Abstract
Shoot branching is inhibited by auxin transported down the stem from the shoot apex. Auxin does not accumulate in inhibited buds and so must act indirectly. We show that mutations in the MAX4 gene of Arabidopsis result in increased and auxin-resistant bud growth. Increased branching in max4 shoots is restored to wild type by grafting to wild-type rootstocks, suggesting that MAX4 is required to produce a mobile branch-inhibiting signal, acting downstream of auxin. A similar role has been proposed for the pea gene, RMS1. Accordingly, MAX4 and RMS1 were found to encode orthologous, auxin-inducible members of the polyene dioxygenase family.
Keywords: Auxin, Arabidopsis, CCD, pea, shoot branching
Variation in shoot branching is an important cause of diversity in plant form. Individual species have a characteristic branching pattern, which can change through the life cycle in response to developmental cues and to environmental conditions (Cline 1991; Beveridge et al. 2003). Branching control therefore requires the integration of many signals, both known and unknown.
Shoot branches arise from axillary meristems that form in the axils of leaves on the primary shoot axis. The axillary meristems themselves initiate leaves to form a bud. Bud growth can arrest but has the potential to reactivate to produce a shoot branch. Removal of the primary shoot apex results in activation of arrested axillary buds. The ability of the shoot apex to repress axillary bud growth is termed apical dominance. Thimann and Skoog (1933) reported that a compound, derived from the shoot apex, and later identified as auxin (indole-3-acetic acid), could inhibit the growth of lateral buds when applied to the stump of a decapitated plant. Subsequent work has provided multiple lines of evidence in support of auxin-mediated bud inhibition in planta. However, a second messenger must relay the auxin signal into the bud because apically derived auxin is not transported into buds (Morris 1977) and exogenous auxin applied directly to buds does not inhibit their growth (Cline 1996).
One model proposes that the effect of auxin on bud growth is mediated by cytokinin. Cytokinin can directly promote bud growth (Cline 1991); transgenic plants with increased auxin levels have reduced cytokinin levels (Eklöf et al. 2000), and cytokinin export from roots increases after decapitation, with this increase being abolished by application of auxin to the decapitated stump (Bangerth 1994). However, there is also good evidence for novel regulators of bud growth downstream of auxin. The ramosus mutants (rms1 to rms5) of pea (for reviews, see Beveridge 2000; Beveridge et al. 2003) have increased lateral branching, but this phenotype can be almost completely rescued by grafting a wild-type (WT) rootstock to an rms1, rms2, or rms5 mutant scion. Such grafting studies show that RMS1 and RMS5 are required for the production of a graft transmissible signal that moves from root to shoot and inhibits branching (Foo et al. 2001; Morris et al. 2001). This mobile signal is unlikely to be auxin or cytokinin because, as well as increased branching, the rms1 and rms5 mutants have reduced root-derived cytokinin and have at least WT auxin levels and transport (Beveridge et al. 1997; Morris et al. 2001). This is exactly the opposite of the prediction for a bushy plant and may be the result of feedback regulation of auxin and cytokinin levels. It is possible that the RMS1/RMS5-dependent long distance signal is a second messenger for auxin. The lateral buds of rms1 shoots can only respond to the inhibitory effects of apical auxin when grafted to WT rootstocks (Beveridge et al. 2000).
To identify genes that regulate bud growth in Arabidopsis, we screened mutagenized populations for plants with increased branching and have identified four loci, mutations at which result in more axillary growth, named max1 to max4 (Stirnberg et al. 2002). In this paper, we describe the phenotype of the max4 mutant and the cloning of the MAX4 gene, and show that this gene is orthologous to RMS1.
