Abstract
Omi/HtrA2 is a mitochondrial serine protease that is released into the cytosol during apoptosis to antagonize inhibitors of apoptosis (IAPs) and contribute to caspase-independent cell death. Here, we demonstrate that Omi/HtrA2 directly cleaves various IAPs in vitro, and the cleavage efficiency is determined by its IAP-binding motif, AVPS. Cleavage of IAPs such as c-IAP1 substantially reduces its ability to inhibit and ubiquitylate caspases. In contrast to the stoichiometric anti-IAP activity by Smac/DIABLO, Omi/HtrA2 cleavage of c-IAP1 is catalytic and irreversible, thereby more efficiently inactivating IAPs and promoting caspase activity. Elimination of endogenous Omi by RNA interference abolishes c-IAP1 cleavage and desensitizes cells to apoptosis induced by TRAIL. In addition, overexpression of cleavage-site mutant c-IAP1 makes cells more resistant to TRAIL-induced caspase activation. This IAP cleavage by Omi is independent of caspase. Taken together, these results indicate that unlike Smac/DIABLO, Omi/HtrA2's catalytic cleavage of IAPs is a key mechanism for it to irreversibly inactivate IAPs and promote apoptosis.
Keywords: Apoptosis, mitochondria, Omi/HtrA2, Smac, IAPs, caspases
Apoptosis is mainly executed by proteolytic activation of caspases. Activated caspases catalytically degrade important intracellular molecules and execute cell death (Cryns and Yuan 1998; Thornberry and Lazebnik 1998). The initial proteolytic cleavage of caspases may occur through the extrinsic cell-surface pathway by the activation of the tumor necrosis family of receptors, and/or from the intrinsic route via the release of a group of apoptotic proteins from the mitochondria to the cytoplasm (Nagata 1997; Wang 2001). Inhibitor of apoptosis (IAP) proteins directly bind and inhibit active caspases through their baculovirus IAP repeat (BIR) domains (Deveraux and Reed 1999; Miller 1999; Verhagen et al. 2001; Salvesen and Duckett 2002). XIAP is the first mammalian IAP protein characterized with caspase inhibitory activity (Deveraux et al. 1997). The BIR3 domain and BIR2 along with its N-terminal linker region of XIAP bind and inhibit caspase-9 and caspase-3, respectively (Takahashi et al. 1998; Deveraux et al. 1999; Sun et al. 1999, 2000). Among the eight IAP proteins in mammals, c-IAP1, c-IAP2, XIAP, Livin α, Livin β, and Ts-IAP also contain a RING zinc-binding motif. Some of them function as a ubiquitin ligase to modify proteins posttranslationally through ubiquitylation (Huang et al. 2000; Yang et al. 2000; Pickart 2001).
Smac/DIABLO and Omi/HtrA2 are two IAP antagonists identified so far in mammals (Du et al. 2000; Verhagen et al. 2000, 2002; Suzuki et al. 2001; Hegde et al. 2002; Martins et al. 2002; van Loo et al. 2002). They both are nuclear-encoded mitochondrial proteins. The cleavage of their mitochondrial-targeting sequences inside mitochondria generates processed active Smac and Omi with new apoptogenic N termini, named the IAP-binding motif. This motif consists of a short stretch of hydrophobic amino acids, AVPI or AVPS, in Smac and Omi, respectively. After its release into the cytosol stimulated by apoptotic triggers, Smac and Omi competitively bind to the BIR domains of IAPs via the IAP-binding motif, so that the BIR-bound caspases are released and reactivated (Liu et al. 2000; Wu et al. 2000; Srinivasula et al. 2001). Although Smac and Omi are both IAP antagonists, they manifest distinct physical characteristics and biochemical activities. The active Smac is a homodimer, whereas Omi is a homotrimer (Chai et al. 2000; Li et al. 2002). Omi is a mitochondrial serine protease, whereas Smac is not (Faccio et al. 2000; Gray et al. 2000). The serine protease domain of Omi is located in the central region of the molecule, and the PDZ domain that regulates its protease activity is located in the C-terminal region. The serine protease activity of Omi is reported to be indispensable for its pro-apoptotic function (Li et al. 2002). However, it is not known whether the serine protease activity of Omi is only important for caspase-independent cell death, or whether it also plays a specific role in antagonizing IAPs. Screening for Omi proteolytic substrates from 40 proteins has revealed that only 4 proteins—α-casein, β-casein, hyaluronidase, and glycoprotein α1 acid—are cleavable by Omi (Gray et al. 2000). None of the four Omi substrates, however, has been reported to be involved in apoptosis.
A recent report by Dr. Arnold Levine's group (Jin et al. 2003) has identified Omi as a p53-targeted gene. p53-mediated Omi up-regulation correlates with c-IAP1 cleavage in etoposide-induced cell death. This c-IAP1 cleavage, as well as the respective apoptotic cell death, can be inhibited by a serine protease inhibitor, indicating that Omi may cleave c-IAP1 (Jin et al. 2003). These results suggest a close correlation of Omi up-regulation with c-IAP1 cleavage. In this study, we independently identified IAPs as a new set of protease substrates of Omi/HtrA2 and characterized the biochemical mechanism for the consequences of c-IAP1 cleavage in cell death.
