Abstract
Commitment to mitosis is regulated by a protein kinase complex called MPF. MPF is inhibited by Wee1-related kinases and activated by Cdc25 phosphatase. MPF activation further boosts Cdc25 and represses Wee1. This feedback control probably involves polo kinase. A dominant cut12.s11 mutation in the Schizosaccharomyces pombe spindle pole body (SPB) component Cut12 both suppresses the conditional lethal mitotic commitment defect of cdc25.22 and promotes premature association of the S. pombe polo kinase, Plo1, with the SPB. We now show that Cut12 associated with Plo1 in two hybrid and immunoprecipitation assays. Plo1 function was required for recognition of the mitotic SPB by the phospho-specific antibody MPM-2. In vivo MPM-2 staining and in vitro kinase assays established that the loss-of-function mutation, cut12.1, reduced mitotic activation of Plo1, whereas the gain-of-function mutation, cut12.s11, promoted higher levels of Plo1 activity than were normally seen in interphase. cut12.s11 could not promote mitotic commitment of cdc25.22 cells when Plo1 function was compromised. Expression of a constitutively active plo1 allele suppressed the mitotic commitment defect of cdc25.22. These data suggest that cut12.s11 suppresses cdc25.22 by promoting Plo1 activity. Furthermore, the delayed mitotic commitment of plo1.ts2 cells suggests that Plo1 is an integral part of the core controls that modulate MPF activation in S. pombe.
Keywords: S. pombe, polo kinase, cut12, mitosis, MPF, centrosome, SPB
Commitment to mitosis is regulated by the activity of a conserved protein kinase complex called MPF (Nurse 1990). Phosphorylation of MPF by protein kinases related to the fission yeast protein kinase Wee1 inhibit its activity. Removal of these phosphates by Cdc25 phosphatase activates MPF and triggers an autocatalytic feedback loop that represses Wee1 and further boosts Cdc25 activity (Hoffmann et al. 1993). Thus, the activation of the bulk of Cdc25 is actually dependent on MPF activity (Kovelman and Russell 1996). Feedback loop activation of Cdc25 involves phosphorylation by MPF itself and one or more additional kinases (Strausfeld et al. 1992; Izumi and Maller 1993, 1995). Polo kinase is a prime candidate for such a Cdc25-activating kinase. It is required for commitment to mitosis in nontransformed human cells and was purified from Xenopus egg extracts as a molecule that bound to, phosphorylated, and activated Cdc25 (Kumagai and Dunphy 1996; Lane and Nigg 1996). The phosphorylation of Cdc25 by Xenopus polo kinase (Plx1) conferred reactivity to the phospho-specific anti-body MPM-2. This reactivity usually occurs on Cdc25 during mitotic commitment (Kuang et al. 1994; Kumagai and Dunphy 1996). Subsequent studies established that depletion of Plx1 from Xenopus oocytes delays mitotic commitment in a way that can be overcome by addition of excess Cdc25C (Qian et al. 1998). Plx1 activity is not required in Xenopus oocyte extracts for MPF activation per se; rather, its presence ensures the abrupt nonlinear kinetics of MPF activation (Karaiskou et al. 1999).
Although less is known about the regulation of Wee1 and the related kinase Myt1, it has been established that both become hyperphosphorylated upon mitotic commitment and that hyperphosphorylation of Wee1 correlated with a reduction in its kinase activity toward p34cdc2/cyclin B (Tang et al. 1993; Honda et al. 1995; Mueller et al. 1995). Polo kinase may inhibit Wee1 during the feedback loop as the budding yeast polo kinase, Cdc5, binds to and phosphorylates the Wee1 kinase Swe1 (Bartholomew et al. 2001).
Studies in fission yeast have suggested that the spindle pole may play a critical role in regulating the commitment to mitosis. The cut12+ gene encodes an essential protein that is required for spindle formation and associates with the cytoplasmic domain of the spindle pole body(SPB; Bridge et al. 1998). Dominant cut12 mutations were isolated in a screen for suppressers of cdc twenty-five (stf). These stf mutants, such as cut12.s11, suppress the conditional lethality of cdc25.22 and permit colony formation of cdc25Δ (Hudson et al. 1990). The fission yeast polo kinase, Plo1, normally displays strong association with the SPB of mitotic but not interphase cells (Mulvihill et al. 1999). In cut12.s11, strong SPB association of Plo1 is seen throughout the cell cycle (Mulvihill et al. 1999; Grallert and Hagan 2002). Thus, the dominant mutation in this SPB component simultaneously induces mitotic behavior of Plo1 and suppresses the cdc25.22 mutation. Both processes are dependent on the activity of the NIMA-related kinase Fin1 (Grallert and Hagan 2002).
We now show that Plo1 binds to Cut12 and that Plo1 function is necessary for the cut12.s11 mutation to promote the transition of the G2/M boundary when Cdc25 function is compromised. A keyrole for Plo1 in regulating mitotic commitment is supported by the observations that both MPF activation and mitotic commitment are delayed in the plo1.ts2 loss-of-function mutant and that induction of the “constitutively active” plo1.S124DT197D mutant allele enables cdc25.22 cells to divide under restrictive conditions. The loss-of-function mutant cut12.1 shows that Cut12 is required for the full activation of Plo1 during mitotic commitment, whereas the gain-of-function mutation cut12.s11 promotes abnormally high levels of Plo1 activity in interphase cells that lack Cdc2 activity. Because Plo1 activation is normally dependent on prior activation of MPF (Tanaka et al. 2001), we discuss these results in the context of a role for the localized activation of polo kinase on the spindle pole in the MPF-activating feedback loop.
Results
Cut12 associates with Plo1
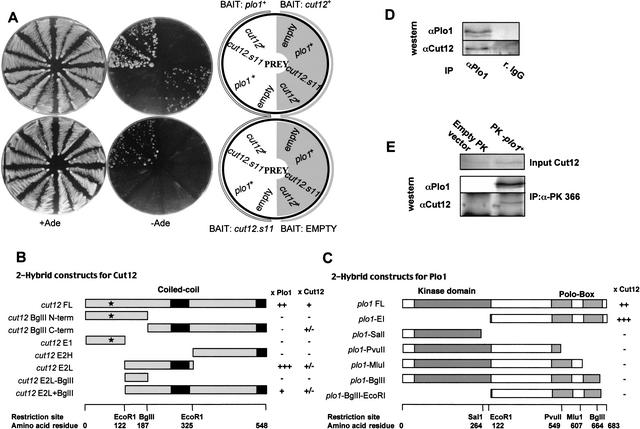
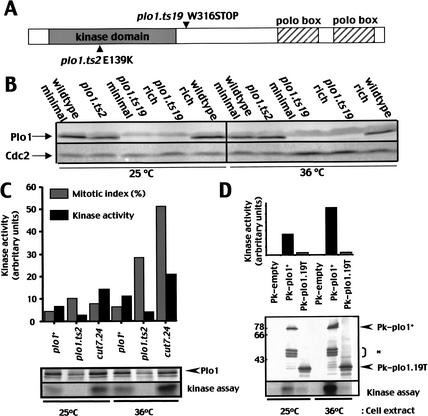
Yeast two-hybrid and immunoprecipitation assays were used to determine whether Plo1 bound directly to Cut12. cut12+ and plo1+ were each inserted into yeast two-hybrid bait and prey vectors (James et al. 1996). A positive reaction was seen when plo1+ was cloned into the bait and cut12+ into the prey plasmid, but not when cut12+ was put in the bait and plo1+ in the prey (Fig. 1A). A reduced interaction in one direction of the assay between partners is an established feature of this assay(Knop and Schiebel 1998). We also detected a weak interaction between Cut12 and itself (Fig. 1A). The generation of sub-clones established that the central region of Cut12 between amino acids 122 and 325 and the C-terminal 361 amino acids of Plo1, which contain the “polo boxes,” were important for the interaction (Fig. 1B,C). We then asked whether Plo1 immunoprecipitates contained Cut12 protein. Cut12 coprecipitated with Plo1 when Plo1 was precipitated with either polyclonal anti-Plo1 antibodies (Fig. 1D), or when an epitope-tagged Plo1 protein was precipitated with antibodies to the tag (Fig. 1E). Thus, Cut12 and Plo1 interact in the two-hybrid assay and are present in common immunoisolated complexes, suggesting that they physically associate with one another.
Figure 1.
Cut12 and Plo1 associated in two-hybrid and immunoprecipitation assays. (A) Yeast strains (PJ69-4A) containing the indicated bait and prey plasmids were streaked on plates lacking or including adenine as indicated. (B) The truncations of the cut12 gene represented in the cartoons were cloned into the prey vector and screened against the full-length plo1+ bait vector. The site of the cut12.s11 mutation is indicated by *. (C) plo1 truncations cloned into the bait vector and screened against the full-length cut12+ prey vector. (B,C) The strength of interaction with the molecule above the column is indicated by + symbols. The - symbol denotes no detectable interaction. The scale below indicates the restriction sites in the context of the amino acid sequence. (D) Immunoprecipitates were prepared from exponentially growing cells using either polyclonal antibodies to Plo1 or a nonspecific, preimmune serum (r.IgG), and the resulting blots were probed with Cut12 antibodies. (E) The monoclonal antibody MAb336, which recognizes the Pk epitope, was used to generate immunoprecipitates from extracts of a wild-type strain containing p41plo1.NPk (Tanaka et al. 2001) and/or an empty Pk vector cultured in the absence of thiamine to induce expression. Blots were probed with the antibodies indicated.