Results and Discussion
Isolation and genetic characterization of the max4 mutants
We have identified a class of Arabidopsis mutants with more axillary branches and placed them in four complementation groups named max1 to max4 (Stirnberg et al. 2002). Four independent recessive alleles were found at the MAX4 locus in the Columbia (Col) ecotype. Two alleles (max4-1 and max4-2) were isolated from the Sainsbury Laboratory Arabidopsis Transposant (SLAT) collection (Tissier et al. 1999), and two alleles (max4-3 and max4-4) were isolated from the AMAZE population (Wisman et al. 1998). There were no apparent differences in the severity of phenotype conferred by these alleles, and the max4-1 allele was chosen for detailed phenotypic analysis, following two rounds of back-crossing to WT.
The max4-1 mutant has increased shoot branching
Mature max4-1 mutant plants grown under a 16-h photoperiod have a bushy appearance at maturity as a result of increased growth of the buds in rosette leaf axils (Fig. 1A). In WT plants, all the cauline nodes and the most apical rosette nodes produced buds that developed into lateral inflorescences and, on average, the uppermost 5.1 ± 0.2 rosette nodes produced an elongated inflorescence >4 mm in length (Fig. 1B). A basipetal gradient of inflorescence lengths was observed, with branches arising in the youngest leaf axils having the greatest mean branch lengths. This gradient continued into older rosette nodes, where buds remained smaller than 4 mm in length. In most plants (n = 7/10) a weak acropetal gradient of bud growth was also observed, although these buds remained very small throughout the life of the plant. Between the acropetal and basipetal gradients were one to three nodes that carried tiny buds, or no bud visible to the naked eye (data not shown).
Figure 1.
Shoot phenotype of max4-1 mutant plants. (A) Wild-type (WT) and max4-1 plants were grown under a 16-h long-day photoperiod for 3 wk. Bar, 1 cm. (B) Lateral bud and inflorescence lengths of WT (closed circles) and max4-1 (open circles). Lateral lengths were measured when the primary shoot apex had ceased activity. Cauline nodes are numbered positively (5 youngest, 1 oldest) and rosette nodes are numbered negatively (-1 youngest, -20 oldest). Node number is aligned so that node 1 is the oldest cauline node. Error bars represent the standard errors of the mean; n = 10. (C) Mean branch numbers originating from the rosette of plants at maturity produced by hypocotyl grafting between WT and max4-1 plants (shoot genotype/root genotype). Error bars represent the standard errors of the mean; n = 4–11.
As in WT, all the cauline nodes of mature max4-1 plants produced buds that developed into elongating inflorescences with the same basipetal gradient (Fig. 1A). However, a greater number of rosette buds developed into inflorescences compared with WT, on average 8.5 ± 0.4 (Fig. 1B). An acropetal gradient of bud growth was also observed in most of the max4-1 plants (n = 8/10), but in contrast to WT, these buds grew out to form short inflorescences. Therefore, both the basipetal and acropetal patterns of bud activity are similar to WT in long-day-grown max4-1 mutants, but the mutant buds are more likely to grow out.
We found no evidence that, unlike the supershoot mutant (Reintanz et al. 2001; Tantikanjana et al. 2001), but similar to the Arabidopsis max1 and max2 mutants (Stirnberg et al. 2001) and the axr1 auxin-resistant mutant (Stirnberg et al. 1999), the max4 mutations affect the number of axillary meristems formed at each node. Rather, the defect appears to be specifically in bud outgrowth. Furthermore, like max1, max2, and axr1, max4 affects both the acropetal and basipetal gradients in a similar way.
MAX4 expression in the roots is sufficient for WT shoot branching
To determine the site of action of MAX4, we performed reciprocal hypocotyl grafting experiments (Turnbull et al. 2002). The self-grafted control plants reproduced branching phenotypes similar to intact controls, indicating that the grafting process does not affect branching (data not shown). Graft combinations with either a WT scion or a WT rootstock showed WT shoot branching patterns (Fig. 1C), indicating that, although the MAX4 gene can act in the shoot to inhibit branching, expression in the root is sufficient for WT shoot branching levels. These data suggest that the MAX4 gene is required for the production of a graft-transmissible inhibitor of shoot branching.
