Fig. 1L, left). In contrast, Hes7 protein is present in the middle PSM (Fig. 1L, right). At phase II (n = 9), when Hes7 transcription moves to the middle PSM (Fig. 1M, left), Hes7 protein moves to the anterior PSM but disappears from the middle and a new signal appears in the posteriormost PSM (Fig. 1M, right). At phase III (n = 10), when Hes7 transcription occurs in the anterior PSM (Fig. 1N, left), Hes7 protein is present in the middle-to-posterior PSM and a weak signal is transiently observed in the anteriormost PSM (Fig. 1N, right). Thus, in all phases, the Hes7 intron signals and Hes7 protein-positive regions are mutually exclusive, indicating that Hes7 transcription occurs only in the Hes7 protein-negative regions. These results suggest that Hes7 transcription is repressed by Hes7 protein.
We also examined the spatial relationship between Hes7 protein and Lfng mRNA, which oscillates in the same phase as Hes7 mRNA but gives narrower bands in the PSM (Bessho et al. 2001b). Similar to the Hes7 intron signals, the Lfng mRNA domains are always exclusive to the expression of Hes7 protein in all phases (Fig. 1O–Q, phase I, n = 7; phase II, n = 8; phase III, n = 13). Thus, Lfng expression also occurs only in the Hes7 protein-negative domains. These results suggest that Hes7 protein represses transcription of both Hes7 and Lfng and thereby makes the two genes cycle together.
The ubiquitin–proteasome pathway rapidly degrades Hes7 protein
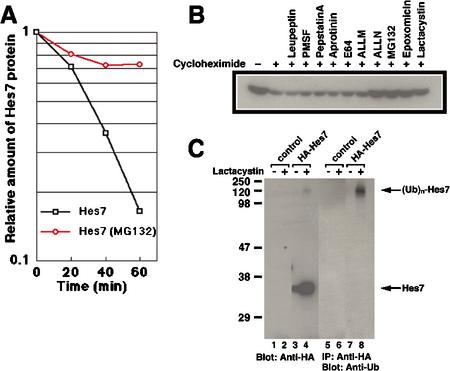
For a 2-h cycle oscillation, Hes7 protein should be unstable. Thus, we next measured the half-life of Hes7 protein. C3H10T1/2 fibroblasts were transfected with the expression vector for Hes7 protein, and, after inhibition of new protein synthesis by cycloheximide, the decay of Hes7 protein was monitored (Fig. 2A). The half-life of Hes7 protein is found to be only 23.1 ± 8.7 min, which may enable a 2-h cycle oscillation. We also examined the instability of Hes7 protein in the PSM. Treatment of the PSM with cycloheximide resulted in more than twofold reduction of the Hes7 protein level within 20 min, indicating that the half-life of Hes7 protein in the PSM may be even shorter than 20 min (data not shown). These results suggest that Hes7 protein could be posttranslationally modified in a PSM-specific manner for more efficient degradation.
Figure 2.
Hes7 protein is degraded by the ubiquitin–proteasome system. (A) C3H10T1/2 cells were transfected with the HA-Hes7 expression vector on the previous day. The cells were cultured in the presence of cycloheximide plus MG132 or cycloheximide only for 30 min and then harvested at the indicated time points. Whole-cell extracts were probed with anti-HA antibody, and the relative immunoreactive activities of the signals were measured. Hes7 protein is degraded with a half-life of 23.1 ± 8.7 min, whereas it is stabilized in the presence of the proteasome inhibitor MG132. (B) Cells were transfected with the HA-Hes7 expression vector on the previous day. The cells were cultured for 2 h with various inhibitors, as indicated above each lane, in the presence (+) of cycloheximide to inhibit new protein synthesis. Cell extracts were probed with anti-HA antibody. Note that Hes7 protein is stabilized by proteasome inhibitors (ALLN, MG132, epoxomicin, and lactacystin) but not by other protease inhibitors (leupeptin, PMSF, pepstatinA, aprotinin, E64, and ALLM). (C) C3H10T1/2 cells were transfected with the HA-Hes7 expression vector and cultured overnight in the presence (+) or absence (-) of lactacystin (20 μM). In the four left lanes, whole-cell extracts were probed with anti-HA antibody. In the presence of lactacystin, Hes7 protein is stabilized and higher-molecular-weight bands appear (lane 4). In the four right lanes, whole-cell extracts were immunoprecipitated with anti-HA antibody and probed with anti-ubiquitin antibody. The high-molecular-weight species are highly reactive to anti-ubiquitin antibody (lane 8).