The association of Cut12 with Plo1 prompted us to ask whether Cut12 function influenced Plo1 activity. We monitored the kinetics of Plo1-associated kinase activity in vitro and in vivo. We used established in vitro Plo1 kinase assays to measure Plo1-specific activity in immunoprecipitates (Tanaka et al. 2001), whereas the in vivo analyses relied on in situ recognition of phospho-epitopes by the monoclonal antibody MPM-2.
Cut12 is required for full activation of Plo1
We monitored Plo1 in vitro kinase activity during mitotic commitment in a cut12.1 mutant strain. cut12.1 cells divide normally at 25°C, but the loss of Cut12 function at 36°C blocks mitosis as cells form monopolar rather than bipolar spindles (Bridge et al. 1998). We used cut7.24 to control for the possibility that any changes we observed were specific consequences of the loss of Cut12 function rather than general features of an early mitotic arrest. cut7+ encodes a BimC-related mitotic motor protein that is required for spindle formation. Like cut12.1, cut7.24 cells are unable to form a mitotic spindle and accumulate early in mitosis with condensed chromosomes (Hagan and Yanagida 1990).
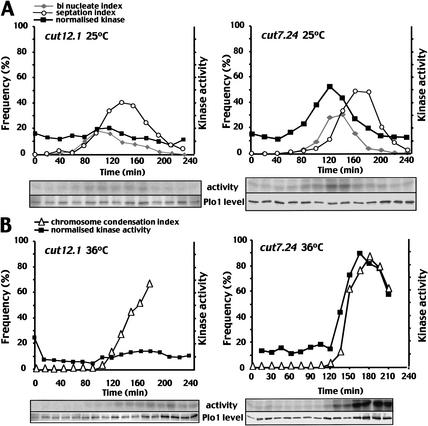
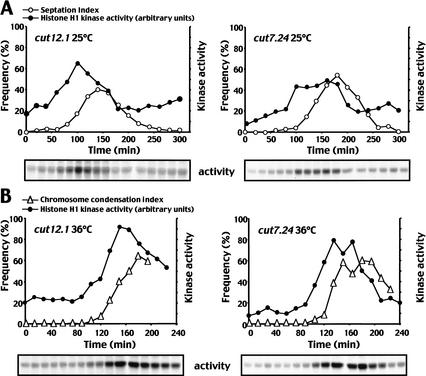
Plo1-specific activity was monitored as the small G2 cells progressed through the cell cycle at either 25°C or 36°C. cut7.24 and cut12.1 both resembled wild-type cells as Plo1-associated kinase activity rose and fell with cell division at 25°C (Fig. 2A; Tanaka et al. 2001). Plo1-associated activity did not rise to the same extent in cut12.1 cells as it did in cut7.24 or wild-type cells (Tanaka et al. 2001). At 36°C, the sharp rise in chromosome condensation indicated that cut12.1 cells entered mitosis synchronously(Fig. 2B). However, this mitotic commitment was not accompanied by the surge of Plo1-associated kinase activity that is normally seen in the wild-type cells (Tanaka et al. 2001), or in the mitotically arrested cells in the control cut7.24 culture. In cut7.24 cells, Plo1-associated activity rose 10 min before the chromosomes condensed and continued to rise as cells accumulated in mitosis (Fig. 2B). No such mitotic stimulation of Plo1 activity occurred in cut12.1 cells (Fig. 2B). Histone H1 kinase assays established that histone H1 kinase activity rose in a similar manner in the cut12.1 and cut7.24 synchronous cultures at both the permissive and restrictive temperatures (Fig. 3A,B). Thus, despite the lack of any increase in Plo1-specific activity at this time, histone H1 kinase activity rose normally as the chromosome condensation indices of the same samples indicated that these cut12.1 cells were accumulating in mitosis (Fig. 3B). This suggested that the inability to promote mitotic Plo1-associated kinase activity in cut12.1 cells was not a simple consequence of an inability to assemble a bipolar spindle, or an inability to commit to mitosis. Rather, the data indicate that Cut12 function was required for full activation of Plo1-associated kinase activity during mitotic commitment.
Figure 2.
Plo1 kinase activity did not rise upon entry into mitosis in cut12.1 mutants. Small G2 cells from mid-log phase cultures of cut12.1 or cut7.24 that had been grown in minimal medium at 25°C were incubated at either 25°C (A) or 36°C (B). Samples were processed for either Plo1 kinase assays or DAPI (chromatin) and calcofluor (cell wall) staining at the times indicated. The top panels show kinase assays, and the bottom panels show the amount of Plo1 in each kinase assay reaction. The septation and binucleate (25°C) or chromosome condensation (36°C) indices (left-hand Y-axis) were plotted alongside the kinase activity per unit Plo1 protein (i.e., specific activity, right-hand Y-axis). Assays were processed in an identical manner with identical cell numbers so that the arbitrary units used to plot the data on the graphs are comparable between the different panels.
Figure 3.
Histone H1 kinase activityrose as cut12.1 cells entered a defective mitosis. Small G2 cells from mid-log phase cultures of cut12.1 or cut7.24 that had been grown in minimal medium at 25°C were incubated at either 25°C (A) or 36°C (B). Samples were processed for either histone H1 kinase assays or DAPI (chromatin) and calcofluor (cell wall) staining at the indicated times. The kinase assays are shown at the bottom of each panel. The septation (25°C) or chromosome condensation (36°C) indices (left-hand Y-axis) were plotted alongside the kinase activity(right-hand Y-axis). The kinase assays were processed in an identical manner with identical cell numbers.
Activation of interphase Plo1 in vitro kinase activity by the cut12.s11 mutation
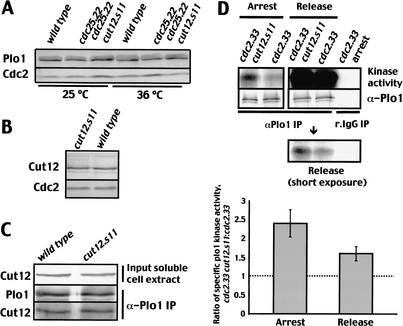
Having established the dependency of Plo1-associated kinase activity on Cut12 function, we next asked whether the dominant cut12.s11 mutant led to any changes in the biochemical characteristics of Plo1. Plo1 levels remain constant throughout the cell cycle and were unaffected by the cut12.s11 mutation (Fig. 4A; Mulvihill et al. 1999). Blotting extracts from an asynchronous cut12.s11 culture revealed a single band at the same position and of the same intensity as wild-type Cut12, indicating that the cut12.s11 mutation did not alter Cut12 levels (Fig. 4B). Nor did cut12.s11 alter the ability of Cut12 to interact with Plo1 in two-hybrid or coimmunoprecipitation assays (Figs. 1A, 4C). We next assayed Plo1-associated kinase activity in Plo1 immunoprecipitates from cut12.s11 cells in which cell cycle progression had been arrested in interphase by incubation of the conditional cdc2 mutation cdc2.33 at 35°C for 2 h. Plo1-associated kinase activity of extracts from arrested cdc2.33 cut12.s11 cells was 2.4-fold (±0.35; n = 6) higher than that of the control cdc2.33 cut12+ cells (Fig. 4D). This established that cut12.s11 increased Plo1 activity in interphase. Moreover the use of the cdc2.33 mutant background established that MPF activity was not required to induce Plo1 activation in cut12.s11 mutant cells. Returning the temperature of the same two populations to 25°C for 30 min promoted mitosis. Kinase assays of these mitotic samples indicated that the cut12.s11 mutation promoted a 1.6 (±0.18; n = 5) increase in Plo1-specific activity during mitosis (Fig. 4D). Because Plo1 activation normally requires the prior activation of p34cdc2 (Tanaka et al. 2001), these data support the hypothesis that cut12.s11 suppresses cdc25.22 because it promotes the activity of the MPF amplification loop kinase Plo1 in interphase cells and there by promotes the autoamplification loop that drives mitotic commitment.
Figure 4.
The cut12.s11 mutation enhanced Plo1 kinase activity. (A) Cells were grown to earlylog phase at 25°C before the culture was split in two. One-half was incubated at 25°C and the other at 36°C for 4 h before protein extracts were processed to blot for Plo1. (B) Western blot showing Cut12 levels in cut12+ and cut12.s11 cells. (C) Western blot of cut12+ and cut12.s11 cultures showing Cut12 levels in the cell extracts (top) that were used to immunoprecipitate Plo1 (middle) and probe for coprecipitated Cut12 (bottom); for controls, see Figure 1D. (D) Plo1 kinase assays of cdc2.33 cut12+ and cdc2.33 cut12.s11 cells that had been grown in minimal medium at 25°C before shifting to 36°C for 2 h (left panel). Cells were then returned to 25°C, and a sample was taken from the mitotic fraction 30 min after the release (right panel). The top panel shows Plo1-associated kinase activity, and the panel directly below it shows the quantityof Plo1 in the immunoprecipitates. Immunoprecipitation was either with antibodies to Plo1 (αPlo1 IP) or nonspecific preimmune antiserum (r.IgG). All lanes in the kinase assays in the top panel were exposed to the same extent, but the middle panel in D shows a lower exposure of the mitotic samples following release from G2 arrest. The bottom panel shows a graph of the ratio of specific Plo1 kinase activity of the double cdc2.33 cut12.s11 mutant to that of the single cdc2.33 mutant.