Auxin responses in the max4-1 mutant
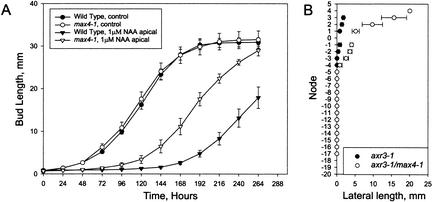
We assayed the response of max4-1 axillary buds to auxin using a split plate assay (Chatfield et al. 2000). Bud outgrowth from excised cauline nodes placed between divided agar sections in a Petri dish is inhibited by apically applied auxin. In this system, without hormone treatment, buds of WT and max4-1 grew out with similar kinetics, with elongation commencing 2 d after node excision (Fig. 2A). Apical auxin inhibited WT bud outgrowth for an average of 6 d, whereas max4-1 mutant buds were partially resistant to apical auxin, being inhibited for an average of only 4 d.
Figure 2.
Altered auxin responses in max4-1 mutants. (A) Lateral inflorescence outgrowth of excised max4-1 and WT nodes in response to the synthetic auxin 1-NAA. The oldest cauline node was excised from WT (closed symbols) or max4-1 (open symbols), and inserted between two agar blocks. The apical agar block contained either no 1-NAA (circles) or 1 μM 1-NAA (triangles). Bud outgrowth was measured every 24 h. Error bars represent standard error of the mean; n = 11 to 19. (B) Lateral bud and inflorescence lengths of axr3—1 (closed circles) and axr3-1, max4-1 double-mutant plants (open circles). Lateral lengths were measured 55 d after sowing, and the mean primary inflorescence height of the axr3-1 and the axr3-1, max4-1 double-mutant plants were not significantly different. Cauline nodes are numbered positively and rosette nodes are numbered negatively. Node number increases with proximity to the shoot apex. Node number is aligned so that node 1 is the oldest cauline node. Error bars represent standard error of the mean; n = 7.
To test further the role of auxin in the max4-1 phenotype, we constructed double mutants between max4-1 and axr3-1. The axr3-1 mutation is a semidominant gain of function mutation resulting in an overresponse to auxin and reduced shoot branching (Cline et al. 2001). The axr3-1, max4-1 double mutant was found to have increased lateral branch lengths in comparison with those of axr3-1 mutant plants (Fig. 2B). This suggests that some of the dominant branch suppressing effects of axr3-1 require MAX4, consistent with the idea that the graft-transmissible MAX4-dependent signal acts downstream of auxin to inhibit branching. In further support of this model, when double mutants were constructed between the loss-of-function auxin-resistant axr1-12 mutant and max4-1, branching levels were no higher than those of the single mutants (data not shown).
MAX4 is not generally required for auxin response because, other than in shoot branching assays, the max4-1 mutation had little or no effect on responses to exogenous auxin or axr3-1 phenotypes (data not shown).
Molecular characterization of the MAX4 gene
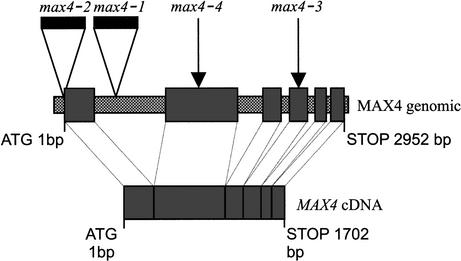
The max4-1 and max4-2 lines were found to contain single transposon insertions that cosegregated with the mutant phenotypes (data not shown). DNA flanking the transposon in max4-1 and max4-2 was isolated by inverse PCR and both amplified fragments were found to be identical in sequence to parts of the same predicted gene (At4g32810; Fig. 3). When At4g32810 was sequenced from the max4-3 and max4-4 alleles, it was found to contain a four-base insertion and a two-base deletion, respectively (Fig. 3). These changes are predicted to result in premature termination of the encoded protein and are consistent with transposon excision footprints (Cardon et al. 1993). The fact that four independent max4 alleles have mutations in this one gene provides strong evidence that this is the MAX4 gene.
