Abstract
Meiosis is thought to require the protein kinase Ime2 early for DNA replication and the cyclin-dependent kinase Cdc28 late for chromosome segregation. To elucidate the roles of these kinases, we inhibited their activities early and late using conditional mutants that are sensitive to chemical inhibitors. Our studies reveal that both Cdc28 and Ime2 have critical roles in meiotic S phase and M phase. Early inhibition of analog-sensitive cdc28-as1 blocked DNA replication, revealing a previously undetected role for Cdc28. Yet Cdc28 was dispensable for one of its functions in the mitotic cell cycle, degradation of Sic1. Late addition of inhibitor to ime2-as1 revealed unexpected roles of Ime2 in the initiation and execution of chromosome segregation. The requirement of Ime2 for M phase is partially explained by its stimulation of the key meiotic transcription factor Ndt80, which is needed in turn for high Cdc28 activity. In accordance with a late role for Ime2, we observed an increase in its activity during M phase that depended on Cdc28 and Ndt80. We speculate that several unique features of the meiotic cell division reflect a division of labor and regulatory coordination between Ime2 and Cdc28.
Keywords: Meiosis, cell cycle, Ndt80, chemical genetics, transcriptional regulation, Saccharomyces cerevisiae
Meiosis is a specialized cell division that produces haploid cells needed for sexual reproduction. Successful completion of meiosis requires several sequential landmark events: DNA replication, recombination and synapsis of homologs, and chromosome segregation. Chromosome segregation occurs in two successive meiotic divisions to yield four haploid meiotic products that are packaged into gametes. Progression through meiosis requires three major decisions. The first is whether to enter the meiotic differentiation program or continue with mitotic proliferation. The second is whether to embark on chromosome preparation, including meiotic S phase, and then recombination and synapsis of homologs. The third decision, whether to proceed into M phase, depends on the successful completion of replication and recombination. Transcription factors and protein kinases are central components of the pathways that control these key decisions. In particular, the transcription factor Ime1 and the kinase Ime2 are thought to participate in the G1–S transition, whereas the transcription factor Ndt80 and the kinase Cdc28 take part in the G2–M transition.
Intertwined with the landmark events of meiosis is a well-characterized transcriptional cascade (Chu et al. 1998; Primig et al. 2000). Ime1 stimulates transcription of the early class of genes and thus is required for entry into the meiotic program and for the meiotic G1–S transition (Vershon and Pierce 2000). An important target of Ime1 is IME2, which encodes a serine–threonine protein kinase (Mitchell et al. 1990). Ime2 is required for meiotic DNA replication (Foiani et al. 1996) and appears to control the G1–S transition by decreasing the level of Sic1 (Dirick et al. 1998), an inhibitor of the cyclin-dependent kinase (CDK) Cdc28 in the mitotic cell cycle (Mendenhall and Hodge 1998). It has been proposed that Ime2-dependent degradation of Sic1 leads to the activation of a CDK-like kinase (possibly Cdc28 itself) associated with the B-type cyclins Clb5 and Clb6, and that this CDK activity triggers meiotic DNA replication (Dirick et al. 1998; Stuart and Wittenberg 1998). Ime2 may play additional roles in initiating DNA replication as well (Guttmann-Raviv et al. 2001). It is also required for normal accumulation of early gene transcripts, destabilization of Ime1, and transcription of NDT80 (Mitchell et al. 1990; Hepworth et al. 1998; Guttmann-Raviv et al. 2002).
Ndt80 stimulates transcription of ∼150 middle genes, including its own gene and genes required for meiotic nuclear divisions (e.g., CLB1) and spore morphogenesis (e.g., SMK1; Chu and Herskowitz 1998; Chu et al. 1998; Hepworth et al. 1998). Cells lacking NDT80 arrest in the pachytene stage of meiotic G2 like cells depleted of Cdc28 activity (Xu et al. 1995), suggesting that Clb activators of Cdc28 are vital targets of Ndt80 regulation. Ndt80 activity appears to be a highly regulated component of the G2–M decision and a target of the pachytene checkpoint. When the pachytene checkpoint is activated by incomplete or defective chromosome preparation, cells arrest before M phase, contain Ndt80 that is under-phosphorylated and less abundant, and lack transcripts from Ndt80-dependent genes (Lydall et al. 1996; Chu and Herskowitz 1998; Hepworth et al. 1998; Tung et al. 2000). Overexpression of NDT80 partially bypasses the checkpoint arrest (Tung et al. 2000; Pak and Segall 2002b).
Although Cdc28 is essential for the G1–S and G2–M transitions in vegetative cells, its role in meiotic progression has been less clear. Cdc28 is clearly essential for the meiotic G2–M transition: cdc28-ts mutants arrest at the pachytene stage of meiotic G2 (Shuster and Byers 1989; Xu et al. 1995), indicating that Cdc28 is required for M phase and dispensable for S phase. As expected, mutants lacking some of the B-type (Clb) cyclins exhibit a similar arrest in G2 (Grandin and Reed 1993; Dahmann and Futcher 1995). The observation that mutants lacking Clb5 and Clb6 fail to initiate meiotic DNA replication (Dirick et al. 1998; Stuart and Wittenberg 1998) suggests that Cdc28 may be required for S phase in meiosis, as it is in mitosis. Another hint that Cdc28 may play a role in meiotic S phase is the activity of the CDK inhibitor Sic1 in preventing meiotic S phase (Dirick et al. 1998). Studies using cdc28-ts and cdc28-degron mutations have, however, failed to support a role for Cdc28 in meiotic S phase (Shuster and Byers 1989; Guttmann-Raviv et al. 2001). Yet these studies are not conclusive, as meiotic experiments with ts mutants cannot be performed at the fully restrictive temperature because elevated temperatures block sporulation even in wild-type strains.
Recently, the mitotic roles of Cdc28 have been studied using a new kind of conditional mutant that is engineered to be sensitive to chemical inhibition. Substitution of a single conserved amino acid creates an analog-sensitive (as) kinase that is uniquely sensitive to inhibition by cell-permeable purine analogs (Bishop et al. 1999, 2000). Treatment with the inhibitor 1-NM-PP1 prevents cdc28-as1 cells from initiating DNA replication or chromosome segregation, depending on the amount of inhibitor added, thus confirming previous conclusions that Cdc28 is required for both S and M phases in the mitotic cell cycle (Bishop et al. 2000). Analog-sensitive mutants can be used to identify late functions of a protein that also acts early in a process and to inhibit a process without perturbing cells by incubation at high temperatures.
Here we describe the roles and interactions of Cdc28, Ime2, and Ndt80 in meiosis, as revealed by analyses of biochemical and cytological markers of meiotic progression in inhibitor-sensitive and other mutants. Our studies demonstrate that Ime2 and Cdc28 function to govern first the G1–S transition and then the G2–M transition and progression through M. Our evidence provides direct support for the proposal that Cdc28 is essential for meiotic S phase, although it plays no role in Sic1 degradation. Ime2 is required for entry into and progression through meiotic M phase, coincident with a second peak in Ime2 kinase activity dependent on Cdc28 and Ndt80. The M-phase requirement for Ime2 can be partially explained by our demonstration that NDT80 transcription depends on Ime2 throughout M phase and is a key factor limiting progression through meiosis I. Additional late functions of Ime2 include phosphorylation of Ndt80 and perhaps other substrates involved in chromosome segregation.
Results
Cdc28 is required for meiotic S phase
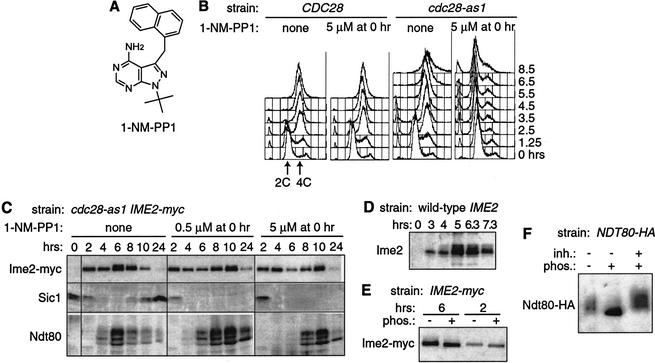
To re-examine whether Cdc28 is necessary for meiotic DNA replication, we exploited the inhibitor-sensitive cdc28-as1 mutant. Previous investigations revealed dose-dependent mitotic cell cycle arrests in cdc28-as1 cells: 0.5 μM 1-NM-PP1 inhibitor causes G2/M arrest; 5 μM causes G1 arrest (Bishop et al. 2000). We found similar arrests in meiosis by adding 1-NM-PP1 (Fig. 1A) to homozygous diploid cdc28-as1 mutants constructed in the fast-sporulating SK1 strain background. Addition of 0.5 μM 1-NM-PP1 to cdc28-as1 cells at the time of transfer to sporulation medium (time 0) did not significantly impair DNA replication but prevented the meiotic divisions and spore formation (data not shown). This G2 arrest is similar to that exhibited by cdc28-ts mutants that arrest in pachytene (Shuster and Byers 1989; Xu et al. 1995). In contrast, treatment of cdc28-as1 cells with high levels of 1-NM-PP1 (5 μM) blocked meiotic DNA replication: the cellular DNA content remained at 2C for more than 8.5 h (Fig. 1B). In the absence of inhibitor, nearly all cells accumulated with 4C DNA content by 3.5 h. The arrest was caused by specific inhibition of Cdc28, because DNA replication was blocked only by the combination of the cdc28-as1 mutation and 5 μM 1-NM-PP1 and not by either condition alone. Several observations indicated that cells arrested in meiosis rather than in the last mitotic division: nearly all cells were in G1 before adding 1-NM-PP1 (unbudded and 2C DNA content), transcription of several meiotic genes occurred normally (data not shown), and cells produced meiosis-specific proteins (see below). Therefore, CDC28 is required for DNA replication in meiosis, just as in the mitotic cell cycle.