Recognition of the mitotic SPB by the phospho-specific antibody MPM-2 is dependent on Plo1 function
Having established that loss or gain of Cut12 function led to a corresponding loss or gain of Plo1 function in in vitro kinase assays, we asked whether this was due to particularly a typical behavior in in vitro assays, or whether it also held true in vivo. We used the monoclonal antibody MPM-2 to monitor in vivo Plo1-dependent events on the SPB. MPM-2 recognizes a mitosis-specific phospho-epitope that is greatly enriched at spindle poles (Davies et al. 1983; Vandre et al. 1984; Lane and Nigg 1996). MPM-2 reactivity of the spindle pole of HeLa cells is abolished by microinjection of antibodies to human polo kinase, Plk1, and phosphorylation by Polo kinase confers MPM-2 reactivity on Cdc25 (Kumagai and Dunphy 1996; Lane and Nigg 1996).
MPM-2 antibodies gave clear cell cycle stage-dependent staining patterns in Schizosaccharomyces pombe (Fig. 5). Signals from the spindle and cytokinetic ring could be seen over a faint nuclear background stain and small dots that could represent kinetochores (Fig. 5, row 4). The optimal conditions for staining particular structures varied. For example, cytokinetic ring staining (Fig. 5, row 7, arrow) was optimal after 8% formaldehyde for 30 min, whereas SPB staining was optimal after 3.7% formaldehyde for 2 min (data not shown). We frequently observed a line of MPM-2 staining between the two SPBs just before anaphase (Fig. 5, row 5, arrow). This may represent a chromosome topologyprotein such as the cohesin subunit and polo kinase target Rad21 (Weitzer and Uhlmann 2002). The strongest staining however, was of mitotic SPBs up to late anaphase. The SPB was identified by costaining with antibodies to the SPB component Sad1. The SPB stained strongly from commitment to mitosis when a single spot of SPB staining was visible (Fig. 5, row 3) until the middle of anaphase B, whereupon it became much fainter. This staining pattern is highly reminiscent of the patterns seen with anti-Plo1 antibodies (Mulvihill et al. 1999).
Figure 5.
MPM2 recognizes the mitotic SPBs and a range of other structures during cell division. Wild-type cells were grown to mid-log phase at 25°C before processing for simultaneous visualization of Sad1, MPM-2, and DAPI staining. The signals for MPM2 (green) are merged with those for Sad1 (red) in the far right panel. The arrows in rows 5 and 7 indicate linear elements in the chromatin and the cytokinetic ring, respectively. All cell cycle stages are shown: interphase (rows 1,2), prophase (row 3), prometaphase (row 4), metaphase (row 5), anaphase B (row 6), telophase (row 7), and G1/cytokinesis (row 8).
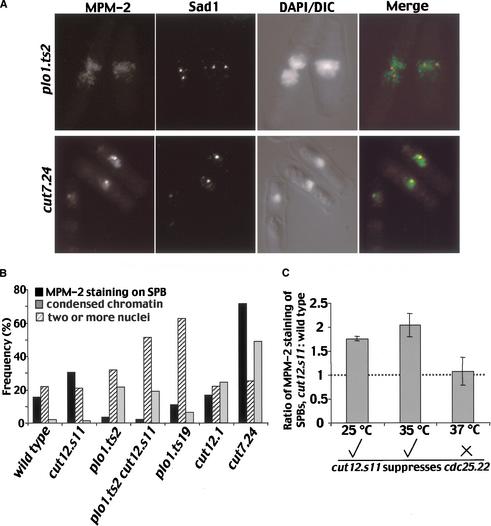
We asked whether MPM-2 reactivity was dependent on Plo1 activity by comparing MPM-2 staining in plo1.ts2 (see below), wild-type, and cut7.24 cultures after the cells had been incubated at 35°C for 4 h to inactivate Plo1 and Cut7, respectively. Both plo1.ts2 and cut7.24 mutants block spindle formation (see below). Because the short fixation times required to see MPM-2 epitopes did not preserve chromatin structure, parallel samples were processed for DAPI staining to determine the frequency of cells with either chromosome condensation or two nuclei in each sample. The chromosome condensation index of 49% indicated the accumulation of cut7.24 cells in mitosis. This mitotic block resulted in a large increase in the proportion of cells in which the SPBs stained with MPM-2 from 11% to 71.5% (Fig. 6A,B). In contrast, even though the chromosome condensation index of 21.5% showed a strong accumulation of plo1.ts2 cells in mitosis, there was no concomitant increase in MPM-2 reactivity of the SPBs (Fig. 6A,B). This established that the MPM-2 reactivity of mitotic SPBs was dependent on the activity of Plo1 kinase.
Figure 6.
MPM2 recognition of mitotic SPBs depended on Plo1 and Cut12 function and was enhanced in cut12.s11 mutants when it could suppress cdc25.22. (A,B) MPM-2/Sad1 staining of plo1.ts2 and cut7.24 cells that had been incubated at 35°C for 4 h as described for Figure 4. (B) A bar chart of the frequency of cells with the features shown in the legend. (C) The ratio of the frequency of MPM-2 recognition of SPBs in cut12.s11 over wild type at the temperatures shown. The ticks and cross underneath the graph indicate whether cut12.s11 can (√) or cannot (×) suppress cdc25.22 at this temperature.
A direct correlation between MPM-2 recognition of the mitotic SPB and loss or gain of Cut12 function
Inactivation of Cut12 by shifting the temperature of a cut12.1 culture from 25°C to 35°C led to an accumulation of cells blocked in mitosis with a chromosome condensation index of 24.5% (Fig. 6B). Despite this accumulation in a phase of the cell cycle during which SPBs become MPM-2 reactive, we detected no concomitant increase in the number of MPM-2 reactive SPBs in this cut12.1 culture (Fig. 6B). This result mirrored the reduction in Plo1 in vitro kinase activity in cut12.1 (Fig. 2B) as it showed that an in vivo readout of Plo1 activity was dependent on Cut12 function. We next asked whether the enhancement of in vitro Plo1-associated kinase activity in cut12.s11 mutants (Fig. 4D) was similarly mirrored by an increase in Plo1-dependent MPM-2 phosphorylation in vivo. Excitingly, it was (Fig. 6B). cut12.s11 led to a 1.8 ± 0.05-fold increase in the number of MPM-2 reactive SPBs at 25°C and 2.0 ± 0.24-fold at 35°C (Fig. 6C). This increase was Plo1-dependent because no MPM-2 staining was seen when Plo1 was inactivated at 35°C in cut12.s11 plo1.ts2 double-mutant cells (Fig. 6B). Most importantly, there was no difference between the number of MPM-2 reactive SPBs of wild-type and cut12.s11 cells at 37°C (Fig. 6C). This is highly significant because it is well established that cut12.s11 is unable to suppress cdc25.22 at this temperature (Hudson et al. 1990; Bridge et al. 1998).
In summary, when cut12.s11 was able to suppress cdc25.22, it also enhanced a Plo1-dependent phosphorylation event on the SPB. When it was no longer able to suppress cdc25.22, it no longer enhanced this phosphorylation event. Furthermore, the phosphorylation event did not happen in the loss-of-function mutant cut12.1. This mutation both blocked the mitotic stimulation of Plo1 in vitro kinase activity and has shown a very strong synthetic enhancement of the cdc25.22 phenotype (Bridge et al. 1998). We can make two key observations from these data. First, there is a striking correlation between Cut12 function and Plo1-dependent phosphorylation events at the SPB. Second, cut12.s11 enhances Plo1 activity under conditions in which it also suppresses cdc25.22.
Does the increase in Plo1 activity in cut12.s11 account for its ability to suppress cdc25.22?
We next wanted to ask whether the correlation between the ability of cut12.s11 to both induce increased Plo1 activity and suppress cdc25.22 reflected a mechanistic link between enhancement of Plo1 activity and the suppression of cdc25.22. In other words, did cut12.s11 suppress cdc25.22 because it increased Plo1 activity in interphase? To address this question, it was necessary to generate conditional plo1 mutants that could be introduced into cut12.s11 cdc25.22 mutants to ask whether compromising Plo1 activity would stop cut12.s11 from promoting mitosis of cdc25.22.
plo1.ts2 and plo1.ts19 have cell division defects
Two temperature-sensitive plo1mutants were generated via a marker switch approach (MacIver et al. 2003). plo1.ts2 has the change E139K in the kinase domain, whereas plo1.ts19 introduced a premature stop codon after the kinase domain, but before the polo boxes, at position 316 (Fig. 7A). The temperature sensitivity of plo1.ts19 was suppressed by expression of the opal nonsense suppressor tRNA sup3.5 (data not shown; Hawthorne and Leupold 1974). Both plo1.ts2 and plo1.ts19 conferred temperature sensitivity for growth on minimal medium. In contrast, the ability of plo1.ts19 cells to form colonies on rich medium at 36°C was indistinguishable from that of wild-type cells. The introduction of a single plo1+ gene at the leu1 locus restored wild-type characteristics to either mutant, indicating that they were recessive, loss-of-function mutations (data not shown). Western blot analysis showed that Plo1 levels in plo1.ts2 cells were not radically different from wild type, whereas the full-length protein appeared to be largely absent from plo1.ts19 on either minimal or rich medium at either 25°C or 36°C (Fig. 7B). Plo1-associated kinase in vitro kinase activity was greatly reduced in extracts from plo1.ts2 that were prepared at either 25°C or 36°C (Fig. 7C).