To identify proteases responsible for Hes7 protein degradation, we used various protease inhibitors to stabilize Hes7 protein. Proteasome inhibitors [N-acetyl-Leu-Leunorleucinal (ALLN), MG132, epoxomicin, and lactacystin] stabilize Hes7 protein, whereas other protease inhibitors [leupeptin, phenylmethylsulfonyl fluoride (PMSF), pepstatinA, aprotinin, E64, and N-acetyl-Leu-Leu-methyoninal (ALLM)] do not (Fig. 2B). In the presence of MG132, Hes7 protein is significantly stabilized and persists even after 60 min (Fig. 2A). These results suggest that Hes7 protein is specifically degraded by the ubiquitin–proteasome system. To confirm this conclusion, we next analyzed whether Hes7 protein is ubiquitinated. The hemagglutinin (HA)-tagged recombinant Hes7 protein (HA-Hes7) was expressed in C3H10T1/2 cells. In the presence of the proteasome inhibitor lactacystin, high-molecular-weight bands (>100 kD) as well as a full-length Hes7 band are detected by Western blotting with anti-HA antibody (Fig. 2C, lane 4). These high-molecular-weight species are highly reactive to anti-ubiquitin antibody (Fig. 2C, lane 8), indicating that HA-Hes7 is polyubiquitinated. These results indicate that Hes7 protein is degraded by the ubiquitin–proteasome system.
Hes7 can repress its own promoter
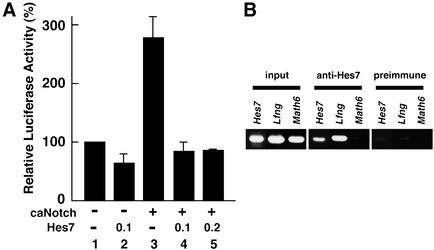
Because Hes7 is a transcriptional repressor (Bessho et al. 2001a), the expression profiles of Hes7 protein and nascent transcript shown above strongly indicate that Hes7 represses its own expression. We thus next asked whether Hes7 directly represses the Hes7 promoter. The Hes7 promoter has a pair of putative binding sites of RBP-J, a Notch signaling mediator, and at least two E-boxes and one N-box, target sequences for Hes7 protein (Bessho et al. 2001a). The luciferase vector under the control of the 0.9-kb Hes7 promoter was cotransfected with the Hes7 expression vector to NIH3T3 cells. Hes7 slightly represses transcription from the Hes7 promoter (Fig. 3A, lane 2). Coexpression of a constitutively active form of Notch (caNotch) up-regulates Hes7 promoter activity (Fig. 3A, lane 3), as previously described (Bessho et al. 2001a). This up-regulation is efficiently inhibited by coexpression of the Hes7 expression vector (Fig. 3A, lanes 4,5). These results demonstrate that Hes7 can override the Notch-induced transcription from the Hes7 promoter, agreeing with the observation that Hes7 transcription does not occur in the Hes7 protein-positive domains. These data also suggest that the 0.9-kb promoter contains the region necessary for cyclic expression of Hes7.
Figure 3.
Analysis of Hes7 and Lfng promoters. (A) The luciferase vector under the control of the Hes7 promoter was cotransfected with or without the expression vector for caNotch and/or the expression vector for Hes7 (0.1 or 0.2μg). The activity of the luciferase vector with the Hes7 promoter alone (lane 1) is taken as 100%. Relative luciferase activities shown with a standard error are the average of three independent experiments performed in duplicate. Hes7 overrides the caNotch-induced activation of the Hes7 promoter (lanes 4,5). (B) ChIP analysis. Anti-Hes7 antibody specifically precipitates the chromatin containing the Hes7 and Lfng promoter regions, but not the Math6 promoter region, from PSM tissues. Preimmune serum does not precipitate these promoters.