Figure 3.
Diagram of MAX4 genomic and MAX4 cDNA showing the relative positions of the transposon-induced mutations. The MAX4 cDNA consists of six exons (larger gray bars) separated by five introns in the genomic DNA (smaller hatched bars). The max4-1 and max4-2 mutations are caused by transposon insertions (black bars) and the max4-3 and max4-4 mutations (arrows) are caused by transposon footprints.
We isolated the MAX4 cDNA by reverse transcription PCR (RT—PCR) of purified polyadenylated mRNA extracted from WT shoot tissue. The resulting cDNA contains a single open reading frame predicted to encode a protein of 570 amino acids (Fig. 3). The cDNA is identical to the coding region of an expressed sequence tag (EST) in the database.
The cDNA was introduced into max4-1 mutant plants under the control of the cauliflower mosaic virus 35S promoter. Seventeen independent T1 plants from these transformations had a WT phenotype, which was stably inherited in the T2 generation (data not shown), confirming that MAX4 is At4g32810. Interestingly, overexpression of MAX4 from the 35S promoter in WT plants had no obvious phenotypic effect (data not shown). This suggests that either MAX4 is not the rate-limiting step in the synthesis of the graft-transmissible substance, or the levels of the substance are regulated posttranscriptionally, or the substance cannot inhibit branching below WT levels.
MAX4 belongs to the polyene chain dioxygenase family
Database searches using the predicted protein sequence of MAX4 (Altschul et al. 1990) show that it is a member of the polyene chain dioxygenase superfamily, and is likely to be localized to plastids (Emanuelsson et al. 2000). A phylogenetic analysis of family members from plants, animals, and bacteria is shown in Figure 4A (constructed using ClustalX 1.8 program; Thompson et al. 1997). MAX4, RMS1 (see below), and a rice sequence (OsMAX4) form a well-supported clade. The abscisic acid (ABA) biosynthetic protein, VP14, falls within another strongly supported clade that includes Arabidopsis carotenoid cleaving dioxygenases (CCDs, also called NCEDs) probably involved in ABA biosynthesis (AtCCD2, AtCCD3, AtCCD5, AtCCD6, and AtCCD9; for review, see Seo and Koshiba 2002). The representatives of the animal RPE65, BETA DIOX1 and BETA DIOX2 proteins, form a third well-supported clade; and a fourth discrete group of proteins with similarity to bacterial lignostibene dioxygenases is represented here by two Sphingomonas paucimobilis proteins. The AtCCD1 protein, which is not involved in ABA biosynthesis but cleaves beta carotene (Schwartz et al. 2001), and AtCCD7 and AtMAX4 are grouped on long branches.
Figure 4.
(A) Phylogenetic analysis of polyene chain dioxygenases. The unrooted phylogenetic tree was generated from multiple sequence alignments, using ClustalX and modified by eye. Positions with gaps were excluded. Numbers at branch forks represent bootstrap values as a percentage of 10,000 bootstraps, and give a confidence limit for grouping together the sequences. Proteins are labeled with a prefix that represents the species origin of the sequence: At, Arabidopsis thaliana; Ps, Pisum sativum, pea; Os, Oryza sativa, rice; Sp, Sphingomonas paucimobilis; h, human; Mm, Mus musculus, mouse. (B) Southern blot analysis of the PsMAX4 gene in WT and isogenic rms1 pea lines. Lanes 2—5 contain EcoR1-digested genomic DNA from rms1-2, Weitor, rms1-3, and Raman, respectively. Lane 1 contains a 1-kb ladder.