Results
Identification of new substrates for the serine protease Omi/HtrA2
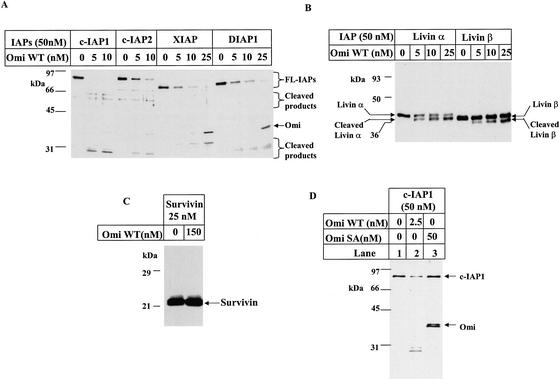
On the basis of the fact that Omi can bind the IAP family of proteins through its N-terminal IAP-binding motif, and enzyme binding to substrate is a prerequisite for the enzymatic reaction to take place, we investigated whether IAP proteins could serve as enzymatic substrates of Omi. We incubated different IAP proteins with wild-type Omi (Omi WT) in vitro and resolved the reaction mixtures by SDS-PAGE, followed by silver staining or Western blotting. As shown in Figure 1, Omi WT could cleave various IAP proteins, including cIAP1, cIAP2, XIAP, DIAP1 (Fig. 1A), Livin α, and Livin β (Fig. 1B). This cleavage activity, however, is absent for Survivin (Fig. 1C). Omi showed the highest cleavage efficiency on c-IAP1 among all of the IAPs tested; we therefore focused on c-IAP1 in the following experiments. In contrast to wild-type Omi, the protease dead mutant Omi SA (Faccio et al. 2000; Gray et al. 2000) completely lost its hydrolytic activity on c-IAP1 (Fig, 1D) and on other IAPs (data not shown). Taken together, c-IAP1, c-IAP2, XIAP, Livin α, Livin β and DIAP1 form a new group of proteolytic substrates of Omi in vitro.
Figure 1.
Omi cleaves IAP proteins. Each IAP protein was incubated with Omi WT or Omi SA for 2 h at 30°C in 40 μL of PBST as described in Materials and Methods. The reaction products were subjected to SDS-PAGE, followed by silver staining for c-IAP1, c-IAP2, XIAP, and DIAP1 (A), or immonostaining for Livin α or Livin β with an anti-Livin antibody (B). (C) Survivin was incubated with excessive amounts of Omi WT, and the result was immunoblotting using an anti-Survivin antibody. (D) c-IAP1 was incubated with WT (lane 2) or Omi SA (lane 3), and the cleavage was detected by silver staining.
Omi/HtrA2 cleavage of c-IAP1 is dependent on its IAP-binding motif and regulated by the PDZ domain
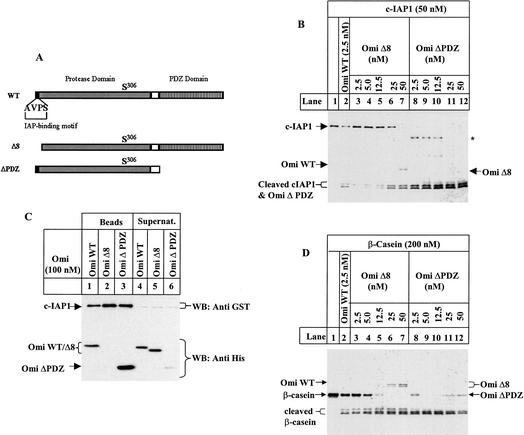
We examined whether Omi binding to IAPs was required for its IAP cleavage by use of an IAP-binding-deficient mutant Omi missing the N-terminal eight amino acid residues, Omi Δ8 (Li et al. 2002). Omi WT at 2.5 nM cleaved 50 nM c-IAP1 (molar ratio at 1:20) to near completion (Fig. 2B, lane 2). In contrast, Omi Δ8 protein at 2.5 nM barely cleaved any c-IAP1 (Fig. 2B, lane 3). The same cleavage activity did not occur unless a 10-fold higher concentration of Omi Δ8 (25 nM) was used (molar ratio at 1:2; Fig. 2B, lane 6). Thus, the proteolytic efficiency of this IAP-binding-deficient Omi was ∼10-fold less than that of wild-type Omi regarding c-IAP1 cleavage. This implied that the difference in IAP cleavage efficiency between Omi WT and Omi Δ8 might be due to the difference in their IAP-binding affinity. Consistently, a GST-based pull-down assay showed that Omi WT bound to c-IAP1, whereas Omi Δ8 did not (Fig. 2C, lanes 1,2, respectively). In contrast to c-IAP1, Omi WT and Omi Δ8 equally efficiently cleaved β-casein (Fig. 2D, lanes 2,3), indicating that deleting the first eight amino acids does not diminish Omi protease activity against substrates in general, and the AVPS-mediated binding determines specific Omi cleavage of IAP. The PDZ domain attenuates Omi protease activity on β-casein, probably through restriction of substrate accessibility to the protease active site (Li et al. 2002). Omi ΔPDZ also showed a better cleavage activity on c-IAP1 (Fig. 2B), and this was at least in part due to its higher binding efficiency to c-IAP1 (Fig. 2C, lane 3).
Figure 2.
Omi/HtrA2 cleavage of c-IAP1 is dependent on its AVPS IAP-binding motif. (A) Schematic representation of wild-type and mutant Omi. WT, wild-type Omi; Δ8, Omi with the N-terminal eight residues deleted; ΔPDZ, Omi lacking the PDZ domain. (B,D) c-IAP (B) or β-casein (D) was incubated with wild-type Omi (lane 2), Omi Δ8 mutant (lanes 3-7) or Omi ΔPDZ mutant (lanes 8-12) in PBST for 2 h at 30°C. The c-IAP1 cleavage was revealed by sliver staining. The asterisk in B indicates a cleavage product produced exclusively by Omi ΔPDZ proteolysis of c-IAP1. (C) GST-fused c-IAP1 (50 nM) was preincubated with 100 nM of wild-type and mutant Omi for 20 min at 4°C in 50 μL of PBST, and then incubated with 20 μL of Glutathione Sepharose beads for 30 min at 4°C. The beads were precipitated by centrifugation and washed with 1.4 mL × 3 of PBST. The proteins bound to the beads and left in the supernatant were mixed separately with SDS sample loading buffer and resolved by SDS-PAGE. The upper part of the filter was immunoblotted for GST and the lower part for Penta-His.