Figure 7.
Plo1 kinase activitywas severely reduced in plo1.ts2 and plo1.ts19. (A) A cartoon indicating the position of the mutations in the plo1 gene. (B) Mid-log-phase cultures of cells grown in either rich or minimal medium were prepared for Western blot analysis to detect Plo1 or p34cdc2 at the temperatures indicated. (C,D) Plo1 kinase assays of wild-type cells containing the plasmids indicated on the graphs after growth to mid-log phase in minimal medium at the indicated temperatures. The level of Plo1 kinase activity per unit of kinase in the precipitate (i.e., specific activity) was plotted to generate the graphs.
The premature stop codon in plo1.ts19 removed the epitopes recognized by the polyclonal antibodies that were routinely used for the Plo1 kinase assays (Ohkura et al. 1995). We therefore generated a version of the plo1 gene in which a stop at position 316 (equivalent to the plo1.ts19 mutation site) was directly followed by the nmt1+ termination sequences. The N terminus of this molecule was fused to the Pk tag, and it was expressed from the derepressed weakened nmt1+ promoter of pREP81 (pREP81NPkplo1.19T). Induction of gene expression of the tagged plo1.19T or the wild-type control molecule for 30 h did not affect progression through mitosis (data not shown). Anti-Pk tag antibodies were used to generate immunoprecipitates from strains that had been expressing either tagged plo1.19T or tagged plo1+, alongside the product of the native plo1+ gene for 30 h. The kinase activity associated with the mutant precipitates was barely higher than the empty vector control and much lower than that seen in wild-type extracts. This was true whether the protein was isolated from cells grown at 25°C or 36°C or the assays were done at 30°C or 36°C (Fig. 7D; data not shown). We concluded that the Plo1-dependent kinase activity of both plo1.ts2 and plo1.ts19 was greatly reduced.
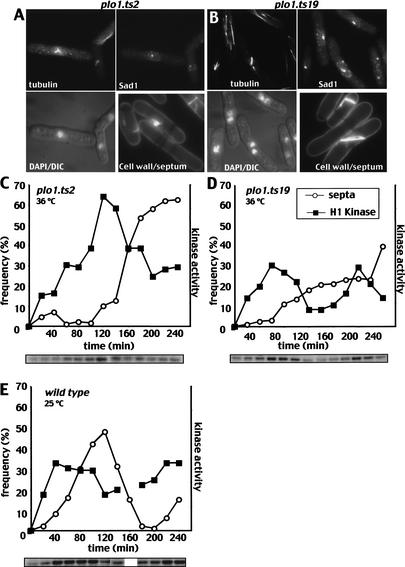
plo1.ts2 strains entered mitosis but did not form spindles as multiple microtubule bundles extended from a single focus of staining for the spindle pole marker Sad1 (Hagan and Yanagida 1995; upper two panels in Fig. 8A; Table 1). Unlike classic “cut” mutants (Hirano et al. 1986), septation did not always follow on from the mitotic arrest. In the cells that did septate, 95.5% of the septa were misshapen (Fig. 8A, lower right panel). plo1.ts19 cells had a range of defects, including monopolar spindles (Fig. 8B, left-hand cell in the series of images; Table 1). Some plo1.ts19 cells did not form a septum and became bi- and multinucleated (Fig. 8B, lower left panel). These deficiencies in septation presumably arose from an inability to activate the septum initiation network (Tanaka et al. 2001).
Figure 8.
Histone H1 kinase activity increased as plo1.ts2 and plo1.ts19 cells underwent defective cell divisions. (A,B) plo1.ts2 and plo1.ts19 cultures were grown to earlylog phase in minimal medium at 25°C before the temperature of the culture was shifted to 36°C for 3 h. One portion was then stained for tubulin, the spindle pole marker Sad1, and DAPI to give the top two and bottom left-hand panels. The cell wall and septum were stained with calcofluor in a second portion. (C–E) H1 kinase and septation profiles of cultures of the indicated strains. After isolation from a culture that was grown in minimal medium at 25°C, small G2 cells were incubated at the temperatures indicated.
Table 1.
Frequencies of mitotic phenotypes (%) in plo1.ts2 and plo1.ts19 after 3 h at 36°C
| Plo1+ | plo1.ts2 | plo1.ts19 | |
|---|---|---|---|
| Mitotic index | 8.5 | 27.0 | 19.5 |
| Proportion of mitotic cells with monopolar spindles | 0 | 94.5 | 50.0 |
| Proportion of binucleate cells in mitosis that have monopolar rather than bipolar spindles | 0 | 5 | 59 |
| Metaphase spindles | 27.0 | 0 | 50.0 |
| Septation index | 12.0 | 34 | 24 |
| Proportion of septa that are disorganized or misplaced | 0 | 95.5 | 86.5 |
| Cells with two or more nuclei | 22 | 10.5 | 70.5 |
plo1.ts2 and plo1.ts19 show full activation of histone H1 kinase
Histone H1 kinase activity gives a good assay of MPF activity and the timing of mitotic commitment in S. pombe (Moreno et al. 1989; Creanor and Mitchison 1994). We therefore used H1 kinase assays of extracts from plo1 mutant cells, synchronized with respect to cell cycle progression, to ask whether the kinetics of mitotic commitment were affected by mutation of plo1+. Both plo1.ts2 and plo1.ts19 cells entered mitosis and activated histone H1 kinase. H1 kinase accumulated at a similar rate over a longer period in plo1.ts2 than in either plo1.ts19 or wild type. The delay in the appearance of septa and the accumulation of cells in mitosis indicated that the defect in spindle formation in plo1.ts2 activated the spindle checkpoint (Fig. 8C–E; Table 1). In contrast, the septation defect of plo1.ts19 did not activate anymitotic checkpoints, so that H1 kinase activity in this strain was periodic and a second wave of activity accompanied a second attempt at division at 36°C (Fig. 8D, 220 min).
Plo1 is required for the suppression of cdc25.22 by cut12.s11
We were now in a position to use plo1.ts2 and plo1.ts19 to ask whether full Plo1 function was required for the dominant cut12.s11 mutation to promote transition of the G2/M boundary when Cdc25 function was compromised. If the suppression of the cell cycle regulator by mutation of the spindle pole component was due to an enhancement of Plo1 function, then compromising Plo1 should compromise the ability of cut12.s11 to suppress cdc25.22. In other words, triple cut12.s11 cdc25.22 plo1.ts mutant cells should not enter mitosis because they arrest cell cycle progression at the G2/M boundary.
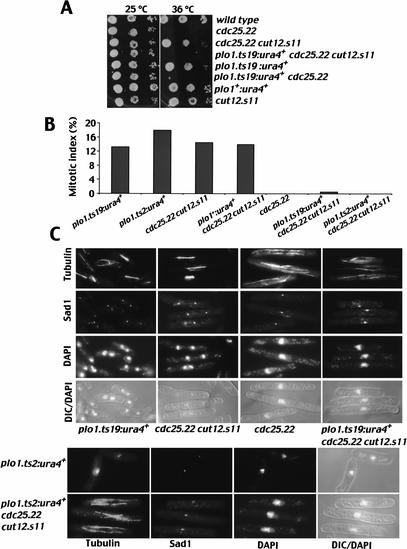
In a first assay, we took advantage of the media dependence of the plo1.ts19 phenotype. On rich medium at 36°C, plo1.ts19 cells formed colonies with a proficiency that was indistinguishable from either wild type or cut12.s11 controls (Fig. 9A). cdc25.22 and cdc25.22 plo1.ts19 cells, on the other hand, could not form colonies on this medium at this temperature, but cdc25.22 cut12.s11 cells could (Hudson et al. 1990; Bridge et al. 1998). Spot test analysis showed that although the plo1.ts19 mutation did not affect colony formation of cut12+, triple cut12.s11 cdc25.22 plo1.ts19 cells were unable to grow (Fig. 9A). This established that mutating plo1 in a way that did not affect cell viability compromised the ability of cut12.s11 to suppress cdc25.22.
Figure 9.
Plo1 function was required for cut12.s11 suppression of cdc25.22. (A) A drop test analysis of the colony-forming potential of the indicated strains at 36°C on YES rich medium. Note that the plo1.ts mutants were generated by a marker switch approach that leaves a ura4+ gene 235 bp upstream in the promoter. This is indicated by plo1.x:ura4+, and the wild-type equivalent allele is plo1+:ura4+. (B,C) Asynchronous early log-phase cultures of the seven strains shown in the figure were shifted from 25°C to 36°C and maintained at 36°C for 3 h before cells were processed for microscopy to simultaneously stain for tubulin, the spindle pole marker Sad1, and DAPI. (B) The mitotic index. (C) Representative images of each strain.