These results suggest the attractive hypothesis that MAX4 encodes a carotenoid-cleaving dioxygenase involved in the synthesis of a mobile branch-inhibiting substance. This substance is very unlikely to be ABA because of the lack of ABA-related phenotypes in max4 mutants (data not shown) and the lack of requirement for ABA in auxin-mediated inhibition of bud outgrowth (Chatfield et al. 2000). Taken together, these data support the hypothesis that MAX4 is involved in the synthesis of a novel hormone that acts downstream of auxin to inhibit shoot branching.
The MAX4 gene is orthologous to the RMS1 gene of pea
The similar phenotypes conferred by max4 and the pea rms mutants prompted us to test whether any of the RMS genes were orthologous to MAX4. We isolated the pea ortholog of MAX4 using degenerate primers based on amino acid alignment between MAX4 and the deduced amino acid sequences from two Medicago truncatula ESTs showing high homology with MAX4 [60% (77/127) and 62% (60/96) identity]. A pea gene was isolated encoding a 561 amino acid protein showing 68% identity with MAX4 across the 518 amino acids at the C terminus. Comparison of the RT—PCR-amplified cDNA and the corresponding genomic DNA revealed five introns for PsMAX4 at the same position as in the Arabidopsis gene and of comparable sizes (740/918; 263/122; 88/110; 78/91; 70/89).
We mapped the PsMAX4 sequence using a recombinant inbred line mapping population (Laucou et al. 1998). PsMAX4 was found to map to the top of linkage group III at same position as RMS1 (Blixt 1976). In an F2 population of 95 individuals (M3T-884 × Torsdag) segregating for rms1, complete cosegregation was observed between PsMAX4 and RMS1. Southern analysis using PsMAX4 as a probe revealed a 12-kb band present for WT progenitors Weitor and Raman and absent for mutant alleles rms1-2 and rms1-3 (Fig. 4B). The map position of PsMAX4, its deletion in two independent rms1 alleles, and the similar phenotypes of max4 and rms1 mutants provide strong evidence that RMS1 and MAX4 are true orthologs.
Expression ofthe MAX4 and RMS1 genes
Transcripts for MAX4 and RMS1 are present at very low levels and are not readily detectable on Northern blots. However, they can be amplified by PCR from all tissues tested (data not shown). These widely distributed but very low expression levels for MAX4 transcripts are supported by Affymetrix gene chip data, publically available from the Nottingham Arabidopsis Stock Centre. To determine more precisely the location of MAX4 transcription, we constructed promoter—GUS fusions using 2.7 kb of DNA upstream of the MAX4 translational start site. This construct was introduced into WT Col plants, and GUS expression was analyzed using the chromogenic substrate X-Gluc. Strong expression was consistently observed in root tips (Fig. 5A). Other tissues such as hypocotyls and petioles occasionally showed very weak expression in some plants from each line (data not shown). Similarly, weak expression was variably observed in nodal sections associated with young axillary buds (Fig. 5C). No GUS staining was observed in the buds themselves, consistent with the remote site of action of MAX4 predicted by the grafting studies.
Figure 5.
Root tip expression of the GUS reporter protein driven by 2.7 kb of DNA upstream of the MAX4 gene. (A) Untreated 5-day-old seedlings. (B) Seedlings transferred to 1 μM 1-NAA for 24 h. (C) Nodal section showing expression close to a young bud, but not in the bud. (D, top panel) RMS1 gene expression in internode 4 of 14-day-old plants, with 5 leaves expanded, that were intact (I) or 6 h after decapitation (D), treated with 0, 500, or 3000 mg/L-1 IAA to the decapitated stump. (Bottom panel) Actin gene expression was monitored as a control.