The cleavage of c-IAP1 by Omi/HtrA2 catalytically potentiates caspase activity
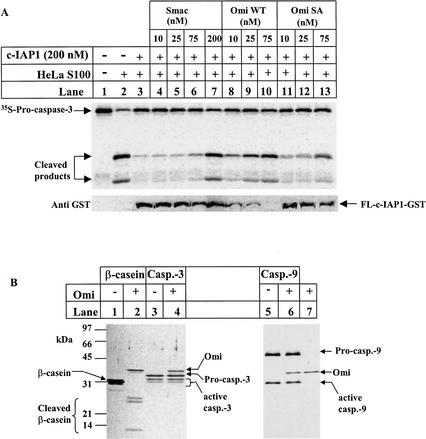
Addition of dATP and cytochrome c to HeLa cell extracts triggers the activation of endogenous caspase-9 (Liu et al. 1996), which can be measured by the cleavage of 35S-labeled procaspase-3 (Fig. 3A, lane 2). This caspase activity was completely inhibited by 200 nM of c-IAP1 (Fig. 3A, lane 3), and this IAP inhibition was relieved by 200 nM of Smac (Fig. 3A, lane 7). In contrast to Smac, this c-IAP1 inhibition was reduced by Omi at 10 nM and relieved at 75 nM, whereas the protease dead mutant Omi just started to reduce the inhibition at 75 nM (Fig. 3A, lanes 8-13). The cleavage of c-IAP1 was further confirmed by Western blotting (Fig. 3A, bottom). Omi did not cleave either the proform or the active form of caspase-9 and caspase-3, as shown by silver staining (Fig. 3B) and fluorogenic caspase substrate assay (data not shown). Thus, the caspase activity was due to Omi cleavage of c-IAP1. Therefore, Smac stoichiometrically antagonizes c-IAP1 through direct binding of its N terminus to IAPs. The binding-directed Omi cleavage of IAPs, on the other hand, is catalytic and irreversible, thereby more efficiently inactivating IAPs.
Figure 3.
c-IAP1 Cleavage by Omi/HtrA2 reduces its caspase inhibitory activity. (A) The c-IAP1 protein (400 nM) was first incubated with Smac, Omi WT, or Omi SA for 2 h at 37°C in 10 μL of Buffer A. These samples were subsequently tested for their caspase reactivation activity by incubating with an equal volume of Hela S100 extracts supplemented with 1 mM MgCl2, 1 mM dATP, 24 ng/μL cytochrome c, 1 mM DTT, and a proper amount of 35S-labeled procaspase-3 for 40 min at 30°C. The reactions were subjected to 13.5% SDS-PAGE, and transferred to the nitrocellulose filter. The filter was first exposed to a phosphor screen for 1 h at room temperature (top panel), and subsequently probed with anti-GST antibody to check c-IAP1 cleavage (bottom panel). (B) Omi does not cleave caspase-3 and caspase-9. Approximately 250 ng of recombinant caspase-3 or caspase-9 was incubated with 50 ng of Omi at 37°C for 2 h in a final volume of 40 μL PBST. Omi cleavage of β-casein was included as a positive control. The reaction mixtures were resolved on a 7.5%-20% gradient gel and visualized by silver staining. All of the samples were run on the same gel. The splitting of the gel into two parts was for the convenience of sample labeling.
The cleavage of c-IAP1 by Omi/HtrA2 reduces its ubiquitin ligase activity on caspase substrates
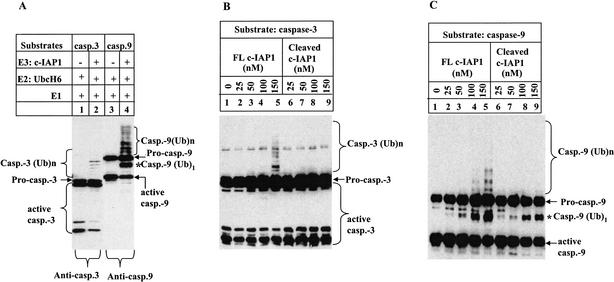
Incubation of caspase-3 and caspase-9, but not their proform, in a reconstituted ubiquitylation system, led to caspase polyubiquitylation in a c-IAP1-dependent manner (Fig. 4A). This result is consistent with the observation that only processed caspases are able to bind to the BIR domains of XIAP (Srinivasula et al. 2001). In this assay system, Omi-cleaved c-IAP1 at 150 nM failed to produce polyubiquitylated caspase-3, whereas the full-length c-IAP1 at the same concentration produced reasonable amounts of polyubiquitylated products (Fig. 4B). The same result was obtained on caspase-9 polyubiquitylation (Fig. 4C). Therefore, c-IAP1 cleavage resulted in a dramatic reduction of its ubiquitin ligase activity on caspase-3 and caspase-9.
Figure 4.
c-IAP1 cleavage by Omi/HtrA2 attenuates its ubiquitin ligase activity on caspase substrates. (A) Establishment of an in vitro assay for c-IAP1 ubiquitin ligase activity. Caspase-3 or caspase-9 at 400 nM was incubated with 200 nM c-IAP1 for 2 h at 30°C in a ubiquitylation system containing 40 mM Tris-HCl (pH 7.5), 50 mM NaCl, 100 nM ubiquitin activating enzyme (E1), 400 nM Ubc H6 (E2), 20 μM ubiquitin, and 2 mM Mg-ATP. The reaction products were subjected to SDS-PAGE and Western blotting for caspase-3 or caspase-9. The asterisk indicates the mono-ubiquitylated (Ub)1-active caspase-9. The poly-ubiquitylated caspase-3 and caspase-9 are denoted by (Ub)n. (B,C) Assay for the ubiquitin ligase activity of c-IAP1 before and after cleavage. c-IAP1 (1500 nM) was first incubated with PBST buffer (FL c-IAP1) or Omi (375 nM; Cleaved c-IAP1) at 37°C for 2 h. The resulting incubation mixture was then assayed for its ubiquitylation activity. A different amount of this incubated c-IAP1 sample was incubated with caspase-3 (400 nM) or caspase-9 (400 nM) using the assay system described in A. The final concentration of c-IAP1 sample in the ubiquitylation assay was 25 to ∼150 nM as indicated. The ubiquitylation was checked by immunoblotting for caspase-3 (B) and caspase-9 (C).