In a second assay, we asked whether the introduction of the plo1.ts19 or plo1.ts2 mutations would affect the ability of cut12.s11 cdc25.22 to enter mitosis at 36°C. Because neither plo1.ts2 nor plo1.ts19 by itself affected the full-scale activation of MPF (Fig. 8), an effect of these mutations on the suppression of cdc25.22 by cut12.s11 would indicate that cut12.s11 was reliant on Plo1 function to suppress cdc25.22. Cells were grown in minimal medium and shifted to 36°C for 3 h. Whereas single plo1.ts2 and plo1.ts19 mutant and double cut12.s11 cdc25.22 mutant cells all entered mitosis (Fig. 9B,C), the single cdc25.22 mutant and both double cdc25.22 plo1.ts and triple cut12.s11 cdc25.22 plo1.ts mutants did not. These latter strains all arrested cell cycle progression with an interphase array of microtubules and no mitotic spindles (Fig. 9B,C). These data established that cut12.s11 required full Plo1 function to transit the G2/M transition when Cdc25 function was compromised.
Activation of Plo1 suppresses the mitotic commitment defect of cdc25.22 cells
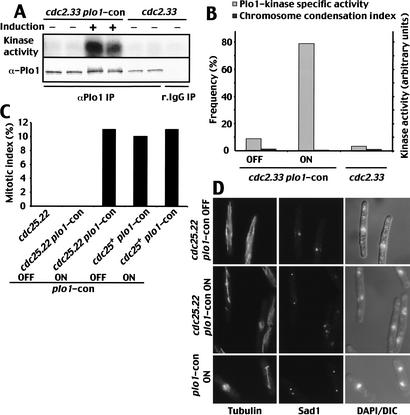
If cut12.s11 does promote suppression of cdc25.22 by boosting Plo1 activity in interphase cells, then it should be possible to suppress cdc25.22 by boosting Plo1 activity independently of the status of Cut12. Gross overproduction of Plo1 protein in cdc25.22 cells promotes cytokinesis events that randomly bisect the nucleus and so presumably activate the DNA damage checkpoint to inhibit mitotic commitment (Ohkura et al. 1995). We therefore sought a less drastic means of enhancing Plo1 activity to address this issue. Mutation of the conserved residues S128 and T201 to aspartic acid in Xenopus Plx results in constitutive activity of this polo kinase (Qian et al. 1999). We therefore introduced the analogous mutations (S124D T197D) into a version of the plo1 gene that was integrated at the leu1 locus under the control of an inducible promoter. The first Plo1 protein to be seen following induction of plo1 alleles from this promoter at the leu1 locus appears at the 13-h time point and takes 7 h to accumulate to the same level as the native protein (Tanaka et al. 2001).
We asked whether plo1.S124DT197D affected interphase Plo1 activity by measuring Plo1 activity in a cdc2.33 mutant in which the plo1.S124DT197D allele was either repressed or induced. Figure 10 shows Plo1 in cdc2.33 cells in which plo1.S124DT197D had been induced at 25°C for 13 h before incubation at the restrictive temperature of 35°C for 2 h had specific kinase activities that were 8.7-fold higher than in the repressed control (Fig. 10A,B). We next asked whether induction of this mutant allele would promote mitosis of cdc25.22 cells at the restrictive conditions. We split a 25°C cdc25.22 plo1.S124DT197D double-mutant culture in two, induced plo1.S1224DT197D expression in one half, and 22.5 h later shifted both cultures to 34°C for 3 h before processing them to stain for microtubules, spindle poles, and chromatin. Whereas cells in which the expression of the constitutively active mutant remained repressed arrested cell cycle progression in interphase, 11% of those in which it had been expressed entered mitosis. This degree of suppression is very similar to the level of suppression of cdc25.22 arising from the presence of the cut12.s11 mutation (Fig. 9B) and established that activation of Plo1 is sufficient to suppress the deficiency in Cdc25 function in cdc25.22 cells.
Figure 10.
Expression of a constitutively active plo1 mutant suppressed the mitotic entry defect of cdc25.22. (A,B) A cdc2.33 plo1.S124DT197D (labeled plo1.con in the figure) culture was grown in minimal medium containing thiamine at 25°C and split in two, and one-half was washed and resuspended in thiamine-free medium to induce plo1.S124DT197D transcription. Thirteen hours later, both halves of the culture were shifted to 35°C for 2 h before being processed for in vitro Plo1 kinase assays (A) and scored for chromosome condensation index by DAPI staining (B). The specific activity per unit protein precipitated was calculated from the gels and blot shown in A and plotted alongside the chromosome condensation index in B. (C–D) A cdc25.22 plo1.S124DT197D culture was grown in minimal medium containing thiamine at 25°C and split in two, and one-half was washed and resuspended in thiamine-free medium to induce plo1.S124DT197D transcription. Then, 22.5 h later, both halves of the culture were shifted to 34°C for 3 h before being processed for immunofluorescence microscopy to visualize microtubules (top left panel), Sad1 (middle panel), or DAPI/DIC (bottom right panel).
Mitotic commitment was delayed and less tightly regulated in plo1.ts2
Did the ability of constitutively active Plo1 kinase to suppress cdc25.22 reflect a genuine requirement for Plo1 during mitotic commitment in an otherwise unperturbed cell cycle? The extended and more random size of plo1.ts2 cells at division suggested that this maybe the case. Despite the fact that these cells are able to enter mitosis, they appear to be doing so in a less efficient, or more random manner (Fig. 8B). We therefore compared the length of mitotic plo1.ts2 cells with wild type and cut7.24 controls. Cells were stained with DAPI, and the length of all cells that had condensed chromatin or anaphase nuclei was recorded (Table 2). Wild-type cells entered the mitotic, constant-length phase (Mitchison and Nurse 1985) at 13.8 ± 0.9 μm, cut7.24 at 15.4 ± 1.11 μm, and plo1.ts2 at 17.6 ± 3.22 μm. Thus, mitotic commitment was later and not as stringently controlled in plo1.ts2 than in either of the control strains. In other words, although mutation of plo1 did not affect the ability to enter mitosis per se, the timing of mitotic commitment was heavily influenced by the status of Plo1.
Table 2.
Cell length at division
| Strain | 25°C | 36°C |
|---|---|---|
| Wild type | 13.5 ± 1.1 | 13.8 ± 0.9 |
| plo1.ts2 | 13.4 ± 1.3 | 17.6 ± 3.22 |
| cut7.24 | 13.2 ± 0.9 | 15.4 ± 1.11 |
| cut12.1 | 14.6 ± 1.3 | 15.0 ± 0.8 |
Cell length measurements (μm) of cells with condensed chromatin or anaphase nuclei were taken after staining with DAPI.
Mutant Plo1 does not associate with the interphase SPB of cut12.s11
We next asked whether plo1.ts2 or plo1.ts19 affected the affinity of Plo1 for the interphase SPB of cut12.s11 cells. For this assay, cells were first incubated in hydroxyurea (HU) for 5 h to generate an interphase population in which cell cycle progression was arrested in early S phase. As described previously, Plo1 did not show strong affinity for the SPB of wild-type cells under these conditions, but did associate with virtually all of the SPBs of cut12.s11 cells at all temperatures (Fig. 11A,B,E; Mulvihill et al. 1999; Grallert and Hagan 2002). At 25°C, plo1.ts2 cut12.s11 resembled cut12.s11 as Plo1 associated with both interphase and mitotic SPBs (Fig. 11A,C). In contrast at 36°C, Plo1 did not associate with interphase SPBs of this double mutant (Fig. 11E). Mixing a cut12.s11 culture with the double mutant so that 25% of the cells in the mixture were cut12.s11 and 75% were cut12.s11 plo1.ts2 established that this lack of staining was not an artifact arising from a processing error as 20% of cells had Plo1 on their SPB (Fig. 11D,E).
Figure 11.
plo1.ts2 conferred temperature sensitivity on the ability of Plo1 to be prematurely recruited to the interphase SPB of cut12.s11 cells. Ten millimolar HU was added to early-log-phase cultures of plo1.ts2, wildtype, cut12.s11, or cut12.s11 plo1.ts2 cells that were growing in minimal medium at 25°C. Three hours later, each culture was split in two and one-half was incubated at 36°C and the other at 25°C for a further 2 h. After this, an extra culture was generated at 36°C by mixing the 36°C cut12.s11 culture with the 36°C plo1.ts2 cut12.s11 culture at a ratio of 1:3. All cultures were then processed for anti-Plo1 immunofluorescence microscopy. (A,B) Representative images from the cut12.s11 culture at 25°C (A) and 36°C (B). (C,D) The mixed culture at 25°C (C) and 36°C (D). (E) A bar chart of the frequency of cells with Plo1 staining of the SPB. Plo1 associated with the SPB of plo1.ts2 cut12.s11 cells at 25°C but not 36°C. The mixed culture (plo1.ts2 cut12.s11 mixed with plo1+ cut12.s11 cells) showed that the lack of staining in the cut12.s11 strain was not due to processing artifacts. The 1:3 ratio of single to double mutants was reflected in the 20% of cells with Plo1 on the SPB. An example of Plo1 staining in the cut12.s11 control cell in this mix is marked by* in D.