To determine whether Hes7 protein interacts with the Hes7 promoter region in vivo, we performed a chromatin immunoprecipitation (ChIP) analysis. Anti-Hes7 antibody, but not preimmune serum, specifically precipitates the chromatin containing the Hes7 promoter region from PSM tissues (Fig. 3B). Similarly, anti-Hes7 antibody specifically precipitates the chromatin containing the Lfng promoter region from PSM tissues (Fig. 3B). These results indicate that Hes7 protein interacts with both Hes7 and Lfng promoter regions in vivo.
Lack of functional Hes7 protein persistently up-regulates Hes7 transcription
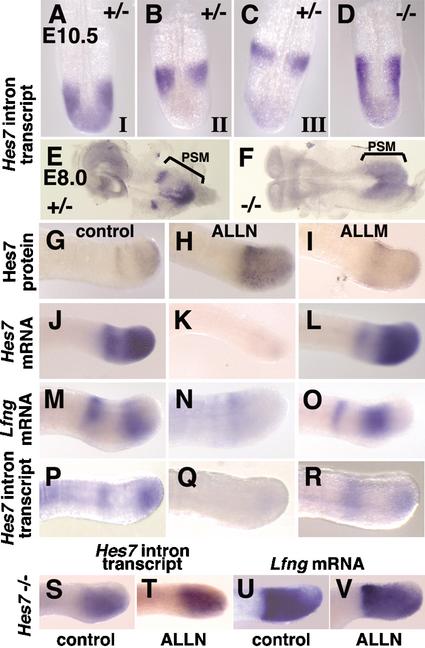
The above results suggest that Hes7 protein periodically represses Hes7 transcription and that this periodic repression leads to cyclic gene expression. If this model is correct, manipulation of Hes7 protein levels should affect cyclic gene expression. To address this issue, we first examined Hes7 transcription in Hes7-null mice. We previously generated Hes7-null mice, which lack exons 2–4 (Bessho et al. 2001b). In these mice, the first exon and intron are intact, and these sequences are expressed under the control of the endogenous Hes7 promoter. As in wild-type embryos (Fig. 1F–H,L–N), the Hes7 intron signals are variable in Hes7+/- embryos at E8.0 and E10.5 (Fig. 4A–C,E), indicating that Hes7 transcription periodically changes in the PSM. In Hes7-/- embryos, however, no such variable Hes7 transcription is observed at E8.0 and E10.5 (Fig. 4D,F). All Hes7-null embryos display homogeneous Hes7 intron signals throughout the PSM. Thus, Hes7 transcription is constitutively activated throughout the PSM in the absence of functional Hes7 protein. This up-regulation is very similar to Lfng expression in Hes7-null embryos, which is also constitutively up-regulated throughout the mutant PSM (Bessho et al. 2001b). Thus, Hes7 protein is required for generation of the Hes7 and Lfng transcription-negative domains.
Figure 4.
Dynamic expression of Hes7 mRNA is affected by Hes7 protein. (A–F) In situ hybridization was performed with the Hes7 intron probe, which detects the expression of the nascent transcript of both mutant and wild-type Hes7 alleles. The expression is dynamic in Hes7+/- embryos at E10.5 (A, phase I, n = 7; B, phase II, n = 7; C, phase III, n = 8) and E8.0 (E, n = 4). In contrast, the nascent transcript is expressed throughout the PSM in Hes7-null embryos both at E10.5 (D, n = 9) and at E8.0 (F, n = 4). (G–V) The caudal part of E10.5 embryos of wild type (G–R) and Hes7-/- (S–V) was cultured for 2 h with 100 μM ALLN, with 100 μM ALLM, or without protease inhibitors, as indicated. The expression patterns of Hes7 protein (G–I), Hes7 mRNA (J–L), Lfng mRNA (M–O,U,V), and Hes7 nascent transcript (P–R,S,T) were examined. Treatment of proteasome inhibitors stabilizes Hes7 protein (H) and down-regulates Hes7 and Lfng transcription (K,N,Q), compared with the control and ALLM-treated explants. In contrast, such repression does not occur in Hes7-/- PSM (S–V).