Mapping Omi/HtrA2 cleavage sites on c-IAP1
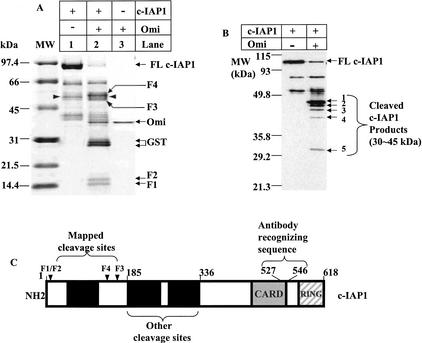
The c-IAP1 fragments generated by Omi proteolysis were subjected to N-terminal sequencing by Edman degradation (Fig. 5, F1-F4). Three preferred cleavage sites were identified after the residues Thr 4, Asn 133, and Leu 161 of c-IAP1 (Fig, 5C, arrowheads), which corresponded to the N-terminal sequencing of F1 and F2 and F3 and F4, respectively. The cleavage sites did not show obvious conservation at the level of amino acid sequences. Although the C-terminal sequence of each fragment is unknown, taking into consideration the N-terminal sequence of each fragment, the estimated size of each fragment, and a total of three cleavage sites, it is likely that F1 is composed of residues 5-133, F2 of residues 5-161, F3 of residues 162-618, and F4 of residues 134-618.
Figure 5.
Mapping Omi/HtrA2 cleavage sites on c-IAP1. (A) Approximately 5 μg of GST-fused c-IAP1 was incubated with 0.4 μg of Omi for 2 h at 30°C. The samples were resolved on a 7.5%-20% linear gradient gel and transferred to a PVDF membrane, followed by Coomassie Brilliant Blue R250 staining. Ten picomoles of each cleaved polypeptide fragments was subjected to N-terminal sequencing by Edman Degradation. Both polypeptide fragments F1 and F2 start with the amino acid sequence ASQRLFPG; F4 starts with SFAHSLSP; and F3 starts with NSRAVEDI. The three cleavage sites are shown in C by arrowheads. The two polypeptides 30 kDin size (lane 2) are GST as determined by N-terminal sequencing. The peptides labeled with arrowheads (lanes 1,2) were c-IAP1 proteolytic products (amino acid sequencing; data not shown) that are likely generated by bacterial proteases. (B) The Omi-cleaved c-IAP1 sample was immunoblotted with an antibody from R&D Systems recognizing the amino acid residue 527-546 of human c-IAP1 as shown in C. The polypeptide fragments F1-F4 in A were no longer detectable by this antibody because of the lack of antibody-recognizing sequences. At least five additional cleavage fragments (1-5) ranging in size from 30 to ∼45 kD were identified on this immunoblot. (C) A diagram of human c-IAP1 labeled with the three mapped and unmapped sites.
The second and third cleavage sites are located in the linker region N-terminal to the BIR2 domain of c-IAP1. Because the binding of the corresponding linker of XIAP-BIR2 is required for its inhibition on active caspase-3, cleavage within this BIR2 linker region of c-IAP1 may have weakened the binding of c-IAP1 to caspases and, consequently, damaged the caspase inhibition of c-IAP1. Although the ubiquitin ligase activity of c-IAP1 relies on its RING domain, the reduced ubiquitin ligase activity suggests that the binding between c-IAP1 and active caspase-3 and caspase-9 might be indispensable for caspase ubiquitylation.
It should be noted that the cleavage on c-IAP1 was not limited to the three mapped sites. Other cleaved fragments that were barely detectable by Coomassie blue staining in Figure 5A were detected by immunoblotting using an antibody against c-IAP1 (Fig. 5B, fragments 1-5). As this antibody could only recognize a portion of the CARD domain (Fig. 5C), and the size of these fragments ranged from 30 to ∼45 kD, the N termini of these fragments could therefore be right within the BIR2 and BIR3 domain, suggesting that extensive cleavage of c-IAP1 by Omi could have further reduced the caspase inhibitory function and ubiquitin ligase activity of c-IAP1. As the amount of Omi is up-regulated by p53 during apoptosis, as demonstrated by Levine's group (Jin et al. 2003), it is likely that cleavage at these sites will become stronger, which results in more potent inhibition of c-IAP1 function.
Mutant c-IAP1 is more resistant to Omi cleavage and better protects cells from apoptosis
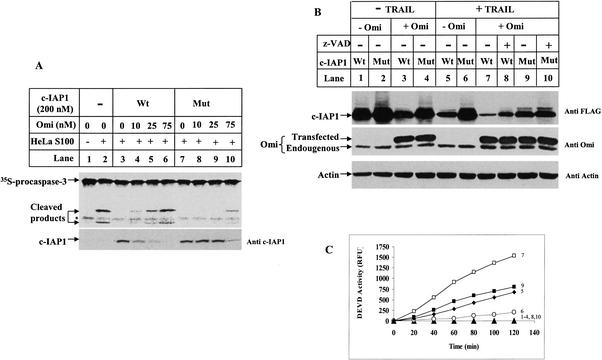
We prepared recombinant c-IAP1 protein mutated at the three preferred cleavage sites and tested its activity in vitro. This mutant c-IAP1 protein still maintained anti-caspase activity (Fig. 6A, top, lane 7); this caspase inhibitory activity, however, could not be relieved by low concentrations of Omi (Fig. 6A, top, lanes 8,9) due to its resistance to Omi cleavage (Fig. 6A, bottom, lanes 8,9). In contrast, Omi at the same concentration already cleaved wild-type c-IAP1 and reactivated caspases (Fig. 6A, lanes 4,5). Omi at 75 or 10 nM resulted in similar amount IAP cleavage and caspase reactivation for mutant and wild-type c-IAP1, respectively (Fig. 6A, lanes 10,4), indicating that mutant c-IAP1 is ∼7.5-fold more resistant to Omi protease.