The inability of a Plo1 mutant protein that has compromised Plo1-associated kinase activity to associate with the interphase SPB of cut12.s11 raised the possibility that Plo1 activity may be critical for the recruitment of this kinase to interphase SPBs of cut12.s11 cells. We therefore used anti-tag antibodies to compare the distribution of wild-type and “kinase dead” (plo1.K65R) molecules that were tagged at their N termini and expressed from the derepressed weakened nmt1+ promoter of pREP81 (Tanaka et al. 2001). Whereas a similarly expressed tagged wild-type control molecule associated with both interphase and mitotic SPBs of cut12.s11, Plo1.K65R, the “kinase dead” mutant protein, only associated with mitotic but not with interphase SPBs (data not shown).
Anti-Pk antibodies were used to localize the truncated Plo1.19T protein that was expressed from pREP81NPkplo1.19T. Although the protein was expressed to similar levels to a full-length control (Fig. 7D; data not shown), no localization was seen at any stage of the cell cycle. This was consistent with the two-hybrid data that indicated that Plo1 associated with Cut12 via the C-terminal 361 amino acids and the failure of other premature truncation proteins to associate with the SPB (Reynolds and Ohkura 2003). We concluded that the efficient association of Plo1 with the interphase SPB of cut12.s11 cells required both Plo1 activity and the C-terminal 367-amino-acid, polo-box-containing, region.
Discussion
Mutations in the spindle pole component Cut12 compensate for compromised Cdc25 function
In fission yeast, Cdc25 is absolutely required for mitotic commitment unless one of the following is mutated: the target p34cdc2, the antagonizing kinase Wee1, or the spindle pole protein Cut12. In the absence of inhibitory kinase activity, newly synthesized p34cdc2 will form an unrestrained complex with cyclin B. Thus, as soon as the Wee1 homolog Mik1 is degraded at the end of S phase (Baber-Furani et al. 2000; Christensen et al. 2000), active MPF will accumulate and drive wee1 cells into mitosis. Cdc25 is not therefore required to dephosphorylate MPF in Wee1 mutants because the active complex forms de novo as fresh cyclin B becomes available to partner p34cdc2. Thus, it is fairly apparent why mutating wee1 or the target cdc2 can alleviate the requirement for Cdc25. It has not been so clear why the mutation of an SPB component should suppress cdc25.22 and support colony formation of cdc25Δ (Hudson et al. 1990).
How could an altered spindle pole suppress cdc25.22?
Several models could account for cut12.s11 suppression of cdc25.22. The site of the activating mutation in cut12.s11 (G71V) lies 11 residues from an MPF consensus phosphorylation site and 4 residues from a MAP kinase consensus site, raising the possibility that the cut12.s11 mutation maymimic the change induced by phosphorylation at one or both of these sites. Assuming that Cut12 was the major rate-limiting MPF substrate for mitotic entry, then cut12.s11 may be have as an activated form of the protein and promote mitosis in the presence of reduced MPF activity. Introduction of plasmid-borne cut12 with mutations in either, or both, the MAP kinase, or MPF consensus sites that maymimic the phosphorylated state failed to suppress cdc25.22. In contrast, plasmids with the cut12.s11, G71V, mutation, either alone or in combination with alanine or glutamic acid mutations at the MPF and MAP kinase consensus sites, did suppress cdc25.22 (K. Tanaka, A.J. Bridge, and I.M. Hagan, unpubl.). It therefore seems unlikely that cut12.s11 is suppressing cdc25.22 because it is mimicking activation of the sole essential MPF target.
MPF associates with the SPB (Alfa et al. 1990; Decottingnies et al. 2001). cut12.s11 could therefore alter the recruitment of MPF to promote mitosis in cdc25.22 cells; however, the lack of any effect of the cut12.s11 or cut12.1 mutations on the distribution of MPF supports the argument against this (Decottingnies et al. 2001; data not shown).
In a further model, Cut12 may influence mitotic commitment in cdc25.22 through premature promotion of the feedback loop to counteract Wee1 function. Given that polo kinase is part of the feedback loop in Xenopus extracts, the simplest way by which Cut12 could influence this regulatory pathway would be by enhancing polo kinase activity.
Direct correlation between Cut12 function and Plo1 kinase activity and the suppression of cdc25.22
We have previouslyshown that cut12.s11 influences the behavior of the fission yeast polo kinase, Plo1. Plo1 recruitment to the SPB is usually restricted to mitosis. In cut12.s11 cells, however, it associates with the SPB throughout the cell cycle (Mulvihill et al. 1999; Grallert and Hagan 2002). We have now established that Cut12 associates with Plo1 in two-hybrid and immunoprecipitation assays. In vitro assays of Plo1 kinase activity showed that Plo1 activity depended on Cut12 function and that the gain-of-function mutation in cut12.s11 enhanced Plo1 activity. Microscopy with MPM-2 antibodies that recognize Plo1-dependent phospho-epitopes suggested that the same held true in vivo. Loss-of-function mutants reduced MPM-2 staining, whereas gain-of-function mutants increased it. Two key observations make it unlikely that the lack of staining in cut12.1 loss-of-function mutants arose from a general, nonspecific defect in SPB structure. First, a gain-of-function mutation in the same gene promotes a Plo1-dependent increase in the MPM-2 reactivity of SPBs. Therefore, the loss of recognition in a loss-of-function mutant (cut12.1) is directly mirrored by a gain of reactivity in a gain-of-function mutation in the same gene (cut12.s11). Second, there is an identical correlation between loss and gain of Cut12 function and the loss and gain of Plo1 activity in in vitro Plo1 kinase assays. Thus, we believe that there is a physical and functional link between Plo1 activity and the function of the spindle pole component Cut12. Crucially, the gain-of-function mutation cut12.s11 was only able to promote Plo1-dependent phosphorylation at the SPB at temperatures at which it was also able to suppress cdc25.22. At 37°C, it was not able to either suppress cdc25.22 or enhance Plo1-dependent phosphorylation of the SPB. This direct correlation between the promotion of Plo1 activity and suppression of cdc25.22 suggested that the suppression of cdc25.22 could be a direct consequence of the increase in Plo1 activity. The requirement for full Plo1 function for mitotic commitment in cut12.s11 cdc25.22 cells suggests that this is, indeed, the case. However, although we cannot rule out the possibility that Plo1 may act in a parallel pathway, the body of data that links Cut12 and Plo1 suggests that a function in a parallel pathway is unlikely.
A critical prediction of a model whereby cut12.s11 suppresses cdc25.22 by promoting Plo1 activity is that promoting Plo1 activity independently of cut12 mutations should also promote mitosis in cdc25.22. This was the case. We could induce the same degree of mitotic commitment in arrested cdc25.22 cells by inducing the expression of a constitutively active plo1 allele in the G2 arrested cells as seen with cut12.s11. In other words, enhancing Plo1 activity promoted mitosis. The contrasting delay in mitotic commitment in plo1.ts2 cells suggests that Plo1 is an integral part of the controls that regulate MPF activity during mitotic commitment in fission yeast.
Why are stf mutations not wee?
If cut12.s11 is activating Plo1 to promote the feedback loop, why does it not make cells divide as soon as Mik1 is destabilized upon completion of S phase and so confer a wee- phenotype? When S. pombe strains are shifted to a poorer or richer nutrient source, there is a transient inhibition of cell division (Fantes and Nurse 1977). This delay in mitotic progression is not seen when wee1 mutants are shifted between nutrient sources (Fantes and Nurse 1977; Young and Fantes 1987). Two interpretations have been offered for this behavior. Either wee1 mutants divide below a size threshold, after which they would be able to respond to the nutrient shift, or the nutrient control, which is seen as size control in experimental cultures, acts through Wee1. It is therefore possible that nutrient control stimulates Wee1 activity until cells have attained the critical mass. If this activation can override feedback loop inhibition, which would be a logical hierarchy for the controls, then a constitutively active feedback loop would only be effective in driving mitotic commitment through suppression of Wee1 function once the nutritional/size controls had ceased to stimulate Wee1. Unfortunately, our lack of understanding of the molecular basis for size or nutrient control does not at present permit a direct test of this hypothesis, but it provides a simple explanation as to how the cut12.s11 could suppress cdc25 mutants through polo kinase without conferring a wee- phenotype.
Plo1 association with the SPB
Association of Plo1 with the SPB is clearly complex. We describe three mutants in plo1 in which the affinity for the interphase SPB of cut12.s11 cells is reduced below the detection levels of our assays. The inability of Plo1.K69R to bind the interphase SPB of cut12.s11 suggests that a Plo1-dependent phosphorylation event is required for Plo1 recruitment/maintenance in cut12.s11. It is presently unclear whether these phosphorylation events would overlap with, or be independent of, the Fin1-dependent phosphorylation events that are required for Plo1 recruitment to interphase SPBs in cut12.s11 (Grallert and Hagan 2002). The association of Plo1.ts2 and Plo1.K69R with mitotic SPBs suggests that Plo1 associates with the SPB in more than one manner and probably has multiple docking sites. This would be consistent both with the association of Plo1 with SPB of mitotic cells lacking cut12 (Mulvihill et al. 1999) and the keyrole played by Plo1 in regulating the SPB-associated SIN (Tanaka et al. 2001). Given the number of roles emerging for polo kinases in diverse systems, such as the physical association with the microtubule nucleating protein Spc72 (Ho et al. 2002), it is likely that Plo1 will dock with multiple SPB-associated molecules.