Figure 1.
Cdc28 is required for meiotic DNA replication. (A) Chemical structure of 1-NM-PP1. (B) Flow cytometry of wild-type CDC28 (KBY316) and mutant cdc28-as1 (KBY442) cells sporulated with or without 5 μM 1-NM-PP1. DNA contents before (2C) and after (4C) DNA replication are indicated. (C) Proteins from cdc28-as1 cells. Protein extracts from KBY442 strain harvested during sporulation with or without 1-NM-PP1 were immunoblotted with antibodies against Ime2-myc (9E10), Sic1, and Ndt80. (D) Untagged Ime2 from sporulating wild-type (KBY259) cells was visualized with antibodies against Ime2. (E) Protein phosphatase treatment of Ime2-myc. Immunoprecipitates from IME2-myc (KBY316) cells harvested after 2 or 6 h of sporulation were divided and treated with (+) or without (-) λ protein phosphatase (phos.) before immunoblotting with the 9E10 antibody. Note that the phosphorylated forms of Ime2-myc are not resolved as well as those of untagged Ime2, because of the increased size of the tagged protein. (F) Protein phosphatase treatment of Ndt80-HA. Immunoprecipitate from NDT80-HA (KBY332) cells sporulated for 6 h was divided and treated with (+) or without (-) λ protein phosphatase (phos.) and phosphatase inhibitors (inh.) before immunoblotting with the 12CA5 antibody.
We next examined the requirement of Cdc28 for accumulation of Ime2, Sic1, and Ndt80 proteins (Fig. 1C). In the absence of 1-NM-PP1, cdc28-as1 cells exhibited wild-type patterns of protein accumulation and phosphorylation (see below; data not shown). When Cdc28 was active, the level of Sic1 declined upon transfer of cells to sporulation medium, remained low at 4and 6 h, and started to increase at 8 h after the meiotic divisions. When Cdc28-as1 was inhibited, Sic1 levels declined normally, demonstrating that Cdc28 is not needed for Sic1 degradation in meiosis, unlike in the mitotic cell cycle (Mendenhall and Hodge 1998).
During an unperturbed meiosis, Ime2 protein levels were high from 1 to 10 h (Fig. 1C; data not shown). Although its known roles are complete by 4–5 h, Ime2 protein levels increased at 5–6 h concomitant with the appearance of forms with decreased electrophoretic mobility, regardless of whether Ime2 was myc-tagged or untagged (Fig. 1C,D). Treatment of immunoprecipitated Ime2-myc with a protein phosphatase suggested that it had a basal level of phosphorylation at all times and became hyperphosphorylated around 6 h (Fig. 1E). Inhibition of Cdc28-as1 had little effect on the appearance of Ime2-myc at 0–4h. Cdc28 was required, however, for the normal increases in the amount and phosphorylation of Ime2-myc at 6 h (Fig. 1C). In the presence of 1-NM-PP1, changes in Ime2-myc protein were reduced and delayed. Additional studies below showed that Cdc28 was important for Ime2 kinase activity at late times.
Ndt80 protein was produced in a relatively narrow temporal window and exhibited three major electrophoretic forms (Fig. 1C). The lowest band comigrated with recombinant Ndt80 expressed in Escherichia coli (data not shown). Treatment of immunoprecipitated Ndt80-HA with a phosphatase collapsed all electrophoretic forms to a single tight band (Fig. 1F), as described previously (Tung et al. 2000). Inhibition of Cdc28-as1 delayed accumulation of Ndt80 by 2–4h in cells arrested in G1 (5 μM 1-NM-PP1) or G2 (0.5 μM). The posttranslational modifications of Ndt80 appeared normal when Cdc28-as1 was inhibited, demonstrating that Cdc28 is dispensable for Ndt80 phosphorylation (Tung et al. 2000). Clearly, DNA replication is not needed for accumulation of phosphorylated Ndt80 (Fig. 1C) or for transcription of many Ndt80-dependent genes (data not shown), as seen also in clb5 clb6 mutants (Stuart and Wittenberg 1998; Sopko et al. 2002). We conclude that Cdc28 activity is required for meiotic DNA replication, contributes to Ime2 hyperphosphorylation, and ensures the normal timing of Ndt80 production.
Ime2 kinase exhibits two peaks of activity
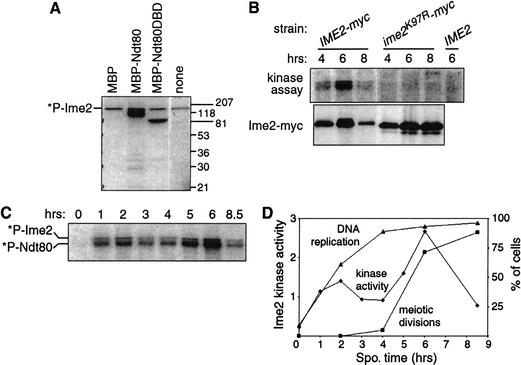
To examine Ime2 kinase activity during meiosis, we developed an assay involving phosphorylation of Ndt80. Ime2-myc immunoprecipitated from meiotic cell extracts phosphorylated chimeric MBP-Ndt80 and MBPNdt80DBD (a DNA-binding fragment of Ndt80; E. Jolly and I. Herskowitz, unpubl.), but not MBP alone (Fig. 2A). Ime2 also phosphorylated itself, as previously reported (Kominami et al. 1993). Additional experiments suggested that Ndt80 is a genuine physiological substrate of Ime2 (see below).
Figure 2.
Ime2 phosphorylates Ndt80 and exhibits a second peak of activity. (A) Activity assay for Ime2. Ime2-myc was immunoprecipitated from KBY316 cells harvested after 5 h of sporulation and incubated with [γ-32P]ATP in the absence (none) or presence of additional proteins purified from E. coli: MBP alone or fused to full-length Ndt80 (MBP-Ndt80) or to a DNA-binding domain of Ndt80 (MBP-Ndt80DBD). Incorporation of radioactivity into Ime2-myc by autophosphorylation is indicated (*P-Ime2). Electrophoretic resolution of phosphorylated forms of Ndt80 was limited by the large MBP tag and the gel conditions. Protein size markers are indicated on the right. (B) Requirement of Ime2 catalytic activity for incorporation of radioactivity. IME2-myc (KBY316), ime2K97R-myc (KBY450), and IME2 (KBY259) cells were processed for the kinase assay with MBP-Ndt80 substrate (top) or for immunoblotting with the 9E10 antibody (bottom). (C) Change in Ime2 activity during meiosis. Cells (KBY316) harvested after 0–8.5 h were assayed for kinase activity using MBP-Ndt80 substrate. Incorporation of radioactivity into MBP-Ndt80 (*P-Ndt80) and Ime2-myc (*P-Ime2) is indicated. (D) Timing of Ime2 activity, meiotic DNA replication, and meiotic divisions. The kinase activity quantified with a PhosphorImager from C is shown in arbitrary units. The percentages of cells containing more than 2C DNA content (DNA replication) or more than one DNA mass (meiotic divisions) are shown.
To determine whether Ime2 is responsible for the phosphorylation of Ndt80, we tested the activity of mutant Ime2 protein with its putative catalytic Lys 97 changed to arginine. Such a mutation abolishes Ime2 function in vivo (Sia and Mitchell 1995; Guttmann-Raviv et al. 2002). Kinase assays with an ime2K97R mutant yielded only background levels of phosphorylation, similar to results with an untagged IME2 strain (Fig. 2B). Thus this assay specifically measures Ime2 kinase activity.
Next we measured changes in Ime2 kinase activity during meiosis using Ndt80. Activity increased dramatically after transfer to sporulation medium, declined after 2 h, and then exhibited a second peak around 6 h before decreasing again (Fig. 2C). Ime2 activity is thus biphasic during meiosis. Samples taken from a single culture and assayed for DNA replication, meiotic divisions, and Ime2 kinase activity revealed that the first activity peak coincided with the start of DNA replication and the second peak coincided with the meiotic divisions (Fig. 2D). The same temporal profile of Ime2 kinase activity was observed using MBP-Ndt80, histone H1, or Ime2 itself as the substrate (data not shown). Concomitant with the second peak, the amount and phosphorylation of Ime2 protein increased (Fig. 1C; see below).