Because MAX4 and RMS1 are predicted to act downstream of auxin, we tested whether the transcription of either gene is affected by auxin. We used RT—PCR to investigate the expression of the RMS1 gene in pea using the classical apical dominance test involving decapitation and replacement of the apex by exogenous auxin (Fig. 5D). Decapitation caused a substantial drop in RMS1 expression within 6 h after treatment. This reduction was not only prevented by replacement of the apex with 500 and 3000 mg/L exogenous IAA, but RMS1 expression was up-regulated compared with intact controls. In contrast, similar experiments in Arabidopsis using both RT—PCR and the promoter::GUS reporter lines failed to detect any up-regulation of MAX4 transcript levels in stem sections in response to apical auxin (data not shown). However, 24 h after transfer of promoter::GUS seedlings to auxin-containing media (1 μM NAA), up-regulation of GUS expression was observed in regions of the root distal to the apparently constitutive root tip expression pattern (Fig. 5B). No such up-regulation was detected following 6 h exposure (data not shown).
These data suggest that auxin may regulate shoot branching partly through transcriptional up-regulation of RMS1/MAX4. In pea, this up-regulation occurs at the node and may be sufficiently rapid to inhibit bud growth in response to apical auxin. In Arabidopsis, however, no up-regulation was detected at the node, despite the fact that this is a major site for AXR1 and auxin action in the regulation of bud growth (Booker et al. 2003). This, combined with the observation that MAX4 action in the root is sufficient for WT branching, suggests an additional role for auxin downstream of the synthesis of the RMS1/MAX4-dependent signal. For example, auxin may regulate the transport of the RMS1/MAX4-dependent signal into axillary buds. A full test for this hypothesis awaits the identification of the novel MAX4/RMS1 dependent, branch-inhibiting signal.
Materials and methods
Plant growth conditions
Arabidopsis plants were grown in 4-cm square compartments (P40, Cookson Plantpak) containing F2 compost treated with Intercept 70WG (both Levington Horticulture) in a growth room with 16 h light (white light at 70 μmole/m-2s-1). Pea seedlings were grown under glasshouse conditions extended to 18 h light (incandescent light providing ∼3 μmole/m-2 s-1 at pot top), as described by Morris et al. (2001).
Phylogenetic analysis
Sequences for phylogenetic analysis were retrieved from the NCBI database (see Supplemental Material). Sequences were aligned with ClustalX software (Thompson et al. 1997). The alignment was analyzed by eye, and regions with a low confidence of alignment were removed using Bioedit software (Hall 1999). The phylogenetic tree was generated using Neighbour Joining and a distance matrix with correction for multiple substitutions. Bootstrapping values were generated with n = 10,000. TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used to visualize the tree.
Gene cloning
The MAX4 gene was isolated using inverse PCR (iPCR) of max4-1 and max4-2 DNA (see Supplemental Material for details). The CaMV35S::MAX4 cDNA construct was made using standard techniques (Sambrook et al. 1989; Supplemental Material). The MAX4 promoter—GUS fusion construct was generated by PCR amplification of a 2.7-kb region upstream of the ATGincluding BamH1 and Xba1 restriction sites in the primers. These sites were used to clone the fragment into the pBI101.1 vector. This plasmid was transformed into Agrobacterium and then into plants using the floral dip method of Clough and Bent (1998). Two typical lines containing a single site of transgene insertion were taken to homozygosity and used for detailed analysis.
Isolation ofthe pea MAX4 homolog PsMAX4
Degenerate primers were designed in consensus regions between the Arabidopsis MAX4 protein and the deduced amino acid sequence of two Medicago truncatula genes showing high homology with MAX4 (see Supplemental Material). These primers were used to amplify PsMAX4 from both cDNA and genomic DNA. The amplification products were used to determine the sequence of the pea cDNA and the positions of introns in the gene (Brunel et al. 1999).