Stabilization of Hes7 protein inhibits transcription of Hes7 and Lfng
We next asked whether Hes7 and Lfng transcription is affected when Hes7 protein is stabilized. To address this issue, we stabilized Hes7 protein in the PSM by proteasome inhibitors. The PSM tissues of E10.5 wild-type embryos were cultured for 2 h in the presence or absence of proteasome inhibitors, MG132 (data not shown), and ALLN (Fig. 4). As a control, the PSM was also treated with ALLM, which does not inhibit proteasome. In the PSM treated with proteasome inhibitors, expression of Hes7 protein is diffusely up-regulated (Fig. 4H), compared with control (Fig. 4G) and ALLM-treated PSM (Fig. 4I). In the PSM treated with proteasome inhibitors, Hes7 mRNA (Fig. 4K), Hes7 intron signals (Fig. 4Q), and Lfng mRNA (Fig. 4N) are all decreased, compared with the control and ALLM-treated PSM (Fig. 4J,L,M,O,P,R). However, in the absence of functional Hes7 protein, Hes7 intron and Lfng mRNA are constitutively expressed even after treatment of the proteasome inhibitor ALLN (Fig. 4S–V), indicating that Hes7 protein is required for proteasome inhibitor-induced repression of Hes7 and Lfng transcription in the wild-type PSM. These results demonstrate that stabilization of Hes7 protein constitutively represses Hes7 and Lfng transcription.
Hes7 negative feedback represents a molecular basis for the segmentation clock
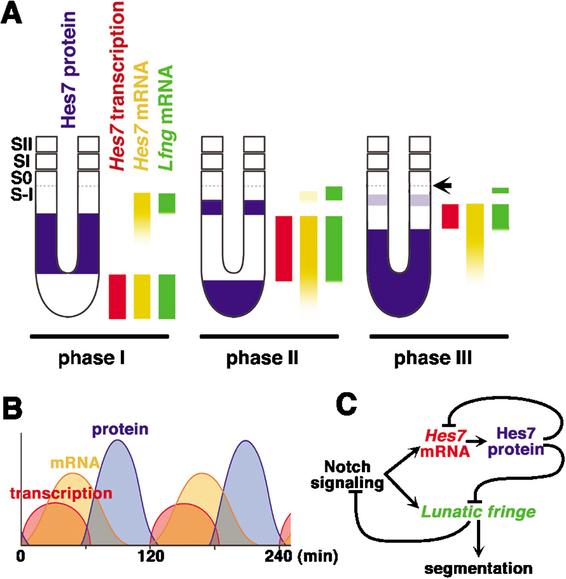
In this study, we show that the Hes7 protein domains and the regions of Hes7 nascent transcript and Lfng mRNA are mutually exclusive in all phases. Furthermore, we find that Hes7 and Lfng transcription is constitutively up-regulated in the absence of Hes7 protein and constitutively down-regulated by stabilization of Hes7 protein. Thus, periodic repression by Hes7 protein establishes the cyclic transcription of Hes7 and Lfng (Fig. 5C). These results indicate that the negative feedback of Hes7 represents a molecular basis for the segmentation clock. This mechanism is the same as that of the recently identified Hes1 oscillator, which is distributed in many cell types (Hirata et al. 2002). We summarize the expression patterns of Hes7 protein, mRNA, and nascent transcript as well as Lfng mRNA in the PSM (Fig. 5A). Based on these spatial expression patterns, we speculate the temporal expression profiles, as follows: Hes7 transcription leads to accumulation of Hes7 mRNA and Hes7 protein but is turned off as soon as Hes7 protein is accumulated, but Hes7 mRNA persists for a while (Fig. 5B). Recent studies revealed that oscillatory expression of Lfng is controlled at the transcriptional level (Cole et al. 2002; Morales et al. 2002). Among several elements in the Lfng promoter, the region 2 (Cole et al. 2002) or the region A (Morales et al. 2002) contains two E-boxes, which are critical for the cyclic expression (Cole et al. 2002). Our data indicate that Hes7, which represses transcription via E-boxes (Bessho et al. 2001a), is likely to regulate the cyclic expression of Lfng through the E-boxes in the region 2/A.