Figure 6.
Cleavage-site mutant c-IAP1 makes cells more resistant to apoptosis. (A) The wild-type (Wt) or mutant (Mut) c-IAP1 proteins at 400 nM were preincubated with Omi for 2 h at 37°C in 10 μL of Buffer A and assayed for their caspase inhibitory activity in HeLa S100 extracts supplemented with dATP and cytochrome c. The caspase-3 cleavage activity was detected on a PhosphorImager (top panel), and cleavage of c-IAP1 was detected on the same filter by an anti-GST antibody (bottom panel). (B) A total of 750 ng of wild-type (Wt) or mutant (Mut) c-IAP1 in p3XFlag-CMV-7 construct was transfected into HeLa cells with or without cotransfection of 100 ng MAVPS Omi-Myc in pcDNA3.1 construct for 3 h using Lipofectamine Plus Reagent. The transfected cells were left untreated (lanes 1-4) or treated with 100 ng/mL TRAIL for 4 h (lanes 5-10). A total of 100 μM z-VAD-fmk was added to the culture medium 2 h before TRAIL treatment (lanes 8,10). Total cell extracts were made, and 20 μg of protein per sample was analyzed by SDS-PAGE and Western blotting. c-IAP1 was detected with an anti-Flag antibody (top panel). Omi was detected with a polyclonal antibody (middle panel) so that both the endogenous (lower band) and exogenously expressed (upper band) Myc-tagged Omi were detected. Immunoblotting for Actin was to show equal sample loadings (bottom panel). The three immunoblotting results were obtained from the same filter. (C) The DEVD activity for the samples in B. The number next to each curve represents the DEVD activity for the same numbered samples in B. The curve that lies on the X-axis (▴) is the DEVD activity for samples in lanes 1-4, 8, and 10 in B.
We next tested whether this mutant c-IAP1 could better block caspase activity in HeLa cells by transfection assays. Overexpression of the cytosolic form of Omi, MAVPS Omi, mimics the already released Omi upon certain apoptotic treatment, and thus, could distinguish the proapoptotic effect of Omi from other apoptotic factors released from mitochondria. The first methionine was likely removed after it was expressed in transfected HeLa cells, as it could bind to c-IAP1 in a pull-down assay (data not shown). Overexpression of MAVPS Omi led to wild-type c-IAP1 cleavage independent of apoptotic stimulus (Fig. 6B, top, cf. lanes 3 and 1), whereas, mutant c-IAP1 was more resistant (Fig. 6B, top, cf. lanes 4 and 2). Under TRAIL treatment, and in the absence of exogenously transfected Omi, the mutant c-IAP1 was more resistant than the wild type to the relatively limited amount of endogenous Omi (Fig. 6B, cf. lanes 5 and 6). This cleavage resistance correlated with a threefold reduced DEVD activity in TRAIL-induced apoptotic cells (Fig. 6C, curves 5,6). When more Omi was present by transfection, mutant c-IAP1 was still more resistant to Omi (Fig. 6B, top, cf. lanes 7 and 9), and this also correlated with a better caspase inhibition (Fig. 6C, cf. curves 7 and 9). In addition, the majority of c-IAP1 cleavage was not by caspases, because it could not be inhibited by the pan caspase inhibitor z-VAD-fmk (Fig. 6B, top, lanes 8,10), which was consistent with the results that Omi did not degrade caspases in vitro (Fig. 3). The mutant c-IAP1 was not absolutely resistant to Omi, because cleavage could also occur at other sites, as demonstrated in Figure 5. Nonetheless, this mutant c-IAP1 already manifested resistance to Omi cleavage and better-inhibited caspases, demonstrating that IAP cleavage by Omi plays an important role in apoptotic progression.
Elimination of endogenous Omi reduces c-IAP1 cleavage and desensitizes cells to apoptosis
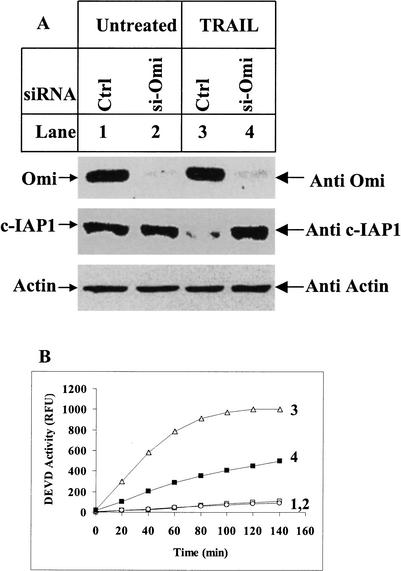
Transfection with small interfering RNA (siRNA) molecules against Omi (si-Omi) effectively eliminated Omi protein expression (Fig. 7A, top), and the corresponding c-IAP1 cleavage under TRAIL treatment was reduced (Fig. 7A, middle, lane 4). The same results were obtained for c-IAP2 and XIAP (data not shown). This reduced IAP cleavage correlated with a two- to approximately threefold lower caspase activity in TRAIL-treated cells (Fig. 7B, curves 3,4). Taken together, Omi cleavage of IAPs happens directly in apoptotic cells and represents an important step in Omi-mediated apoptotic progression.
Figure 7.