Is mitotic commitment controlled from the spindle pole in other eukaryotes?
Microinjection of antibodies against the human polo kinase Plk1 blocks the G2-to-M transition in nontransformed cells, indicating that Plk1 plays a critical role in regulating mitotic commitment in mammalian cells (Lane and Nigg 1996). Both MPF and polo kinase associate with the spindle poles of species as diverse as yeast and man, suggesting that this association maybe a key element in the regulation of mitotic commitment (Bailly et al. 1989; Alfa et al. 1990; Golsteyn et al. 1995). The acceleration of MPF activation upon injection of centrosomes into enucleated Xenopus eggs would certainly support a role for this organelle in regulating MPF activation (Perez-Mongiovi et al. 2000). Significantly, a recent study has highlighted the importance of an interplay between Plk1 and MPF on the human spindle pole during mitotic commitment (Jackman et al. 2003). Phospho-specific antibodies that recognized residues on cyclin B that have been phosphorylated by either Plk or MPF were used in immunofluorescence studies to identify where MPF was initially activated during mitotic commitment. Both phospho-epitopes appeared first on the centrosome, prior to being seen anywhere else in the cell. Most importantly, phosphorylation of cyclin B by Plk was an early mitotic event that preceded the auto-phosphorylation of cyclin B by MPF. Thus, it would appear likely that commitment to mitosis in higher eukaryotes is also initiated and potentially regulated by events on the spindle pole.
A role for the spindle pole in regulating mitotic commitment mirrors a growing body of data that describes an essential role for the spindle pole in regulating mitotic exit in yeast, flies, and humans (Bardin and Amon 2001; Khodjakov and Rieder 2001; Piel et al. 2001). The microtubule organizing center is therefore organizing more than just the microtubule array and probably acts as a site upon which diverse regulatory pathways converge to coordinate cell division controls. This would ensure that a decision to progress to the next stage of the cycle is not simply the result of accumulating a threshold level of regulatory proteins at random points in the cell. Rather, the decision is taken by a critical subset of molecules at the pole before amplification throughout the cell. Regulation from this one critical location makes it easy to ensure full cross-talk between pathways with minimum effort and error.
Materials and methods
Cell culture and strains
The strains used in this study are listed in Table 3. The plo1.ts mutants were generated by a marker switch approach that leaves a ura4+ gene 235 bp upstream in the promoter (MacIver et al. 2003). This is indicated by plo1.x:ura4+, and the wild-type equivalent allele is plo1+:ura4+. Cells were cultured in yeast extract media (YES), appropriately supplemented EMM2 minimal media as described in Moreno et al. (1991). Thiamine-mediated repression of nmt1+ or nmt41, HU treatment, and size selection were according to Tanaka et al. (2001).
Table 3.
Strains used in this study
| Strain number | Genotype | Source |
|---|---|---|
| IH136 | cut7.24 leu1.32 h− | Hagan and Yanagida 1990 |
| IH365 | leu1.32 ura4.d18 h− | Lab stock |
| IH415 | ade6.704 ura4.d18 h− | Lab stock |
| IH422 | ade6.M210/ade6.M216 mei1.102/mei1+ | Lab stock |
| IH571 | cut12.NEGFP leu1.32 ura4.d18 h− | Bridge et al. 1998 |
| IH666 | cut12.s11 leu1.32 h+ | Hudson et al. 1990 |
| IH740 | cdc25.22 cut12.s11 leu1.32 ura4.d18 h+ | Lab stock |
| IH741 | cdc25.22 cut12.s11 ura4.d18 h− | Bridge et al. 1998 |
| IH470 | cdc2.33 ade6.704 leu1.32 ura4.d18 h− | This study |
| IH1899 | cdc2.33 ade12.s11 leu1.32 ura4.d18 h− | This study |
| IH754 | cut12.1 ade6.704 ura4.d18 h− | Bridge et al. 1998 |
| IH1625 | plo1.ts2:ura4+ leu1.32 ura4.d18 h− | This study |
| IH1636 | plo1.ts19:ura4+ leu1.32 ura4.d18 ade6.M210 h− | This study |
| IH1963 | cdc25.22 leu1.32 ura4.d18 h− | Fantes 1981 |
| IH2122 | plo1.ts19:ura4+ cdc25.22 cut12.s11 ura4.d18 h− | This study |
| IH2156 | plo1.ts2:ura4+ cdc25.22 cut12.s11 ura4.d18 | This study |
| IH2311 | plo1.ts2:ura4+ cut12.s11 ura4.d18 | This study |
| IH2444 | ura4.d18 leu1.32 h− pREP81 PK-N empty vector | This study |
| IH2230 | ura4.d18 leu1.32 h− pREP81-PK-N-plo19T | This study |
| IH2445 | ura4.d18 leu1.32 h− pREP81 PK-N plo1+ | This study |
| IH2514 | plo1.ts19:ura4+ leu1::plo1+ ura4.d18 | This study |
| IH2515 | plo1.ts2:ura4+ leu1::plo1+ ura4.d18 | This study |
| IH2550 | cut12.s11 leu1.32 h+ pREP81-Pk-N-plo1+ | This study |
| IH2551 | cut12.s11 leu1.32 h+ pREP81-PK-N-ploK69R | This study |
| IH2613 | plo1+:ura4+ cdc25.22 cut12.s11 ura4.d18 leu1.32 | This study |
| IH2893 | leu1::plo1.S124DT197D(nmt41):ura4+ leu1.32 ade6.704 ura4.d18 | This study |
| IH2899 | leu1::plo1.S124DT197D(nmt41):ura4+ cdc25.22 ura4.d18 leu1.32 | This study |
| IH2928 | leu1::plo1.S124DT197D(nmt41):ura4+ cdc2.33 ura4.d18 ade6 | This study |
Molecular genetics
The Quick Change Mutagenesis (Stratagene) approach along with appropriate primers was used to introduce a stop codon and an NdeI restriction site at amino acid position 316 of the plo1+ reading frame in pHN180 to generate pPLO316TNdeI. The NsiI–BglII restriction fragment from pPLO316TNdeI was inserted into p81NPkplo1+ to generate pPK-Nplo316NdeRep81 before subcloning of the 316-amino-acid NdeI fragment from this plasmid into pRep81-Pk-N to generate pRep81-PK-Nplo19T. Two-hybrid screening using the restriction fragments indicated in Figure 1B and C was according to James et al. (1996). Then S124D and T197D were changed into HA-tagged plo1 in pINT5-41 to make a constitutively active allele of plo1 referred to as plo1.S124DT197D or plo1.con. The NdeI–SmaI fragment of plo1-con, lacking the HA tag, and the SmaI–SmaI plo1.con fragment were cloned into pINT6-41 (Drummond and Hagan 1998). The subsequent plo1.con-containing NotI fragment was integrated into the leu1 locus of IH415 with plo1.con under the control of the nmt41 promoter.
Microscopy
Plo1 and anti-α-tubulin immunofluorescence procedures were as in Grallert and Hagan (2002) and used anti-Sad1 AP9.2, TAT1, and HN184 (Woods et al. 1989; Hagan and Yanagida 1995; Ohkura et al. 1995). For MPM-2, staining fixation in 3.75% fresh formaldehyde was for 2 min. At least 200 cells were counted for each time point.
Biochemistry
Protein extracts for Western blotting samples were prepared following precipitation with trichloro acetic acid (TCA). A total of 5 × 107 cells were washed and resuspended in stop buffer (Moreno et al. 1991), and TCA was added to a final concentration of 20%. After lysis in a Ribolyser (Hybaid), cells were resuspended in SDS sample buffer. Western blotting with anti-plo1 HN184 sera (Ohkura et al. 1995) and Plo1 kinase assays with HN184 were performed as described previously with the modifications outlined below (Tanaka et al. 2001). For assays following precipitation with MAb336 (Serotec) antibody against the Pk epitope, or to detect coimmunoprecipitation of Plo1 and Cut12, Dynabeads (Dynal) rather than Sepharose beads were used. The antibody was covalently coupled to beads according to the manufacturer's instructions. Following the immunoprecipitation, the immunocomplex was washed once in WB1 (50 mM HEPES at pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100), twice in WB2 (50 mM HEPES at pH 7.5, 1 mM EDTA), and resuspended in CK buffer (Tanaka et al. 2001) before being run on a gel or used in a kinase assay. Cut12 was detected with antibodies as described in Bridge et al. (1998). Histone H1 kinase assays were performed according to the protocol of Creanor and Mitchison (1994).
Acknowledgments
We thank Keith Gull and Hiro Ohkura for antibodies; members of the Cell Division group and Elmar Schiebel for stimulating discussions; and Alan Bridge for critical reading of the manuscript. This work was supported by Cancer Research UK.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.256003.