Figure 5.
Summary of spatial and temporal expression patterns of Hes7 and model of the mechanism for the segmentation clock. (A) Spatial relationship of Hes7 protein, Hes7 transcription, Hes7 mRNA, and Lfng mRNA in the mouse PSM. Hes7 and Lfng transcription does not occur in the Hes7 protein-positive domains in all three phases. The prospective somite boundary at phase III is indicated by an arrow. (B) Temporal relationship of Hes7 protein, Hes7 mRNA, and Hes7 transcription in PSM cells. (C) A model of the mechanism for the segmentation clock based on the negative feedback loop of Hes7. Periodic repression by Hes7 protein is essential for cyclic expression and synchronization of Hes7 and Lfng mRNA.
Dale et al. (2003) recently demonstrated that Lfng represses its own expression through modulation of the Notch pathway in chick, and concluded that the negative feedback loop of Lfng is a molecular basis for the segmentation clock. It is likely that Lfng also constitutes a negative feedback loop in mouse, because the expression domains of the lacZ gene, which is knocked into the Lfng allele, become wider in the Lfng-null mutant mouse (Zhang and Gridley 1998). However, although Hes7 is required for the dynamic expression of Lfng (Bessho et al. 2001b), Lfng is not required for the dynamic expression of Hes7 (Y. Takahashi and Y. Saga, pers. comm.). Thus, Hes7 is an upstream regulator of Lfng oscillation, and the negative feedback loop of Lfng could be involved in refinement of cyclic expression or keeping accuracy of the 2-h cycle. It was recently reported that Wnt signaling is also involved in the segmentation clock (Aulehla et al. 2003). Interestingly, in hypomorphic mutants for Wnt3a, Lfng oscillation is lost, suggesting the cross-talk between Wnt and Notch signaling. However, the relationship between Hes7 oscillation and Wnt signaling remains to be determined.
Materials and methods
Generation of the anti-Hes7 antibody, whole-mount immunochemistry, and whole-mount in situ hybridization
A full-length Hes7 protein with an additional six histidine residues in the N-terminal region was expressed in Escherichia coli and immunized to guinea pigs. The serum was collected and used as an anti-Hes7 polyclonal antibody. Whole-mount immunochemistry was performed as described previously (Tomita et al. 2000). Briefly, embryos were fixed with 4% paraformaldehyde in PBS at 4°C for 3 h and treated with 0.1% H2O2 overnight. Then, the embryos were incubated with anti-Hes7 antibody (1/100 diluted) at 4°C for 3–5 d, and next with peroxidase-conjugated anti-guinea pig IgG (Chemicon) at 4°C overnight. The peroxidase deposits were visualized by 4-chloro-1-naphthol. Whole-mount in situ hybridization was performed, as described previously (Bessho et al. 2001b). Hes7 nascent transcripts were detected with the entire first intron of the Hes7 gene (1.0 kb).
Measurement of protein half-life and treatment of protease inhibitors
The HA-Hes7 expression vector was transfected to C3H10T1/2 cells. Next day, the culture medium was changed with or without 100 μM leupeptin, 1 mM PMSF, 50 μg/mL pepstatin A, 50 μg/mL Aprotinin, 100 μM E64, 100 μM ALLM, 100 μM ALLN, 100 μM MG132, 10 μM Epoxomicin, or 100 μM lactacystin. Cells were harvested and subjected to Western blot with anti-HA-peroxidase (3F10, rat monoclonal, Roche, 1/5000).
Ubiquitination experiment
C3H10T1/2 cells were transfected with the HA-Hes7 expression vector, and ubiquitination was examined as previously described (Hirata et al. 2002).
Promoter analysis
The luciferase assay was done as previously described (Bessho et al. 2001a).