Mapping of PsMAX4
For mapping the PsMAX4 sequence, a cleaved amplified polymorphic sequence (CAPS) marker was designed to detect a DraI restriction site polymorphism between the two parents of our mapping population of 139 recombinant inbred lines (Laucou et al. 1998); Térèse and line K586 (isogenic to Torsdag; see Supplemental Material). This CAPS marker was also used for cosegregation analysis between PsMAX4 and the pea branching gene RMS1 in a population of 95 F2 individuals from a cross between the rms1-10 mutant line M3T-884, obtained from Térèse by EMS mutagenesis (Rameau et al. 1997; Symons and Murfet 1997), and Torsdag. Linkage analysis was carried out using the MAPMAKER/EXP 3.0 computer program (Lincoln et al. 1992). Southern blot analysis of the two radiation-induced rms alleles was carried out using standard techniques (see Supplemental Material).
Ps-MAX4 expression study in pea
Fourteen-day-old cv. Torsdag plants (five leaves expanded, counting acropetally from the cotyledonary node as zero) were left intact or were decapitated below the apex (internode 5) and IAA in lanolin was applied to the decapitated stump, as described by Beveridge et al. (2000). Six hours after treatment, internode 4 was collected. Total RNA was extracted using a modification of the hot-phenol method (Kreig 1996). RT—PCR and Southern blotting were used to determine the abundance of RMS1 transcripts in these samples. For details, see Supplemental Material.
Acknowledgments
We thank Yi Li and Sandie Baldauf for help with phylogenetic analysis, Petra Stirnberg for helpful discussions, and Stephen Day for critical reading of the manuscript. The AtNCED7 sequence was kindly provided by Don McCarty. This work was supported by the Biotechnology and Biological Sciences Research Council of the UK, BioGemma. UK, the European Commission, Union Nationale Interprofessionnelle des Plantes riches en Protéines, Génoplante, and the Australian Research Council. E.F. was funded by an Australian Postgraduate Award. We thank the horticultural technicians at York for expert plant care.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.256603.
Footnotes
Supplemental material is available at http://www.genesdev.org.
References
- Altschul S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bangerth F. 1994. Response of cytokinin concentration in the xylem exudate of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment, and relationship to apical dominance. Planta 194: 439–442. [Google Scholar]
- Beveridge C.A. 2000. Long-distance signaling and a mutational analysis of branching in pea. Plant Growth Regul. 32: 193–203. [Google Scholar]
- Beveridge C.A., Symons, G.M., Murfet, I.C., Ross, J.J., and Rameau, C. 1997. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signals. Plant Physiol. 115: 1251–1258. [Google Scholar]
- Beveridge C.A., Symons, G.M., and Turnbull, C.G.N. 2000. Auxin inhibition of decapitation-induced branching is dependent on graft-transmissible signals regulated by genes Rms1 and Rms2. Plant Physiol. 123: 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge C.A., Weller, J.L., Singer S.R., and Hofer, J.M.I. 2003. Axillary meristem development: Budding relationships between networks controlling flowering, branching and photoperiod responsiveness. Plant Physiol. 131: 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt S. 1976. Linkage studies in Pisum. XV. Establishing the Rms-gene and the linkage of Rms and Fas in chromosome 3. Agri. Hort. Genet. 34: 83–87. [Google Scholar]
- Booker J.P., Chatfield, S.P., and Leyser, O. 2003. Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell 15: 495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel D., Froger, N., and Pelletier, G. 1999. Development of amplified consensus genetic markers (ACGM) in Brassica napus from Arabidopsis thaliana sequences of known biological function. Genome 42: 387–402. [PubMed] [Google Scholar]
- Cardon G.H., Frey, M., Saedler, H., and Gierl, A. 1993. Mobility of the maize transposable element En/Spm in Arabidopsis thaliana. Plant J. 3: 773–784. [PubMed] [Google Scholar]
- Chatfield S.P., Stirnberg, P., Forde, B.G., and Leyser, H.M.O. 2000. The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 24: 159–169. [DOI] [PubMed] [Google Scholar]
- Cline M.G. 1991. Apical dominance. Bot. Rev. 57: 318–358. [Google Scholar]
- Cline M.G. 1996. Exogenous auxin effects on lateral bud outgrowth in decapitated shoots. Ann. Bot. 78: 255–266. [Google Scholar]
- Cline M.G., Chatfield, S.P., and Leyser, H.M.O. 2001. NAA restores apical dominance in the axr3-1 mutant of Arabidopsis thaliana. Ann. Bot. 87: 61–65. [Google Scholar]
- Clough S.J. and Bent, A.F. 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Eklöf S., Åstot, C., Sitbon, F., Moritz, T., Olsson, O., and Sandberg, G. 2000. Transgenic tobacco plants co-expressing Agrobacterium iaa and ipt genes have wild-type hormone levels but display both auxin and cytokinin-overproducing phenotypes. Plant J. 23: 279–284. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen, H., Brunak, S., and Heijne, G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Foo E., Turnbull, C., and Beveridge, C.A. 2001. Long distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol. 126: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41: 95–98. [Google Scholar]
- Kreig P.A. 1996. A lab guide to RNA isolation, analysis and synthesis. Wiley-Liss, Chichester, UK.