The second peak of Ime2 activity depends on Ndt80 and Cdc28
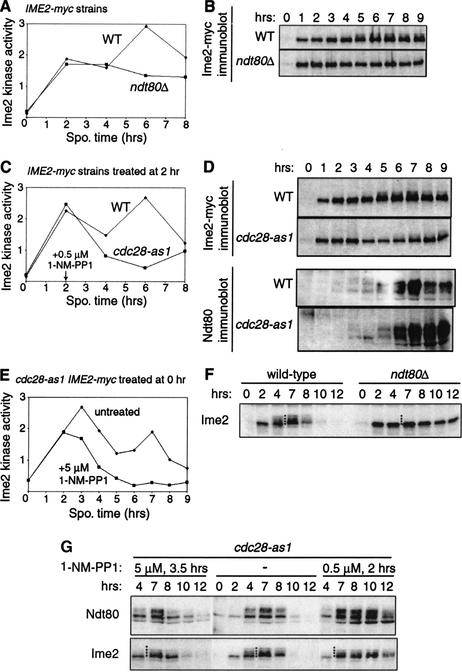
To determine if the second peak occurs in meiotic G2 or M, Ime2 kinase activity was examined in IME2-myc strains arrested in meiotic G2 because of cdc28 or ndt80 mutations (Shuster and Byers 1989; Xu et al. 1995). Deletion of NDT80 did not impair Ime2 activity at early times in meiosis but prevented the second peak of activity (Fig. 3A). Immunoblotting of samples from the same cultures showed that the modified forms of Ime2-myc seen in the wild-type strain after 5 h were absent in the ndt80Δ mutant (Fig. 3B; see also Fig. 3F). Hence, the late changes in Ime2 activity and phosphorylation depended on Ndt80.
Figure 3.
The second peak of Ime2 activity occurs after pachytene. (A,B) Dependence of Ime2 kinase activity and protein on NDT80. Mutant ndt80Δ (KBY430) and wild-type (WT, KBY316) cells were harvested for the Ime2 kinase assay (A) and immunoblotting with 9E10 (B). (C–E) Requirement of Cdc28 for Ime2 activity and for Ime2 and Ndt80 proteins. Mutant cdc28-as1 (KBY442) and wild-type (WT, KBY316) strains were sporulated and treated with 0.5 μM 1-NM-PP1 at 2 h (C,D) or 5 μM 1-NM-PP1 at 0 h (E). Samples were examined by the Ime2 kinase assay (C,E) or immunoblotting to visualize Ime2-myc and Ndt80 (D). (F,G) Modification of untagged Ime2 in wild-type (KBY259), ndt80Δ (KBY427), and cdc28-as1 (KBY439) cells. The cdc28-as1 mutant was treated with 1-NM-PP1 as indicated. Multiple forms of Ime2 are indicated by a column of dots placed between 4and 7 h.
We used the cdc28-as1 mutant to examine the dependence of Ime2 activity on Cdc28 (Bishop et al. 2000). Arrest of cells in G2 by addition of 0.5 μM 1-NM-PP1 at 2 h reduced and delayed the second peak of Ime2-myc activity (Fig. 3C). Parallel immunoblot analysis showed delayed and reduced accumulation of hyperphosphorylated Ime2-myc (Fig. 3D; see also Fig. 3G) but did not diminish the production of phosphorylated Ndt80. To determine if the early peak of Ime2 activity also requires Cdc28, we assayed kinase activity in cdc28-as1 cells treated with high (5 μM) 1-NM-PP1 at time 0. Although DNA replication was blocked, Ime2 kinase activity increased substantially by 2 h before declining (Fig. 3E). Indirect measures of Ime2 activity confirmed that Cdc28 was not essential for the early functions of Ime2: degradation of Sic1 and production of phosphorylated Ndt80 depended on Ime2 (see below) yet occurred even when Cdc28 was strongly inhibited early in meiosis (Fig. 1C). Thus, Cdc28 is required for Ime2 activity late but not early in meiosis.
To better resolve the forms of Ime2, we evaluated phosphorylation of Ime2 in strains with untagged Ime2 rather Ime2-myc. In the wild-type strain (Fig. 3F) and the untreated cdc28-as1 strain (Fig. 3G), a single form of Ime2 was observed early in meiosis and additional reduced-mobility forms of Ime2 appeared at 4h. The events of sporulation occurred earlier in this experiment than in previous experiments because of day-to-day variation. Deletion of NDT80 prevented the late changes in Ime2 (Fig. 3F). In contrast, arrest of cdc28-as1 cells in G2 delayed the accumulation of reduced-mobility forms of Ime2 (from 4h to 7 h) and caused subtle but reproducible changes in Ime2 mobility (Fig. 3G), suggesting that Cdc28 is required for some modifications of Ime2 and dispensable for others. Thus, Ndt80 may stimulate late changes in Ime2 by Cdc28-dependent and Cdc28-independent mechanisms.
The dependence of Ime2 activity on NDT80 and CDC28 demonstrates that the second peak in kinase activity occurs after pachytene. In addition, we observed a correspondence between the second peak of activity, increased amounts of Ime2, and altered Ime2 phosphorylation. A peak in Ime2 activity during the meiotic divisions suggests that Ime2 may play a role late in sporulation. We used ime2-as1 mutants to investigate such a role.
Mutant Ime2-as1 can be specifically inhibited
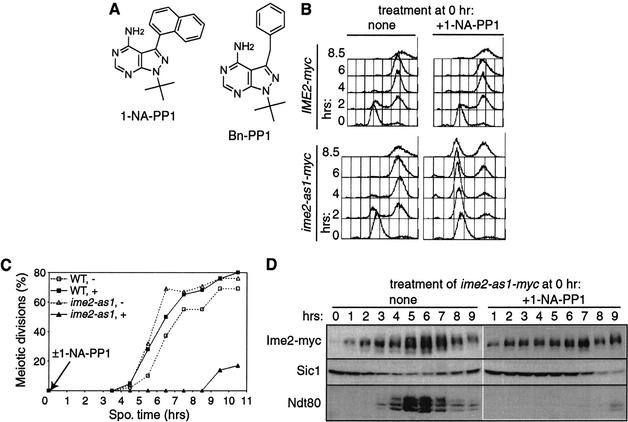
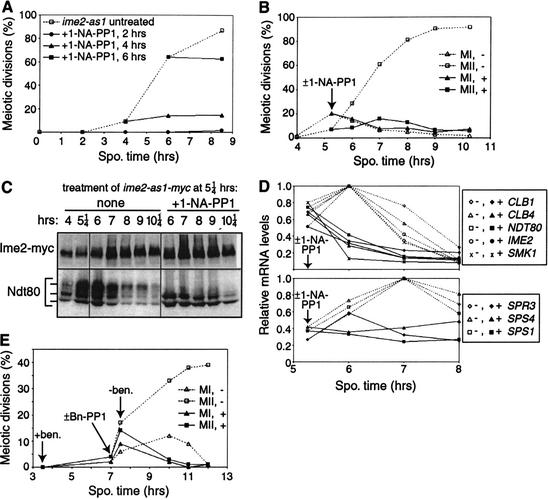
An Ime2 kinase sensitive to the inhibitors 1-NA-PP1 and Bn-PP1 was created by mutating Met 146 to glycine (Fig. 4A). In the absence of inhibitor, a homozygous diploid ime2-as1 mutant appeared normal during vegetative growth, meiotic DNA replication (Fig. 4B), meiotic nuclear divisions (Fig. 4C), and spore formation (81.4% ime2-as1 asci vs. 81.8% IME2 asci). Addition of 20 μM 1-NA-PP1 or 10 μM Bn-PP1 severely impaired sporulation of an ime2-as1 strain but had no effect on a wild-type strain. Mutant ime2-as1 cells treated with 20 μM 1-NA-PP1 at 0 h exhibited a delay in DNA replication of more than 6.5 h (Fig. 4B), similar to an ime2K97R kinase-inactive mutant (data not shown) and almost as severe as ime2Δ strains (Foiani et al. 1996; Dirick et al. 1998). Inhibition of Ime2-as1 also blocked the meiotic divisions (Fig. 4C) and impaired spore formation.
Figure 4.
The ime2-as1 mutation confers sensitivity to 1-NA-PP1. (A) Chemical structures of 1-NA-PP1 and Bn-PP1. (B–D) Wild-type (WT, KBY316) and mutant ime2-as1 (KBY518) cells sporulated in the presence (+) or absence (-) of 20 μM 1-NA-PP1 were harvested for flow cytometry (B), DAPI staining (C), and immunoblotting (D). (C) Meiotic divisions are shown as the percentage of cells with more than one DAPI-staining body. (D) Protein extracts were immunoblotted with antibodies against Ime2-myc (9E10), Sic1, and Ndt80.