ChIP analysis
ChIP analysis was carried out as previously described (Shang et al. 2000). The PSM cells of E10.5 embryos were cross-linked with 1% formaldehyde at 37°C for 10 min, sonicated, and incubated with anti-Hes7 antiserum (1/200) or normal guinea pig serum (1/200) for 12 h. Immune complexes were incubated with protein A Sepharose beads (Amersham), which were then washed 6 times, and incubated with 100 μg/mL Proteinase K for DNA extraction. DNA was analyzed by PCR using specific primers for the Hes7 promoter (5′-ACTTAGACTCTCTTCCCCT GATAATCTCCT-3′ and 5′-TAGATGCCCAGCTCAAGGACCCCG GCACGA-3′), the Lfng promoter (5′-ATAGCAGGTAGGACTTTTTCC TTGTCCTTG-3′ and 5′-GGAGTGGGATATGGTGGTTCCCAGGCT TCT-3′), and the Math6 promoter (5′-CACAGCCAGGCGCGCGGCGT GCAGAGTGAT-3′ and 5′-GCTTCCAAGTCCAATCGGAAAGTTTTA TAG-3′).
Acknowledgments
We thank D. Ish-Horowicz, O. Pourquié, Y. Saga, and Y. Takahashi for critical reading of the manuscript; A. Gossler, B. Herrmann, Y. Saga, and Y. Takahashi for communicating their unpublished results; and J. Hatakeyama, R. Shigemoto, and Y. Shinkai for technical advice. This work was supported by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Japan Society for the Promotion of Science.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding authors.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1092303.
References
- Aulehla A. and Johnson, R.L. 1999. Dynamic expression of Lunatic fringe suggests a link between notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev. Biol. 207: 49–61. [DOI] [PubMed] [Google Scholar]
- Aulehla A., Wehrle, C., Brand-Saberi, B., Kemler, R., Gossler, A., Kanzler, B., and Herrmann, B.G. 2003. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 4: 395–406. [DOI] [PubMed] [Google Scholar]
- Bessho Y., Miyoshi, G., Sakata, R., and Kageyama, R. 2001a. Hes7: A bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells 6: 175–185. [DOI] [PubMed] [Google Scholar]
- Bessho Y., Sakata, R., Komatsu, S., Shiota, K., Yamada, S., and Kageyama, R. 2001b. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes & Dev. 15: 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S.E., Levorse, J.M., Tilghman, S.M., and Vogt, T.F. 2002. Clock regulatory elements control cyclic expression of Lunatic fringe during somitogenesis. Dev. Cell 3: 75–84. [DOI] [PubMed] [Google Scholar]
- Cooke J. 1998. A gene that resuscitates a theory—Somitogenesis and a molecular oscillator. Trends Genet. 14: 85–88. [DOI] [PubMed] [Google Scholar]
- Dale J.K., Maroto, M., Dequeant, M.L., Malapert, P., McGrew, M., and Pourquié, O. 2003. Periodic Notch inhibition by Lunatic fringe underlies the chick segmentation clock. Nature 421: 275–278. [DOI] [PubMed] [Google Scholar]
- Dale K.J. and Pourquié, O. 2000. A clock-work somite. Bioessays 22: 72–83. [DOI] [PubMed] [Google Scholar]
- Dunwoodie S.L., Clements, M., Sparrow, D.B., Sa, X., Conlon, R.A., and Beddington, R.S. 2002. Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129: 1795–1806. [DOI] [PubMed] [Google Scholar]
- Evrard Y.A., Lun, Y., Aulehla, A., Gan, L., and Johnson, R.L. 1998. Lunatic fringe is an essential mediator of somite segmentation and patterning. Nature 394: 377–381. [DOI] [PubMed] [Google Scholar]
- Forsberg H., Crozet, F., and Brown, N.A. 1998. Waves of mouse Lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr. Biol. 8: 1027–1030. [DOI] [PubMed] [Google Scholar]
- Henry C.A., Urban, M.K., Dill, K.K., Merlie, J.P., Page, M.F., Kimmel, C.B., and Amacher, S.L. 2002. Two linked hairy/Enhancer of split-related zebrafish genes, her1 and her7, function together to refine alternating somite boundaries. Development 129: 3693–3704. [DOI] [PubMed] [Google Scholar]