- Laucou V., Haurogné, K., Ellis, N., and Rameau, C. 1998. Genetic mapping in pea. 1. RAPD-based genetic linkage map of Pisum sativum. Theor. Appl. Genet. 97: 905–915. [Google Scholar]
- Lincoln S., Daly, M., and Lander, E.S. 1992. Constructing genetic maps with MAPMAKER/EXP 3.0. In Whitehead Institute technical Report, 3rd ed. Whitehouse Technical Institute, Cambridge.
- Morris D.A. 1977. Transport of exogenous auxin in two-branched dwarf pea seedlings (Pisum sativum L.). Planta 136: 91–96. [DOI] [PubMed] [Google Scholar]
- Morris S.E., Turnbull, C.G.N., Murfet, I.C., and Beveridge, C.A. 2001. Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol. 126: 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameau C., Bodelin, C., Cadier, D., Grandjean, O., Miard, F., and Murfet, I.C. 1997. New ramosus mutants at loci Rms1, Rms3 and Rms4 resulting from the mutation breeding program at Versailles. Pisum Genet. 29: 7–12. [Google Scholar]
- Reintanz B., Lehnen, M., Reichelt, M., Gershenzon, J., Kowalczyk, M., Sandberg, G., Godde, M., Uhle, R., and Palme, K. 2001. bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13: 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schwartz S.H., Qin, X., and Zeevaart, J.A.D. 2001. Characterization of a novel carotenoid cleavage dioxygenase from plants. J. Biol. Chem. 276: 25208–25211. [DOI] [PubMed] [Google Scholar]
- Seo M. and Koshiba, T. 2002. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7: 41–48. [DOI] [PubMed] [Google Scholar]
- Stirnberg P., Chatfield, S.P., and Leyser, H.M.O. 1999. AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 121: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., Sande, K., and Leyser, H.M.O. 2002. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Symons G.M. and Murfet, I.C. 1997. Inheritance and allelism tests on six further branching mutants in pea. Pisum Genet. 29: 1–6. [Google Scholar]
- Tantikanjana T., Yong, J.W.H., Letham, D.S., Griffith, M., Hussain, M., Ljung, K., Sandberg, G., and Sundaresan, V. 2001. Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes & Dev. 15: 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann K.V. and Skoog, F. 1933. Studies on the growth hormone of plants. III. The inhibiting action of the growth substance on bud development. Proc. Nat. Acad. Sci. 19: 714–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D. 1999. Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11: 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C.G.N., Booker, J.P., and Leyser, H.M.O. 2002. Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 32: 255–262. [DOI] [PubMed] [Google Scholar]
- Wisman E., Guillermo, C., Franz, P., and Saedler, H. 1998. The behavior of the autonomous maize element En/Spm in Arabidopsis thaliana allows efficient mutagenesis. Plant Mol. Biol. 37: 989–999. [DOI] [PubMed] [Google Scholar]