When Ime2-as1 was inhibited, Ime2 protein accumulated normally but did not exhibit wild-type hyperphosphorylation at 5–7 h (Fig. 4D). Sic1 degradation and Ndt80 accumulation were delayed by about 6 h (Fig. 4D). In summary, treatment of ime2-as1 cells with 1-NA-PP1 at time 0 caused meiotic defects similar to those of an ime2Δ mutant (Foiani et al. 1996; Dirick et al. 1998; Hepworth et al. 1998). Treatment with Bn-PP1 appeared to inhibit Ime2-as1 slightly better than 1-NA-PP1 and had little effect on wild-type cells (data not shown). We conclude that 1-NA-PP1 and Bn-PP1 inhibit Ime2-as1 efficiently and specifically.
Ime2 is required for the meiotic nuclear divisions
To determine the roles of Ime2 in meiosis, we inhibited the ime2-as1 strain at various times and measured cytological and molecular markers of meiotic progression. Treatment of ime2-as1 cells with 20 μM 1-NA-PP1 blocked the meiotic divisions (Fig. 5A). Even when 1-NA-PP1 was added at 4or 6 h, after DNA replication was completed and when some nuclei had begun to divide, no additional meiotic divisions were observed. To study the role of Ime2 at late times, we added 1-NA-PP1 at 5.25 h and assessed meiotic divisions (Fig. 5B), Ime2 and Ndt80 proteins (Fig. 5C), and mRNA levels (Fig. 5D) from a single culture. Before addition of 1-NA-PP1, 7% of cells had undergone both MI and MII and thus contained four bright DAPI-stained bodies of approximately equal size. Inhibition of Ime2-as1 reduced subsequent meiotic divisions 10-fold: an additional 9.0% of cells manifested a typical MII pattern in contrast to 84.5% in the absence of inhibitor (Fig. 5B). If only MII were inhibited, cells would accumulate after MI with two DAPI bodies. Instead, the number of MI cells decreased when Ime2 was inhibited (Fig. 5B) or remained constant (data not shown). Hence, Ime2 activity is essential for MI and may be necessary for MII as well.
Figure 5.
Ime2 is required for the meiotic nuclear divisions. (A) Meiotic divisions in ime2-as1 (KBY518) cells treated with 20 μM 1-NA-PP1 at 2, 4, or 6 h of sporulation. The percentage of cells in each sample with more than one DAPI-staining body is shown. (B–D) Sporulating ime2-as1 (KBY518) cells were treated with (+) or without (-) 20 μM 1-NA-PP1 at 5.25 h and harvested at 4–10.25 h of sporulation for analysis. (B) DAPI-stained cells were scored for the occurrence of meiosis I (MI, 2 DAPI bodies) and meiosis II (MII, 4 DAPI bodies). (C) Protein extracts were immunoblotted with antibodies against Ime2-as1-myc (9E10) and Ndt80. (D) Northern blots were probed to visualize IME2; NDT80; the middle sporulation genes CLB1, CLB4, SMK1, SPS1, SPS4, and SPR3; and the loading control PFY1; then quantitated. Normalized transcript levels are shown. Before 1-NA-PP1 was added, transcript levels of all genes (except PFY1) had increased at least 30-fold relative to the 0-h time point. (E) Sporulating ime2-as1 (KBY518) cells were incubated with 60 μg/mL benomyl at 3.5 h and treated with (+) or without (-) 10 μM Bn-PP1 at 7 h. Cells were washed free of benomyl at 7.5 h and resuspended in medium with or without Bn-PP1. DAPI-stained cells were scored as in B.
Inhibition of Ime2-as1 at 5.25 h had a dramatic effect on phosphorylation of Ndt80 and transcription of Ndt80-dependent genes. After addition of 1-NA-PP1, the uppermost form of Ndt80 disappeared within 15 min, and the mobility of Ime2 increased slightly (Fig. 5C; data not shown). The transcriptional consequences of inhibiting Ime2 were revealed by examination of transcripts from IME2, NDT80, and six middle sporulation genes—;CLB1, CLB4, SMK1, SPS1, SPS4, and SPR3 (Fig. 5D). Cells had high levels of these transcripts by 5.25 h before 1-NA-PP1 was added. Transcript levels continued to increase and peaked at 6 or 7 h in untreated cells, but plateaued or decreased in cells treated with 1-NA-PP1. Ime2-deficient cells had 25%–50% of the normal amounts of transcripts at 6–7 h. We conclude that continued activity of Ime2 throughout M phase is necessary for maintenance of phosphorylated Ndt80 and for full transcription of middle sporulation genes. Because Ndt80 is essential for transcription of all genes tested except IME2 (Chu and Herskowitz 1998; Chu et al. 1998; Hepworth et al. 1998), our observations suggest that Ime2 may augment Ndt80 activity. The M-phase requirement for Ime2 may be partially due to its actions on Ndt80.
To temporally delimit the late role(s) of Ime2, we tested if Ime2 activity was required after metaphase I using a benomyl release experiment. Sporulating ime2-as1 cells were treated with the microtubule-depolymerizing drug benomyl (60 μg/mL added at 3.5 h) to cause arrest in metaphase I. Half of the culture was treated with 10 μM Bn-PP1 at 7 h to inhibit Ime2-as1 activity for 30 min, after which cells were washed and resuspended in benomyl-free sporulation medium with or without Bn-PP1. When Ime2 was inhibited, no meiotic divisions occurred after release from benomyl arrest, unlike the untreated strain in which nearly 40% of cells completed both meiotic divisions by 12 h (Fig. 5E). Similar results were obtained after treatment of the cdc28-as1 mutant with 1-NM-PP1 (data not shown), confirming that Cdc28 is required for meiosis II (Shuster and Byers 1989). Thus, Ime2, like Cdc28, was required after metaphase I.
Ime2 plays late meiotic roles in addition to promoting NDT80 transcription
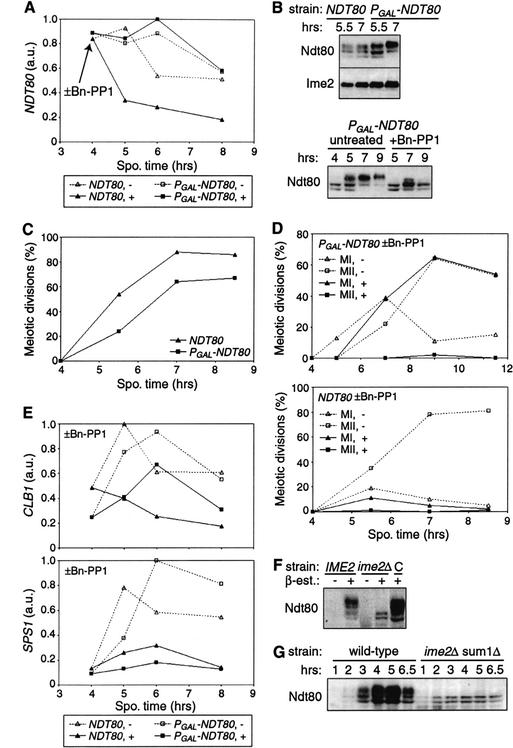
The finding that activity of Ime2 during the meiotic divisions was required for normal levels of NDT80 and other middle gene transcripts (Fig. 5D) raised the possibility that the primary late role of Ime2 is to promote transcription of NDT80. To test whether Ime2 plays additional roles in meiotic divisions, we replaced the native upstream regulatory sequence of chromosomal NDT80 with a regulatory region that allowed inducible, Ime2-independent transcriptional control. The GAL1,10 UAS was placed upstream of NDT80 and controlled by a chimeric transcription factor, Gal4-ER, that activates transcription only in the presence of estradiol (Picard 1999). Addition of 1 μM β-estradiol to PGAL-NDT80 GAL4-ER cells at 3 h stimulated production of NDT80 mRNA (Fig. 6A) and Ndt80 protein (Fig. 6B) to nearly wild-type levels. By 8.5 h, 49% of the PGAL-NDT80 GAL4-ER cells had completed meiosis II, and an additional 18% had completed meiosis I, compared with 81% and 5%, respectively, for an NDT80 strain (Fig. 6C). Ndt80 production was sufficient for 20% of cells to form mature asci after 40 h (compared with 81% wild-type asci). In the absence of β-estradiol, these cells were unable to execute the meiotic divisions or spore formation.
Figure 6.