Elimination of endogenous Omi makes cells more resistant to apoptosis. (A) siRNA oligonucleotides against Omi (si-Omi) was transfected twice into HeLa cells with Luciferase GL2 siRNA duplex as control (Ctrl). All of the siRNA-transfected samples were treated with 2 μM proteasome inhibitor MG132 before being subjected to 250 ng/mL TRAIL treatment for 4 h. MG132 was used to block proteasome-mediated c-IAP1 degradation. Ten micrograms of protein per sample were subjected to immunoblotting for endogenous Omi and c-IAP1. Immunoblotting for Actin was to show equal sample loadings. The three immunoblotting results were obtained from the same filter. (B) DEVD activity assay for the samples in A. The number next to each curve represents the DEVD activity for the same numbered samples in A. The addition of MG-132 did not induce caspase activity as shown in curve 1.
Discussion
In this study, we demonstrate a novel mechanism for the proapoptotic protease Omi/HtrA2 to sensitize cell death as through catalytic cleavage and irreversible inactivation of the IAPs. Being the best substrate among all the IAPs tested, c-IAP1 cleavage produces stronger caspase activity and sensitizes cells to apoptosis through undermining its caspase inhibitory activity and caspase degradation-promoting function. This IAP cleavage is not by caspases, but rather an independent upstream step from caspase activation. The recent report by Dr. Levine's group also supports our findings (Jin et al. 2003). They have shown that c-IAP1 is cleaved in etoposide-induced cell death, and a serine protease inhibitor completely blocks this death. They further demonstrate that Omi/HtrA2 is up-regulated by p53, and could therefore be the protease to cleave c-IAP1. Our results have demonstrated the direct evidence for Omi cleavage on c-IAP1 in apoptotic cells and in vitro. In addition, we have elucidated the biochemical mechanism for this cleavage consequence on modulating caspase activity. A compound inhibitor has been reported recently to inhibit Omi-induced cell death (Cilenti et al. 2003). It could support our findings if this inhibitor functions through preventing IAP cleavage.
Omi cleavage of IAPs is not a random event, but rather is involved in certain IAP-regulated apoptotic processes. Only those BIR domain-containing proteins that are potent caspase inhibitors (c-IAP1, c-IAP2, XIAP, Livin α, Livin β, and DIAP1) are enzymatic substrates of Omi. Being a BIR domain-containing protein that mainly functions in cytokinesis (Li et al. 1998), Survivin fails to be cleaved by Omi. Moreover, this AVPS motif-mediated binding to IAPs is essential for Omi to cleave IAPs, but it is not required for Omi to cleave β-casein, suggesting that this motif-directed enzyme-substrate recognition and binding specifically regulates IAP proteolysis. This regulatory mechanism is likely to play a critical role in vivo in determining Omi's cleavage specificity for the substrates directly involved in apoptosis. Our results have also demonstrated a fundamental mechanistic difference between Omi and Smac in counteracting IAPs. Smac antagonizes IAPs exclusively by stoichiometric binding. Omi also binds to IAPs; this binding-directed catalytic proteolysis of IAPs is the key mechanism for Omi to irreversibly reduce IAP inhibition and efficiently promote cell death.
In contrast to the IAP cleavage products observed in our in vitro assay, we failed to detect c-IAP1 cleavage products in live cells. The N-terminal Asparagine residue in the large cleavage fragment of c-IAP1 (see Fig. 5 legend) is a destabilizing residue known to cause rapid polypeptide degradation by proteasome through the N-end rule pathway (Varshavsky 1996). Drosophila DIAP1 has recently been reported to be degraded in this manner after caspase cleavage (Ditzel et al. 2003). We therefore suspect that this c-IAP1 fragment bearing the N-terminal Asparagine generated by Omi cleavage may also be subject to this specific degradation, and this could be the reason why we cannot observe the cleaved c-IAP1 products. This possibility is currently under investigation.
It is necessary to pinpoint the physiological roles of Omi. Recent reports suggest that Omi is regulated by translation under conditions of heat shock or ER stress (Gray et al. 2000). The enzymatic activity of Omi is substantially enhanced in kidney ischemia/reperfusion in mice (Faccio et al. 2000). It would be interesting to investigate whether Omi indeed cleaves IAPs and whether caspase activity is, in fact, elevated under such stress conditions. If so, this would provide insight into understanding the role of apoptosis in the pathology of such stress conditions. Some answers will wait for the gene-targeted knockout studies of Omi in mice. It is of importance to examine whether Omi knockout mice manifest certain developmental defects as the result of reduced IAP cleavage. Regardless of the precise mechanism of this IAP cleavage by Omi in vivo, discrimination in different upstream signals may allow the cells to take a different route to inactivate IAPs. This study focuses on Omi cleavage of c-IAP1; the mechanism is likely to be of quite general significance, given the conserved functional composition among IAP molecules. Future work will be done to distinguish the pathways utilized by Omi and Smac in response to various upstream signals.
Materials and methods
Antibodies
Monoclonal anti-c-IAP1 antibody was purchased from Pharmingen; polyclonal antibody against the amino acid residues 527-546 of human c-IAP1 from R&D Systems; polyclonal anti-caspase-3 and monoclonal anti-Survivin and caspase-9 from R&D Systems; HRP conjugated anti-GST antibody, anti-c-Myc, and anti-Flag M2 antibodies from Sigma; HRP conjugated anti Penta-His antibody from QIAGEN; monoclonal anti-Livin antibody from IMGENEX; monoclonal anti-Actin from Santa Cruz Biotechnology. Polyclonal antisera against Omi and Smac were obtained from rabbits immunized with recombinant Omi and Smac proteins by Rockland Immunochemicals, Inc.
Generation of cDNA constructs
The cDNA for the mature form of Omi was PCR amplified and subcloned into the pET21b vector to generate C-terminal hexa-His-tagged construct. The point mutation and various deletion mutations of Omi were generated by PCR and subcloned similarly into pET21b. The cDNAs for human Livin α and Livin β were subcloned into the pQE30 vector to generate N-terminal hexa-His-tagged constructs. Human c-IAP1, c-IAP2, XIAP, and Drosophila DIAP1 cDNAs were subcloned into pGEX-4T-2 to generate GST fusion proteins. The p3XFlag-CMV-7 vector was used to express N-terminal 3XFlag tagged c-IAP1 in mammalian cells. The pcDNA 3.1(-) vector was used to express C-terminal c-Myc (EQKLISEEDL)-tagged mature form or Ser 306 → Ala mutant form Omi (starting from MAVPS).