References
- Alfa C.E., Ducommun, B., Beach, D., and Hyams, J.S. 1990. Distinct nuclear and spindle pole body populations of cyclin-cdc2 in fission yeast. Nature 347: 680–682. [DOI] [PubMed] [Google Scholar]
- Baber-Furani B.A., Rhind, N., Boddy, M.N., Shanahan, P., Lopez-Girona, A., and Russell, P. 2000. Regulation of mitotic inhibitor Mik1 helps to enforce the DNA damage checkpoint. Mol. Biol. Cell 11: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly E., Doree, M., Nurse, P., and Bornens, M. 1989. p34cdc2 is located in both nucleus and cytoplasm—Part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J. 8: 3985–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A.J. and Amon, A. 2001. MEN and SIN: What's the difference? Nat. Rev. Mol. Cell Biol. 2: 815–826. [DOI] [PubMed] [Google Scholar]
- Bartholomew C.R., Woo, S.H., Chung, Y.S., Jones, C., and Hardy, C.F. 2001. Cdc5 interacts with the Wee1 kinase in budding yeast. Mol. Cell. Biol. 21: 4949–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge A.J., Morphew, M., Bartlett, R., and Hagan, I.M. 1998. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes & Dev. 12: 927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen P.U., Bentley, N.J., Martinho, R.G., Nielsen, O., and Carr, A.M. 2000. Mik1 levels accumulate in S phase and may mediate an intrinsic link between S phase and mitosis. Proc. Natl. Acad. Sci. 97: 2579–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creanor J. and Mitchison, J.M. 1994. The kinetics of H1 kinase activation during the cell cycle of wild type and wee mutants of the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 107: 1197–1204. [DOI] [PubMed] [Google Scholar]
- Davies F.M., Tsao, T.Y., Fowler, S.K., and Rao, P. 1983. Monoclonal antibodies to mitotic cells. Proc. Natl. Acad. Sci. 80: 2962–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottingnies A., Zarzov, P., and Nurse, P. 2001. In vivo localisation of fission yeast cyclin-dependent kinase cdc2p and cyclin B cdc13p during mitosis and meiosis. J. Cell Sci. 114: 2627–2640. [DOI] [PubMed] [Google Scholar]
- Drummond D.R. and Hagan, I.M. 1998. Mutations in the bimC box of Cut7 indicate divergence of regulation within the bimC family of kinesin related proteins. J. Cell Sci. 111: 853–865. [DOI] [PubMed] [Google Scholar]
- Fantes P. and Nurse, P. 1977. Control of cell size at division in fission yeast by growth-modulated size control over nuclear division. Exp. Cell Res. 107: 377–386. [DOI] [PubMed] [Google Scholar]
- Golsteyn R.M., Mundt, K.E., Fry, A.M., and Nigg, E.A. 1995. Cell-cycle regulation of the activity and subcellular-localization of Plk1, a human protein-kinase implicated in mitotic spindle function. J. Cell Biol. 129: 1617–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A. and Hagan, I.M. 2002. Schizosaccharomyces pombe NIMA-related kinase Fin1, regulates spindle formation and an affinity of Polo for the SPB. EMBO J. 21: 3096–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I. and Yanagida, M. 1990. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature 347: 563–566. [DOI] [PubMed] [Google Scholar]
- ____. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129: 1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne D.C. and Leupold, U. 1974. Suppressor mutations in yeast. Curr. Topics Micro. Immuno. 64: 1–47. [DOI] [PubMed] [Google Scholar]
- Hirano T., Funahashi, S., Uemura, T., and Yanagida, M. 1986. Isolation and characterization of Schizosaccharomyces pombe cut mutants that block nuclear division but not cytokinesis. EMBO J. 5: 2973–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae bymass spectrometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Hoffmann I., Clarke, P.R., Marcote, M.J., Karsenti, E., and Draetta, G. 1993. Phosphorylation and activation of human cdc25-C bycdc2-cyclin B and its involvement in the self amplification of MPF at mitosis. EMBO J. 12: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Tanaka, H., Ohba, Y., and Yasuda, H. 1995. Mouse p87Wee1 kinase is regulated by M-phase specific phosphorylation. Chrom. Res. 3: 300–308. [DOI] [PubMed] [Google Scholar]
- Hudson J.D., Feilotter, H., and Young, P.G. 1990. stf1: Non wee mutations epistatic to cdc25 in the fission yeast Schizosaccharomyces pombe. Genetics 126: 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T. and Maller, J.L. 1993. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol. Biol. Cell 4: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1995. Phosphorylation and activation of Xenopus Cdc25 phosphatase in the absence of Cdc2 and Cdk2 kinase activity. Mol. Biol. Cell 6: 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M., Lindon, C., Nigg, E.A., and Pines, J. 2003. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 5: 143–148. [DOI] [PubMed] [Google Scholar]
- James P., Halladay, J., and Craig, E.A. 1996. Genomic libraries and a host strain designed for highly efficient two hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaiskou A., Jessus, C., Brassac, T., and Ozon, R. 1999. Phosphatase 2A and Polo kinase, two antagonistic regulators of Cdc25 activation and MPF auto-amplification. J. Cell Sci. 112: 3747–3756. [DOI] [PubMed] [Google Scholar]
- Khodjakov A. and Rieder, C.L. 2001. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153: 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M. and Schiebel, E. 1998. Receptors determine the cellular localization of a γ-tubulin complex and there by the site of microtubule formation. EMBO J. 17: 3952–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovelman R. and Russell, P. 1996. Stockpiling of Cdc25 during a DNA-replication checkpoint arrest in Schizosaccharomyces pombe. Mol. Cell. Biol. 16: 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang J., Ashorn, C.L., Gonzalea-Kuyvenhoven, M., and Penkala, J.E. 1994. cdc25 is one of the MPM-2 antigens involved in the activation of maturation-promoting-factor. Mol. Biol. Cell 5: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A. and Dunphy, W.G. 1996. Purification and molecular-cloning of Plx1, a Cdc25-regulatorykinase from Xenopus egg extracts. Science 273: 1377–1380. [DOI] [PubMed] [Google Scholar]
- Lane H.A. and Nigg, E.A. 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135: 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver F.H., Glover, D.M., and Hagan, I.M. 2003. A `marker switch' approach for targeted mutagenesis of genes in Schizosaccharomyces pombe. Yeast 20: 587–594. [DOI] [PubMed] [Google Scholar]
- Mitchison J.M. and Nurse, P. 1985. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 75: 357–376. [DOI] [PubMed] [Google Scholar]
- Moreno S., Hayles, J., and Nurse, P. 1989. Regulation of p34cdc2 protein-kinase during mitosis. Cell 58: 361–372. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar, A., and Nurse, P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth. Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Mueller P.R., Coleman, T.R., and Dunphy, W.G. 1995. Cell-cycle regulation of a Xenopus Wee1-like kinase. Mol. Biol. Cell 6: 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill D.P., Petersen, J., Ohkura, H., Glover, D.M., and Hagan, I.M. 1999. Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol. Biol. Cell 10: 2771–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503–508. [DOI] [PubMed] [Google Scholar]
- Ohkura H., Hagan, I.M., and Glover, D.M. 1995. The conserved Schizosaccharomyces pombe kinase Plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes & Dev. 9: 1059–1073. [DOI] [PubMed] [Google Scholar]
- Perez-Mongiovi D., Beckhelling, C., Chang, P., Ford, C.C., and Houliston, E. 2000. Nuclei and microtubule asters stimulate maturation/M phase promoting factor (MPF) activation in Xenopus eggs and egg cytoplasmic extracts. J. Cell Biol. 150: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M., Nordberg, J., Euteneuer, U., and Bornens, M. 2001. Centrosome dependent exit of cytokinesis in animal cells. Science 291: 1550–1553. [DOI] [PubMed] [Google Scholar]
- Qian Y.W., Erikson, E., Li, C., and Maller, J.L. 1998. Activated polo-like kinase Plx1 is required at multiple points during mitosis in Xenopus laevis. Mol. Cell. Biochem. 18: 4262–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y.W., Erikson, E., and Maller, J.L. 1999. Mitotic effects of a constitutively active mutant of the Xenopus polo-like kinase Plx1. Mol. Cell. Biol. 19: 8625–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N. and Ohkura, H. 2003. Polo boxes form a single functional domain that mediates interactions with multiple proteins in fission yeast polo kinase. J. Cell Sci. 116: 1377–1387. [DOI] [PubMed] [Google Scholar]
- Strausfeld U., Labbe, J.C., Fesquet, D., Cavadore, J.C., Picard, A., Sadhu, K., Russell, P., and Doree, M. 1992. Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by a human CDC25 protein. Nature 351: 242–245. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Petersen, J., MacIver, F.H., Mulvihill, D.P., Glover, D.M., and Hagan, I.M. 2001. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J. 20: 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z.H., Coleman, T.R., and Dunphy, W.G. 1993. 2 distinct mechanisms for negative regulation of the Wee1 protein-kinase. EMBO J. 12: 3427–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandre D.D., Davis, F.M., Rao, P.N., and Borisy, G.G. 1984. Phosphoproteins are components of mitotic microtubule organizing centers. Proc. Natl. Acad. Sci. 81: 4439–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S. and Uhlmann, F. 2002. Chromosome segregation: Playing polo in prophase. Dev. Cell 2: 381–382. [DOI] [PubMed] [Google Scholar]
- Woods A., Sherwin, T., Sasse, R., Macrae, T.H., Baines, A.J., and Gull, K. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal-antibodies. J. Cell Sci. 93: 491–500. [DOI] [PubMed] [Google Scholar]
- Young P.G. and Fantes, P.A. 1987. Schizosaccharomyces pombe mutants affected in their division response to starvation. J. Cell Sci. 88: 295–304. [DOI] [PubMed] [Google Scholar]