Increased expression of NDT80 partially rescues the meiotic division defect of Ime2-deficient cells. (A–E) Transcription from PGAL-NDT80 was induced at 3 h by addition of 0.5 μM β-estradiol to sporulating NDT80 ime2-as1 (KBY518) and PGAL-NDT80 GAL4-ER ime2-as1 (KBY649) cells. At 4 h, cultures were split and treated with 10 μM Bn-PP1 (+) or not (-). (A) Amount of NDT80 transcript shown in arbitrary units (a.u.). (B) Immunoblots showing production and phosphorylation of Ndt80 protein. Ime2 is shown as a loading control. (C) Graph of meiotic divisions in cells with wild-type or inducible NDT80. The percentage of cells in each sample with more than one DAPI-staining body is shown. (D) Impaired meiotic divisions after inhibition of Ime2-as1. DAPI-stained cells were scored for the occurrence of meiosis I (MI, 2 DAPI bodies) and meiosis II (MII, 4DAPI bodies). (E) Transcript levels of representative middle genes. The CLB1 profile shown is similar to that of CLB4, CLB5, SMK1, and SPO77. The SPS1 profile is similar to that of SPR3 and SPS4. (F) Phosphorylation of Ndt80 in an ime2Δ strain with inducible NDT80. IME2 (KBY373) and ime2Δ (KBY387) strains containing inducible NDT80 (PGAL-NDT80 GAL4-ER) were sporulated for 6 h in the absence (-) or presence (+) of 5 μM β-estradiol and harvested for immunoblotting with Ndt80 antibodies. A heterozygous control strain (C, KBY375) contained both wild-type and inducible NDT80 genes. (G) The immunoblot shows phosphorylation of Ndt80 in wild-type (KBY316) and ime2Δ sum1Δ mutant (KBY619) cells.
To examine if Ime2-independent production of Ndt80 was sufficient to rescue the meiotic division defect of Ime2-deficient cells, ime2-as1 PGAL-NDT80 GAL4-ER cells were sporulated, treated with β-estradiol at 3 h, then inhibited with 10 μM Bn-PP1 at 4h after DNA replication was complete. Samples were collected to examine Ndt80 protein, meiotic divisions, and middle gene transcription. Under these conditions, Ime2 was required for phosphorylation of Ndt80, but was not essential for accumulation of NDT80 mRNA and Ndt80 protein (Fig. 6A,B). A reproducible reduction in Ndt80 protein levels was observed in the absence of Ime2 activity, despite similar levels of NDT80 mRNA (see also Fig. 6F). Yet this reduced amount of Ndt80 protein was similar to the amount of Ndt80 found in unperturbed wild-type cells (data not shown). Inhibition of Ime2 dramatically reduced the incidence of meiosis II but had little effect on meiosis I (Fig. 6D). No asci were observed after 48 h. Thus, Ime2-independent PGAL-NDT80 expression rescued the severe meiosis I defect caused by inhibition of Ime2 (Figs. 6C, 5B). This demonstrates that NDT80 transcription was a limiting factor in progression from pachytene through meiosis I. However, high levels of Ndt80 were not sufficient for execution of a second meiotic division, indicating that Ime2 plays additional roles.
Because genetic evidence suggested that Ime2 antagonizes the transcriptional repressor Sum1 (Pak and Segall 2002a), we tested if deletion of Sum1 would further promote meiotic divisions when Ime2-as1 was inhibited. Deletion of Sum1 failed to rescue the meiosis defects of ime2-as1 GAL-NDT80 GAL4-ER and ime2-as1 NDT80 cells (data not shown), indicating meiotic defects are not caused by a failure to inactivate Sum1. Ime2 might be required additionally for phosphorylation of Ndt80 or for Ndt80-independent functions.
We next measured mRNA levels from representative middle genes (CLB1, CLB4, CLB5, SPS1, SPS4, SPR3, SMK1, and SPO77) in PGAL-NDT80 cells (Fig. 6E). When Ime2 was active, levels of middle gene transcripts in PGAL-NDT80 cells treated with estradiol were similar to those in wild-type NDT80 cells, but accumulation was delayed by about an hour. When Ime2-as1 was inhibited, induced expression from PGAL-NDT80 increased the amounts of many middle gene transcripts (CLB1, CLB4, CLB5, SMK1, and SPO77) to a level intermediate between that measured in untreated and treated NDT80 cells, but had little effect on other middle genes (SPS1, SPS4, and SPR3). Under these conditions the amounts of NDT80 transcript and Ndt80 protein were comparable to those observed in unperturbed wild-type cells, yet the uppermost form of Ndt80 was absent (Fig. 6A,B). Whereas lower forms of Ndt80 clearly were capable of promoting transcription, Ime2-dependent phosphorylation of Ndt80 was correlated with increased middle gene transcription.
Because Ime2 can phosphorylate Ndt80 in vitro (Fig. 2) and Ime2 activity is needed for maintenance of phosphorylated Ndt80 and for full middle gene transcription (Figs. 5, 6), we wanted to investigate further the requirements for and consequences of Ndt80 phosphorylation. Ime2-dependent phosphorylation of Ndt80 was examined in ime2Δ strains that produce Ndt80 protein by virtue of GAL-NDT80 induction (see above) or deletion of SUM1 (Pak and Segall 2002a). Wild-type IME2 and mutant ime2Δ strains containing estradiol-inducible NDT80 were sporulated in the presence or absence of estradiol for 6 h. The uppermost band of Ndt80 protein was absent, and the amount of Ndt80 was reduced in the ime2Δ mutant (Fig. 6F), yet examination of Ndt80 activity in these strains using an MSE-lacZ reporter (Hepworth et al. 1995) suggested that Ime2-dependent phosphorylation of Ndt80 was not essential for its ability to activate transcription. A β-galactosidase filter assay showed qualitatively similar reporter gene activity in ime2Δ and IME2 cells in which Ndt80 was induced (data not shown). Similarly, the double mutant sum1Δ ime2Δ produced Ndt80 lacking the uppermost form (Fig. 6F), and was able to turn on transcription of at least some middle sporulation genes (Pak and Segall 2002a).
In summary, a variety of strains with reduced Ime2 activity (ime2-as1 or ime2Δ) support the same conclusions. Ime2 was required for accumulation of the uppermost form of Ndt80. This Ime2-dependent phosphorylation of Ndt80 augmented but was not essential for transcription of middle sporulation genes.
Discussion
Progression through meiosis in Saccharomyces cerevisiae requires a transcriptional cascade and the actions of two key kinases, Cdc28 and Ime2. We used a chemical genetic approach to potently inhibit Cdc28 and Ime2 kinases at various times of meiosis and thereby identify additional roles for these kinases. Our demonstration that Cdc28 is necessary for meiotic DNA replication and dispensable for Sic1 degradation supports the idea that Ime2 has assumed some but not all of the functions of Cdc28 in meiotic S phase. We found that Ime2, like Cdc28, is needed for the G2–M transition and for execution of two sequential rounds of chromosome segregation. This late requirement for Ime2 is partly due to its stimulation of NDT80 transcription throughout M phase. Additional roles of Ime2 may include boosting middle gene transcription by phosphorylation of Ndt80 or promoting functions independent of Ndt80.
Use of inhibitor-sensitive mutants to identify early and late roles of Cdc28 and Ime2
By using protein kinases designed to be sensitive to chemical inhibitors (Bishop et al. 2000), we found that Cdc28 and Ime2 have dual roles in meiosis, first, in promoting DNA replication, and later, in promoting nuclear divisions. We demonstrated that Cdc28 is essential for meiotic S phase (Fig. 1), in contrast to previous studies with temperature-sensitive cdc28 mutants (Shuster and Byers 1989; Guttmann-Raviv et al. 2001). Because sporulation is a naturally temperature-sensitive process, prior experiments were limited to intermediate, semirestrictive temperatures that allowed wild-type strains to sporulate but may not have fully inactivated mutant Cdc28 proteins. With the cdc28-as1 mutant, more inhibitor was required to block early events than late events in both the mitotic and meiotic cell divisions (Bishop et al. 2000; data not shown), consistent with our proposal that the ts mutations did not reduce Cdc28 activity sufficiently to block early events. Using an ime2-as1 mutant, we demonstrated that Ime2 is required for the meiotic divisions (Fig. 5) in addition to its established early functions. These requirements for Ime2 may correspond to its two peaks of activity (Fig. 2). The second peak is temporally correlated with increased levels and hyperphosphorylation of Ime2 (Fig. 3), raising the possibility of dual regulation: at late times, cells may produce more molecules of Ime2 and phosphorylation may increase the stability or activity of each molecule of Ime2.
Cdc28E: The requirement of Cdc28 for meiotic DNA replication fits well with previous observations that known regulators of Cdc28 are critical for meiotic S phase. Because cells lacking Clb5 and Clb6 fail to undergo meiotic DNA replication (Stuart and Wittenberg 1998), it is likely that the early activity of Cdc28 (Cdc28E) is executed in partnership with Clb5 and Clb6. Cdc28E/Clb5,6 is the presumptive target of negative regulation by Sic1 (Dirick et al. 1998). Cdc28E may regulate proteins at the replication origin in meiosis as in mitosis (Toone et al. 1997) and may also promote the initiation of meiotic recombination (Smith et al. 2001). Cdc28E is not needed for the meiotic decline in Sic1 protein levels (Fig. 1), indicating that Ime2 has usurped this function performed by Cdc28 in the mitotic cell cycle (Dirick et al. 1998) and that the early activity of Ime2 (Ime2E) is independent of Cdc28E.