Protein expression and purification
Hexa-His-tagged Omi and Survivin were expressed in Escherichia coli strain BL21 (DE3), and Livin α and Livin β were expressed in JM 109 and purified with Ni-NTA Sepharose affinity chromatography. The GST-fused c-IAP1, c-IAP2, XIAP, and DIAP1 were expressed in E. coli strain BL21 (DE3) and purified with Glutathione Sepharose affinity chromatography, followed by Superdex 200 gel-filtration chromatography. The protein concentrations were determined by the modified Bradford method (Zor and Selinger 1996).
Omi/HtrA2 serine protease activity assay
Proteins were incubated with Omi in PBST containing 20 mM Pi (pH 7.4), 100 mM NaCl, 0.5 mM EDTA, 0.05% Tween 20, and 1 mM DTT or in Buffer A containing 20 mM HEPES (pH 7.4), 10 mM KCl, and 1.5 mM MgCl2 for 2 h at 30°C or 37°C. The reaction mixture was subjected to SDS-PAGE and analyzed by Western blotting or Silver or Coomassie blue staining.
Assay for DEVD activity
The assay contained 30 μg of cell extracts and 40 μM of fluorogenic substrate Ac-DEVD-AMC. The cleavage of Ac-DEVD-AMC was measured kinetically in RFU by excitation at 380 nm and emission at 460 nm.
Multisite mutagenesis of c-IAP1
PCR mutagenesis was used to substitute the amino acids flanking the three Omi cleavage sites on c-IAP1. The mutated c-IAP1 had the K3TAS6 replaced with QMGA, R132NSF135 with QMGH, and P160LNA163 with EHMA. The DNA with expected multipoint mutations was inserted into the plasmid pGEX-4T-2 through EcoRI/NotI sites, or into the p3XFlag-CMV-7 through HindIII/SalI sites. The mutant constructs were confirmed by DNA sequencing.
RNA interference
Plated in 6-well plates at 1.0 × 105 cells/well for 1 d, HeLa cells were transfected twice in a 24-h interval with double-stranded siRNA (1000 ng/well) using GeneSilencer reagent (Gene Therapy Systems) according to the manufacturer's instructions. After the second transfection, the cells were treated with 2 μM MG132 for 1 h before being exposed to TRAIL treatment, and processed for SDS-PAGE and Western blotting. Oligonuleotides were from Dharmacon and had the following sequences: si-Omi sense, 5′-GGGGAGUUUGUUGUUGCCAdTdT-3′ and si-Omi antisense, 5′-UGGCAACAACAAACUCCCCdTdT-3′. Luciferase GL2 siRNA duplex was used as control.
Acknowledgments
We thank Donna di Natale for valuable editorial help in this manuscript preparation. We also thank Arrie Morris, who now works at KCMO Public Health Department, for excellent technical support, and the members in the laboratory of Drs. Joan and Ronald Conaway for helpful discussions on ubiquitylation. We thank Dr. Yigong Shi (Princeton University) for kindly providing DIAP1 cDNA. We acknowledge the people at the HHMI Biopolymer Laboratory/W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University for amino acid sequencing. This work was supported by the Stowers Institutional Funding to Chunying Du.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ”advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1097903.
References
- Chai J., Du, C., Wu, J.W., Kyin, S., Wang, X., and Shi, Y. 2000. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature 406: 855–862. [DOI] [PubMed] [Google Scholar]
- Cilenti L., Lee, Y., Hess, S., Srinivasula, S., Park, K.M., Junqueira, D., Davis, H., Bonventre, J.V., Alnemri, E.S., and Zervos, A.S. 2003. Characterization of a novel and specific inhibitor for the pro-apoptotic protease Omi/HtrA2. J. Biol. Chem. 278: 11489–11494. [DOI] [PubMed] [Google Scholar]
- Cryns V. and Yuan, J. 1998. Proteases to die for. Genes & Dev. 12: 1551–1570. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L. and Reed, J.C. 1999. IAP family proteins—suppressors of apoptosis. Genes & Dev. 13: 239–252. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L., Takahashi, R., Salvesen, G.S., and Reed, J.C. 1997. X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388: 300–304. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L., Leo, E., Stennicke, H.R., Welsh, K., Salvesen, G.S., and Reed, J.C. 1999. Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J. 18: 5242–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel M., Wilson, R., Tenev, T., Zachariou, A., Paul, A., Deas, E., and Meier, P. 2003. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell. Biol. 5: 467–473. [DOI] [PubMed] [Google Scholar]
- Du C., Fang, M., Li, Y., Li, L., and Wang, X. 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102: 33–42. [DOI] [PubMed] [Google Scholar]
- Faccio L., Fusco, C., Chen, A., Martinotti, S., Bonventre, J.V., and Zervos, A.S. 2000. Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J. Biol. Chem. 275: 2581–2588. [DOI] [PubMed] [Google Scholar]
- Gray C.W., Ward, R.V., Karran, E., Turconi, S., Rowles, A., Viglienghi, D., Southan, C., Barton, A., Fantom, K.G., West, A., et al. 2000. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur. J. Biochem. 267: 5699–5710. [DOI] [PubMed] [Google Scholar]
- Hegde R., Srinivasula, S.M., Zhang, Z., Wassell, R., Mukattash, R., Cilenti, L., DuBois, G., Lazebnik, Y., Zervos, A.S., Fernandes-Alnemri, T., et al. 2002. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J. Biol. Chem. 277: 432–438. [DOI] [PubMed] [Google Scholar]