- Hirata H., Yoshiura, S., Ohtsuka, T., Bessho, Y., Harada, T., Yoshikawa, K., and Kageyama, R. 2002. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298: 840–843. [DOI] [PubMed] [Google Scholar]
- Holley S.A., Geisler, R., and Nüsslein-Volhard, C. 2000. Control of her1 expression during zebrafish somitogenesis by a Delta-dependent oscillator and an independent wave-front activity. Genes & Dev. 14: 1678–1690. [PMC free article] [PubMed] [Google Scholar]
- Holley S.A., Julich, D., Rauch, G.J., Geisler, R., and Nüsslein-Volhard, C. 2002. her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development 129: 1175–1183. [DOI] [PubMed] [Google Scholar]
- Jiang Y.J., Aerne, B.L., Smithers, L., Haddon, C., Ish-Horowicz, D., and Lewis, J. 2000. Notch signalling and the synchronization of the somite segmentation clock. Nature 408: 475–479. [DOI] [PubMed] [Google Scholar]
- Jouve C., Palmeirim, I., Henrique, D., Beckers, J., Gossler, A., Ish-Horowicz, D., and Pourquié, O. 2000. Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development 127: 1421–1429. [DOI] [PubMed] [Google Scholar]
- Leimeister C., Dale, K., Fischer, A., Klamt, B., Hrabe de Angelis, M., Radtke, F., McGrew, M.J., Pourquié, O., and Gessler, M. 2000. Oscillating expression of c-Hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial signaling through multiple interacting bHLH factors. Dev. Biol. 227: 91–103. [DOI] [PubMed] [Google Scholar]
- McGrew M.J., Dale, J.K., Fraboulet, S., and Pourquié, O. 1998. The Lunatic fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr. Biol. 8: 979–982. [DOI] [PubMed] [Google Scholar]
- Morales A.V., Yasuda, Y., and Ish-Horowicz, D. 2002. Periodic Lunatic fringe expression is controlled during segmentation by a cyclic transcriptional enhancer responsive to notch signaling. Dev. Cell 3: 63–74. [DOI] [PubMed] [Google Scholar]
- Oates A.C. and Ho, R.K. 2002. Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development 129: 2929–2946. [DOI] [PubMed] [Google Scholar]
- Palmeirim I., Henrique, D., Ish-Horowicz, D., and Pourquié, O. 1997. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91: 639–648. [DOI] [PubMed] [Google Scholar]
- Pourquié O. 2001. Vertebrate somitogenesis. Annu. Rev. Cell Dev. Biol. 17: 311–350. [DOI] [PubMed] [Google Scholar]
- Pourquié O. and Tam, P.P. 2001. A nomenclature for prospective somites and phases of cyclic gene expression in the presomitic mesoderm. Dev. Cell 1: 619–620. [DOI] [PubMed] [Google Scholar]
- Saga Y. and Takeda, H. 2001. The making of the somite: Molecular events in vertebrate segmentation. Nat. Rev. Genet. 2: 835–845. [DOI] [PubMed] [Google Scholar]
- Sawada A., Fritz, A., Jiang, Y., Yamamoto, A., Yamasu, K., Kuroiwa, A., Saga, Y., and Takeda, H. 2000. Zebrafish Mesp family genes, mesp-a and mesp-b are segmentally expressed in the presomitic mesoderm, and Mesp-b confers the anterior identity to the developing somites. Development 127: 1691–1702. [DOI] [PubMed] [Google Scholar]
- Serth K., Schuster-Gossler, K., Cordes, R., and Gossler, A. 2003. Transcriptional oscillation of Lunatic fringe is essential for somitogenesis. Genes & Dev. 17: 912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Hu, X., DiRenzo, J., Lazar, M.A., and Brown, M. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103: 843–852. [DOI] [PubMed] [Google Scholar]
- Shermoen A.W. and O'Farrell, P.H. 1991. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell 67: 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., Moriyoshi, K., Nakanishi, S., Guillemot, F., and Kageyama, R. 2000. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 19: 5460–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N. and Gridley, T. 1998. Defects in somite formation in lunatic fringe-deficient mice. Nature 394: 374–377. [DOI] [PubMed] [Google Scholar]