Cdc28L: A later activity of Cdc28 (Cdc28L) is required for the G2–M transition (Shuster and Byers 1989) but not for transcription of the middle sporulation genes. We observed that inhibition of Cdc28-as1 did not reduce transcript levels of middle genes (data not shown), consistent with prior reports using a cdc28-ts mutant and mutants lacking several CLB genes (Hepworth et al. 1998; Stuart and Wittenberg 1998; Tung et al. 2000), and did not alter the level or phosphorylation of Ndt80 (Figs. 1, 3). Cdc28L is important for the second peak in Ime2 kinase activity (Ime2L) but is probably not the kinase directly responsible for late changes in Ime2 phosphorylation (Fig. 3). Cdc28L may trigger and coordinate chromosome distribution by phosphorylating some of the same Cdc28 substrates found in the mitotic cell cycle; for example, proteins needed for formation of the bipolar spindle and for activity of the anaphase-promoting complex (APC). Cdc28L may also be necessary for disassembly of the synaptonemal complex and completion of recombinational crossovers (Shuster and Byers 1989; Allers and Lichten 2001).
Ime2E: Prior studies have identified a variety of processes that require early action of Ime2: promotion of S phase by facilitating degradation of Sic1 (Dirick et al. 1998), transcription of the early genes (Mitchell et al. 1990), and destabilization of Ime1 (Guttmann-Raviv et al. 2002). Ime2 appears to have completely assumed responsibility for regulating Sic1 degradation. Ime2 is required for the initial transcription of NDT80 (Hepworth et al. 1998), probably as a consequence of its negative regulation of Sum1 (Fig. 6G; Pak and Segall 2002a).
Ime2L: Ime2L is needed to negotiate the G2–M transition (Fig. 5A; data not shown), to promote events after metaphase I (Fig. 5E), and to execute meiosis II (Fig. 6C). In addition to the role of Ime2E in initiating transcription of NDT80 early in meiosis, Ime2L is needed for its continued transcription throughout the meiotic divisions (Fig. 5D). When Ime2 is inhibited, the low NDT80 transcript level is insufficient to promote meiosis I (Fig. 6). This observation demands a revision of the simple model in which Ime2 initiates NDT80 transcription, and thereafter Ndt80 assumes all responsibility for transcription of its own gene (Chu and Herskowitz 1998; Pak and Segall 2002a). Perhaps the strength of Ndt80 autoactivation is insufficient to maintain high NDT80 mRNA levels without assistance from a parallel Ime2-dependent activation pathway. Alternatively, Ime2 may play an important posttranscriptional role in stimulating the ability of Ndt80 to activate transcription of its own gene, either by altering the behavior of individual Ndt80 molecules or by increasing the translation or stability of Ndt80 protein.
The requirement of Ime2 for the meiotic divisions cannot wholly be explained by its stimulation of NDT80 transcription. The meiosis II defect of cells that contain a normal amount of NDT80 transcript and lack Ime2 activity might be caused by a failure of Ime2 to phosphorylate Ndt80 or other substrates (Fig. 6). Ime2 governs the phosphorylation of Ndt80 in meiotic cells (Figs. 5, 6) and efficiently phosphorylates Ndt80 in vitro (Fig. 2), suggesting that Ndt80 is a physiological substrate of Ime2. During preparation of this paper, Sopko et al. (2002) also reported the stimulation of Ndt80 phosphorylation by Ime2 and proposed that phosphorylation stimulated the transcriptional activity of Ndt80. Yet Ime2-dependent phosphorylation of Ndt80 is dispensable for meiosis I and for transcription of some middle genes (Fig. 6). It is possible that the meiosis II defect of Ime2-deficient cells results from inadequate transcription of one or more key middle genes when Ndt80 is improperly phosphorylated. Ime2-dependent phosphorylation of Ndt80 may increase its stability (and thereby augment its steady-state levels) or may stimulate its specific activity (and thereby enhance the transcriptional activation function of each Ndt80 molecule). Alternatively, Ime2-dependent phosphorylation of Ndt80 may be needed for a nontranscriptional role of Ndt80 in meiosis II, or Ime2 might be necessary for additional functions independent of Ndt80, for example, regulation of proteolysis (Bolte et al. 2002).
Many of the genes with the greatest similarity to IME2 are expressed exclusively in meiotic cells, like IME2. Two IME2 orthologs in Saccharomyces pombe, mde3 and pit1, are important for timing of the meiotic divisions and are essential for spore morphogenesis (Abe and Shimoda 2000). Mammals have a “male germ cell-associated kinase,” Mak, that is homologous to Ime2. Although its function is unknown, Mak is found specifically in primary spermatocytes during late meiotic G2 (Jinno et al. 1993; Shinkai et al. 2002). The identification of a late role for Ime2 provides a further rationale for examining functional parallels between Ime2 and Mak.
The program of meiosis: decisions and positive feedback loops
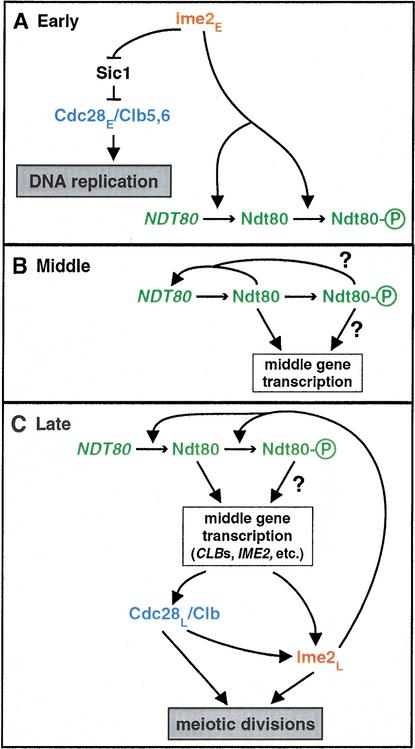
Identification of early and late modes of Cdc28 and Ime2 action leads us to propose an integrated scheme for meiotic development (Fig. 7). We divide the meiotic program into four stages: Initiation, Early, Middle, and Late. Our focus here begins after the initiation of meiosis triggered by Ime1, which induces transcription of IME2 and other early genes (Mitchell et al. 1990; Kupiec et al. 1997). We note that our designation of Early, Middle, and Late stages does not correspond to the transcriptional classes bearing the same names (Chu et al. 1998). In the Early stage (Fig. 7A), leading to the G1–S transition, Ime2E promotes the degradation of Sic1 (Fig. 4D; Dirick et al. 1998), which allows Cdc28E/Clb5,6 to trigger replication initiation (Fig. 1B). Ime2E also promotes transcription of NDT80 (Hepworth et al. 1998; Pak and Segall 2002a) and phosphorylation of Ndt80 (Figs. 1, 5, 6) independent of DNA replication.
Figure 7.
Pathway of meiotic progression. See text for details. Cdc28 and Ime2 are required for both early (DNA replication) and late (nuclear divisions) processes in meiosis. (A) Early: An early form of Ime2 (Ime2E) antagonizes Sic1 and promotes Ndt80 activity at both transcriptional and posttranslational levels. An early activity of Cdc28 (Cdc28E) is essential for meiotic DNA replication. (B) Middle: Ndt80 turns on transcription of its own gene in an autoactivation loop that helps boost transcription of the middle sporulation genes. (C) Late: Late activities of Cdc28L and Ime2L are necessary for the meiotic divisions. Ndt80 and Cdc28L stimulate activity, accumulation, and hyperphosphorylation of Ime2L. In turn, Ime2L is needed to maintain high levels and phosphorylation of Ndt80, to promote full transcription of middle genes, and thus to ensure adequate Cdc28L/Clb kinase activity. Ime2L may also play an NDT80-independent role in the meiotic divisions.
In the Middle stage (Fig. 7B), leading to the G2–M transition, the amount and activity of Ndt80 rise, thus initiating transcription of middle genes that encode functions necessary for M phase and beyond. In an autoactivation loop, Ndt80 activates transcription of its own gene (Chu et al. 1998), which is necessary for normal accumulation of NDT80 transcript (Pak and Segall 2002a). Ime2 continues promoting NDT80 transcription either directly or via its phosphorylation of Ndt80 (Fig. 6). The basally phosphorylated forms of Ndt80 can activate transcription, and the Ime2-dependent hyperphosphorylated form might have increased transcriptional activity or stability, or may perform additional functions (Figs. 5, 6; Pak and Segall 2002a; Sopko et al. 2002).