- Huang H., Joazeiro, C.A., Bonfoco, E., Kamada, S., Leverson, J.D., and Hunter, T. 2000. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 275: 26661–26664. [DOI] [PubMed] [Google Scholar]
- Jin S., Kalkum, M., Overholtzer, M., Stoffel, A., Chait, B.T., and Levine, A.J. 2003. CIAP1 and the serine protease HTRA2 are involved in a novel p53-dependent apoptosis pathway in mammals. Genes & Dev. 17: 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ambrosini, G., Chu, E.Y., Plescia, J., Tognin, S., Marchisio, P.C., and Altieri, D.C. 1998. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396: 580–584. [DOI] [PubMed] [Google Scholar]
- Li W., Srinivasula, S.M., Chai, J., Li, P., Wu, J.W., Zhang, Z., Alnemri, E.S., and Shi, Y. 2002. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 9: 436–441. [DOI] [PubMed] [Google Scholar]
- Liu X., Kim, C.N., Yang, J., Jemmerson, R., and Wang, X. 1996. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86: 147–157. [DOI] [PubMed] [Google Scholar]
- Liu Z., Sun, C., Olejniczak, E.T., Meadows, R.P., Betz, S.F., Oost, T., Herrmann, J., Wu, J.C., and Fesik, S.W. 2000. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature 408: 1004–1008. [DOI] [PubMed] [Google Scholar]
- Martins L.M., Iaccarino, I., Tenev, T., Gschmeissner, S., Totty, N.F., Lemoine, N.R., Savopoulos, J., Gray, C.W., Creasy, C.L., Dingwall, C., et al. 2002. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J. Biol. Chem. 277: 439–444. [DOI] [PubMed] [Google Scholar]
- Miller L.K. 1999. An exegesis of IAPs: Salvation and surprises from BIR motifs. Trends Cell. Biol. 9: 323–328. [DOI] [PubMed] [Google Scholar]
- Nagata S. 1997. Apoptosis by death factor. Cell 88: 355–365. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70: 503–533. [DOI] [PubMed] [Google Scholar]
- Salvesen G.S. and Duckett, C.S. 2002. Apoptosis: IAP proteins: Blocking the road to death's door. Nat. Rev. Mol. Cell. Biol. 3: 401–410. [DOI] [PubMed] [Google Scholar]
- Srinivasula S.M., Hegde, R., Saleh, A., Datta, P., Shiozaki, E., Chai, J., Lee, R.A., Robbins, P.D., Fernandes-Alnemri, T., Shi, Y., et al. 2001. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410: 112–116. [DOI] [PubMed] [Google Scholar]
- Sun C., Cai, M., Gunasekera, A.H., Meadows, R.P., Wang, H., Chen, J., Zhang, H., Wu, W., Xu, N., Ng, S.C., et al. 1999. NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature 401: 818–822. [DOI] [PubMed] [Google Scholar]
- Sun C., Cai, M., Meadows, R.P., Xu, N., Gunasekera, A.H., Herrmann, J., Wu, J.C, and Fesik, S.W. 2000. NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP. J. Biol. Chem. 275: 33777–33781. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Imai, Y., Nakayama, H., Takahashi, K., Takio, K., and Takahashi, R. 2001. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell 8: 613–621. [DOI] [PubMed] [Google Scholar]
- Takahashi R., Deveraux, Q., Tamm, I., Welsh, K., Assa-Munt, N., Salvesen, G.S., and Reed, J.C. 1998. A single BIR domain of XIAP sufficient for inhibiting caspases. J. Biol. Chem. 273: 7787–7790. [DOI] [PubMed] [Google Scholar]
- Thornberry N.A. and Lazebnik, Y. 1998. Caspases: Enemies within. Science 281: 1312–1316. [DOI] [PubMed] [Google Scholar]
- van Loo G., van Gurp, M., Depuydt, B., Srinivasula, S.M., Rodriguez, I., Alnemri, E.S., Gevaert, K., Vandekerckhove, J., Declercq, W., and Vandenabeele, P. 2002. The serine protease Omi/HtrA2 is released from mitochondria during apoptosis. Omi interacts with caspase-inhibitor XIAP and induces enhanced caspase activity. Cell Death Differ. 9: 20–26. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. 1996. The N-end rule: Functions, mysteries, uses. Proc. Natl. Acad. Sci. 93: 12142–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen A.M., Ekert, P.G., Pakusch, M., Silke, J., Connolly, L.M., Reid, G.E., Moritz, R.L., Simpson, R.J., and Vaux, D.L. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102: 43–53. [DOI] [PubMed] [Google Scholar]
- Verhagen A.M., Coulson, E.J., and Vaux, D.L. 2001. Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol. 2: REVIEWS3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen A.M., Silke, J., Ekert, P.G., Pakusch, M., Kaufmann, H., Connolly, L.M., Day, C.L., Tikoo, A., Burke, R., Wrobel, C., et al. 2002. HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J. Biol. Chem. 277: 445–454. [DOI] [PubMed] [Google Scholar]
- Wang X. 2001. The expanding role of mitochondria in apoptosis. Genes & Dev. 15: 2922–2933. [PubMed] [Google Scholar]
- Wu G., Chai, J., Suber, T.L., Wu, J.W., Du, C., Wang, X., and Shi, Y. 2000. Structural basis of IAP recognition by Smac/DIABLO. Nature 408: 1008–1012. [DOI] [PubMed] [Google Scholar]
- Yang Y., Fang, S., Jensen, J.P., Weissman, A.M., and Ashwell, J.D. 2000. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 288: 874–877. [DOI] [PubMed] [Google Scholar]
- Zor T. and Selinger, Z. 1996. Linearization of the Bradford protein assay increases its sensitivity: Theoretical and experimental studies. Anal. Biochem. 236: 302–308. [DOI] [PubMed] [Google Scholar]