The Late stage encompasses the initiation and execution of M phase (Fig. 7C). Our studies of Cdc28, Ime2, and Ndt80 revealed several molecular and functional interdependencies. Ndt80 promotes transcription of many genes required for chromosome segregation and spore morphogenesis, including CLB1 and CLB3–CLB6, which encode activators of Cdc28 (Chu and Herskowitz 1998). Three observations suggest that Ndt80 also promotes IME2 transcription, thus closing a positive feedback circuit controlling transcription of IME2 and NDT80. First, expression of Ndt80 in vegetative cells induces transcription of IME2 (Chu et al. 1998). Second, the IME2 promoter contains a putative Ndt80-binding site in a small region previously shown to be necessary for autoregulation of IME2 transcription (Bowdish and Mitchell 1993), indicating Ndt80 may mediate this autoactivation. Third, IME2 transcription is initiated like an early gene but peaks and declines like a middle gene (Fig. 5D; Bowdish and Mitchell 1993; Primig et al. 2000). In addition, Ndt80 may stimulate Ime2L activity by augmenting Cdc28L activity or by inducing transcription of a gene that encodes or regulates the kinase that hyperphosphorylates Ime2L. The consequences of hyperphosphorylation for Ime2 activity and stability remain to be determined. Ime2L activity is needed during M phase for transcription of NDT80 and CLBs, and thus presumably for high Cdc28L activity (Figs. 5, 6). Cdc28L and Ime2L are both required for progression beyond pachytene and for meiotic events after metaphase I (Fig. 5).
We speculate that Ime2L is both a target and regulator of Cdc28L and Ndt80 in a positive regulatory circuit, perhaps analogous to a complex positive feedback loop involving CDK and MAPK kinases in Xenopus oogenesis (Nebreda and Ferby 2000).This hypothetical circuit may serve two functions, (1) to amplify signals and yield rapid changes in Cdc28L and Ime2L activity levels that may govern commitment to meiotic chromosome segregation; and (2) to link together Cdc28L and Ime2L activities to coordinate the events of the meiotic divisions. Coordination of Cdc28L and Ime2L activities would be particularly important to address the challenges introduced by a proposed division of labor between the kinases, in which several of the mitotic functions of Cdc28 may be assumed in meiosis by Ime2. This scheme, which incorporates early and late modes of Cdc28 and Ime2 activities, provides a framework on which to place additional signaling inputs, mechanistic details, and molecular targets of these protein kinases. Our discoveries of roles and interdependencies of several key regulators of meiotic progression raise many questions whose detailed answers will give added insight into meiotic regulation.
Materials and methods
Strains
All strains were in the SK1 strain background (Padmore et al. 1991) and were constructed using standard yeast genetic methods. Homozygous diploid strains were constructed by making the desired haploids, back-crossing, sporulating to obtain the opposite mating type, and mating appropriate segregants. The genotypes of all strains were confirmed by diagnostic PCR reactions and, when appropriate, sequencing of PCR products. All strains were derived from haploid segregants of the homozygous diploid KBY259 (ho::hisG lys2 ura3 leu2::hisG his3-11,15 trp1ΔFA::hisG), which was constructed from a standard SK1 strain (Chu and Herskowitz 1998) by allelic replacement with the his3-11,15 allele from a W303 strain and the designer trp1ΔFA allele (Chu and Herskowitz 1998; Horecka and Jigami 1999). Strains were homozygous diploids unless noted otherwise, and only the relevant genotypes are shown. Additional details are available upon request.
Chromosomal deletion of genes and introduction of C-terminal epitope tags were accomplished as described (Wach et al. 1994; Longtine et al. 1998). Homozygous diploid strains with tagged alleles were KBY316 (IME2-13myc:TRP1) and KBY332 (NDT80-3HA:HIS3). The IME2-myc and NDT80-HA genes were fully functional: wild-type, IME2-myc/ime2Δ, and NDT80-HA/ndt80Δ strains exhibited similar kinetics and efficiency of meiotic DNA replication, meiotic divisions, spore formation, and meiosis-specific protein accumulation.
The ndt80 and cdc28 mutations were introduced into wild-type (KBY259) and IME2-13myc:TRP1 (KBY316) strain back grounds. KBY427 (ndt80Δ::HIS3Cg) and KBY430 (ndt80Δ::HIS3Cg IME2-13myc:TRP1) were constructed by precise replacement of the NDT80 coding region with HIS3 from Candida glabrata (Kitada et al. 1995). KBY439 (cdc28-as1) and KBY442 (cdc28-as1 IME2-13myc:TRP1) were made by loop in-loop out of pJAU01 containing the cdc28-as1 (F88G) allele (Bishop et al. 2000). KBY619 (ime2Δ::LEU2 sum1Δ::HIS3) was constructed by sequential deletion of two genes; IME2 was deleted using a fragment from the plasmid pAM412 (Mitchell et al. 1990), and SUM1 was deleted by precise replacement of the SUM1 coding region with a PCR product containing HIS3 from C. glabrata (Kitada et al. 1995).
Mutant ime2 was generated by site-directed mutagenesis of IME2 in pKB44 (Transformer Kit, Clontech; or QuikChange Kit, Stratagene) and sequenced to verify the introduction of desired mutations and absence of additional mutations. The ime2-as1 strain KBY518 (ime2-M146G-13myc:TRP1) was made by loop in-loop out of pKB69, a pRS306 derivative containing ime2-M146G. The kinase-inactive ime2 mutant KBY450 (ime2-K97R-13myc:TRP1) was generated using a different two-step allelic replacement method. First, the central portion of IME2-myc (nucleotides 281–1160) was replaced with URA3 from Kluyveromyces lactis to yield KBY443 (MATa ime2ΔM::URA3Kl-13Myc:TRP1; Wach et al. 1994). Second, KBY443 was transformed with a restriction fragment from pKB46 containing ime2K97R and grown on 5-FOA. Replacement of URA3Kl with mutant ime2 was confirmed by colony PCR. Mutant chromosomal loci were characterized by sequencing of PCR products amplified from genomic DNA templates.
Ime2-independent expression of NDT80 was achieved when strains containing two chromosomal modifications were treated with estradiol. For Figure 6F, the homozygous diploid strain KBY373 (ndt80::TRP1-PGAL-NDT80 his3::PADH1-GAL4(848).ER::HIS3) was constructed by insertion of the GAL1,10 UAS upstream of the chromosomal NDT80 coding region (Longtine et al. 1998) and insertion of GAL4(848).ER into the chromosomal his3 locus by transformation with pKB33, an integrating version of pHCA/GAL4(848).ER (Picard 1999). GAL4(848).ER encodes full-length Gal4fused to the hormone-binding domain of a human estrogen receptor; in this chimera, Gal4is active only in the presence of β-estradiol (Picard 1999). KBY387 (ime2Δ::LEU2Cg ndt80::TRP1-PGAL-NDT80 his3::pADH1-GAL4(848).ER::HIS3) contains a precise replacement of the IME2 ORF with LEU2 from C. glabrata (Wach et al. 1994). KBY375 is similar to KBY373 except it is heterozygous at the NDT80 locus (NDT80/ndt80::TRP1-PGAL-NDT80).
To examine whether NDT80 expression rescued the meiotic division defects of Ime2-deficient cells (Fig. 6A–E), it was necessary to modify the source of Gal4-ER. The ADH1 promoter controlling GAL4(848).ER transcription in the strains above was strongly down-regulated in acetate medium. To produce more Gal4-ER, and consequently more Ndt80, GAL4(848).ER was subcloned into a plasmid containing the GPD1 regulatory region (Mumberg et al. 1995). PGPD1-GAL4(848).ER was further subcloned into the URA3 integrating vector pRS306 to yield pKB80. The homozygous diploid ime2-as1 strain with inducible NDT80, KBY649 (ime2-M146G-13myc:TRP1 ndt80::TRP1-PGAL1-NDT80 ura3::PGPD1-GAL4(848).ER::URA3), was made by first crossing strains to create KBY611 (MATa ime2-M146G-13myc:TRP1 ndt80::TRP1-PGAL1-NDT80), followed by integration of pKB80 at the ura3 locus and mating to an appropriate partner.
Cultures were sporulated in 1% potassium acetate plus 0.02% raffinose at a density of 3.6 × 107 cells/mL using a standard regimen (Padmore et al. 1991; Grether and Herskowitz 1999).
PP1-analog inhibitors and β-estradiol
The inhibitors 1-NA-PP1 and 1-NM-PP1 were synthesized and handled as described (Bishop et al. 1999), except that the synthesis of 1-NA-PP1 used DMF instead of ethanol as the solvent for the reaction between 1-napthyl(methoxy)methylidenemalononitrile and tert-butylhydrazine. The inhibitor 3-benzyl-1-tert-butyl-1H-pyrazolo[3,4-d]pyrimidin-4-ylamine (called Bn-PP1) was synthesized using a modified procedure (Bishop et al. 1999). Inhibitors were added from 10–20 mM stock solutions in DMSO; control cultures were treated with an equal volume of DMSO.
A 5 mM stock solution of β-estradiol (Sigma) was made in ethanol and stored at -20°C until use. Estradiol treatment of wild-type cells had no effect on meiotic progression.
Monitoring DNA replication and meiotic divisions
Cells were fixed by adding formaldehyde to 4.6% (to assay meiotic divisions) or by resuspending cell pellets in 70% ethanol plus 50 mM Tris-HCl (pH 7.5; to assay DNA replication) and incubating them at 4°C for 1–14 d. DNA replication was evaluated by flow cytometry as described (Piatti et al. 1995) using Sytox Green (Molecular Probes) to stain DNA and a Becton Dickinson FACSort with CellQuest software. The meiotic divisions were examined by staining DNA with DAPI (Grether and Herskowitz 1999).
The chromosomal DNA in ime2-as1 cells treated with 1-NA-PP1 after DNA replication became increasingly diffuse over time. After addition of 1-NA-PP1 to ime2-as1 cells at 5.25 h, 60%–75% of the cells harvested at 8 h exhibited stringy, faint, speckled DAPI staining. In some cultures, up to half of these cells contained three to four small, nonuniform DAPI bodies against a speckled background, which introduced ambiguity in scoring. A cell was scored as MII if it had four DAPI bodies that were nearly as large as in wild-type MII cells, regardless of whether it contained speckled DAPI staining. These aberrant DAPI patterns depended on Ndt80, because under the same treatment conditions ndt80Δ ime2-as1 cells exhibited a single, tight, bright DAPI-staining body, just like untreated ndt80Δ cells. These observations indicate that although cells lacking Ime2 activity fail to undergo proper chromosome segregation, they are not simply arrested in G2 or M phase.
Immunoblotting
Protein extracts prepared using TCA precipitation were separated by SDS-PAGE electrophoresis as described (O'Rourke and Herskowitz 1998). Sic1 gels were 12% polyacrylamide (37.5:1 bis), and Ime2/Ndt80 gels were 7.5%. Typically, each lane contained proteins from the equivalent of 8.1 × 106 cells. After electrophoretic transfer to nitrocellulose membrane, lanes were examined for equal protein loading by PonceauS staining (Sigma). Immunological recognition and ECL visualization were performed according to the manufacturer's instructions (Amersham). Primary antibodies were mouse monoclonal α-myc 9E10 and α-HA 12CA5 (Covance) and rabbit polyclonal antisera raised against Sic1 (Skowyra et al. 1997), Ime2 (Sia and Mitchell 1995), and Ndt80. Ndt80 antibodies were generated by immunizing rabbits with the C-terminal third of Ndt80 (amino acids 409–627) fused to a his6 tag and purified from E. coli.
Ime2 kinase assay
To measure the kinase activity of Ime2-myc, 10–30 mL of sporulating cells was collected by centrifugation, frozen in liquid nitrogen, and stored at -80°C. Chemicals were purchased from Sigma or Boehringer Mannheim, unless indicated otherwise. All steps before the kinase reaction were done at 0°C–4°C and were adapted from previously described protocols (Grether and Herskowitz 1999). Cells were mixed with 500–600 μL of 0.5-mm glass beads (BioSpec Products) and 500 μL of lysis buffer (50 mM HEPES at pH 7.4, 75 mM KCl, 1 mM EGTA, 1 mM MgCl2, 0.1% NP-40, 50 mM NaF, 50 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM PMSF, 8.8 μg/mL aprotinin, 4μg/mL anti-pain, 0.1 μg/ml pefabloc SC, 2 μg/mL pepstatin A, 1 μg/mL chymostatin, 1 mM benzamidine, and 2 μg/mL leupeptin), then lysed using a mini bead beater (Biospec Products). After centrifugation, protein concentrations of clarified extracts were measured using the Bradford assay (Bio-Rad). Equal amounts of protein from each sample (2–6 mg) were mixed with 20–30 μL of a 1:1 slurry of 9E10 beads (Covance) for 1.5 h. Ime2-myc immunoprecipitates were washed three times in lysis buffer and once in kinase buffer (20 mM HEPES at pH 7.4, 100 mM KCl, 10 mM MgCl2), before resuspension in 25 μL of kinase buffer containing 5 μCi [γ-32P]ATP (Amersham), 10 μM ATP, and 10 μg of protein substrate (described below). After incubation at 30°C for 30 min, reactions were stopped with 12 μL of 4× sample loading buffer, boiled for 5 min, and separated by 12% SDS-PAGE. Gels were fixed for 20 min in three changes of 25% methanol plus 7.5% glacial acetic acid, dried, and exposed to X-ray film or a PhosphorImager screen. The amount of radiolabeled phosphate incorporated into substrate proteins (including Ime2-myc itself) was measured with a PhosphorImager and ImageQuant software (Molecular Dynamics). Kinase activity is shown in arbitrary units. Initial experiments determined that these conditions were within the linear range of the assay.
The substrates used in this study were chimeras with the maltose-binding protein (MBP). MBP was expressed alone from pMAL-c2 (New England Biolabs), fused to the N terminus of full-length Ndt80 (encoded by pKB32, a derivative of pMAL-c2), or fused to the N terminus of a DNA-binding domain of Ndt80 (DBD, amino acids 51–350, pEJ151). MBP, MBP-Ndt80, and MBP-Ndt80DBD were expressed in E. coli and purified as recommended by the manufacturer (New England Biolabs).
Phosphatase treatments
Immunoprecipitation of Ime2-myc and Ndt80-HA were performed as in the kinase assay, except that HA11 beads (Covance) were used for Ndt80-HA. Immunoprecipitates were washed three times in lysis buffer and once in prephosphatase buffer (50 mM Tris-HCl at pH 7.5, 0.1 mM EDTA, 2 mM MnCl2, 0.1 mg/mL BSA). Beads were resuspended in 50 μL of reaction buffer (50 mM Tris-HCl at pH 7.5, 0.1 mM EDTA, 5 mM DTT, 2 mM MnCl2, 0.01% Brij35) with 50 U of λ protein phosphatase (New England Biolabs). Control reactions omitted phosphatase or additionally contained phosphatase inhibitors (50 mM NaF, 2 mM vanadate). After incubation at 30°C for 30 min, beads were collected by centrifugation, resuspended in 30 μL of 1.5× protein loading buffer, boiled for 5 min, separated by 7.5% SDS-PAGE, and analyzed by immunoblotting with 9E10 or 12CA5 antibodies.
Analysis of mRNA transcripts
The amounts of mRNAs were determined by Northern blotting. Total cellular RNA was extracted by the hot acid phenol method (Spellman et al. 1998) and examined using standard protocols for formaldehyde-agarose gel electrophoresis, transfer to nitrocellulose, and detection of individual mRNA species (Chu and Herskowitz 1998). Northern blots were hybridized sequentially with radiolabeled probes corresponding to IME2; NDT80; the middle sporulation genes CLB1, CLB4, SMK1, SPS1, SPS4, and SPR3; and the loading control PFY1. The radioactivity of each band was quantified using a PhosphorImager and divided by the PFY1 signal to account for loading differences. To more easily compare the timing of accumulation of the different transcripts and the differences between cultures, the loading-corrected values for a given transcript were then normalized to the maximal value measured for that transcript. The peak value of each transcript is defined as 1. The genes shown in Figure 4peaked at 6 or 7 h, whereas the early genes peaked at 4h (data not shown).
Acknowledgments
We thank L. Huang, E. Jolly, A. Mitchell, V. Nguyen, M. Tyers, and J. Ubersax for technical help, plasmids, and reagents. We are indebted to L. Huang, Y. Kassir, J. Li, A. Nicolas, and D. Morgan for helpful discussions, and B. Byers, L. Dirick, J. Gerton, K. Jaglo, A. Mitchell, and D. Morgan for comments on the manuscript. Funding was provided by postdoctoral fellowships (to K.R.B.) from the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (DRG-1443) and the California Division of the American Cancer Society; and by the NIH grant GM59256 (to I.H.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734solely to indicate this fact.
Corresponding author.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1101503.
References
- Abe H. and Shimoda, C. 2000. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4and a genome-wide search for its target genes. Genetics 154: 1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T. and Lichten, M. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Bishop A.C., Kung, C.Y., Shah, K., Witucki, L., Shokat, K.M., and Liu, Y. 1999. Generation of monospecific nanomolar tyrosine kinase inhibitors via a chemical genetic approach. J. Am. Chem. Soc. 121: 627–631. [Google Scholar]
- Bishop A.C., Ubersax, J.A., Petsch, D.T., Matheos, D.P., Gray, N.S., Blethrow, J., Shimizu, E., Tsien, J.Z., Schultz, P.G., Rose, M.D., et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395–401. [DOI] [PubMed] [Google Scholar]
- Bolte M., Steigemann, P., Braus, G.H., and Irniger, S. 2002. Inhibition of APC-mediated proteolysis by the meiosis-specific protein kinase Ime2. Proc. Natl. Acad. Sci. 99: 4385–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowdish K.S. and Mitchell, A.P. 1993. Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S. and Herskowitz, I. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1: 685–696. [DOI] [PubMed] [Google Scholar]
- Chu S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P.O., and Herskowitz, I. 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- Dahmann C. and Futcher, B. 1995. Specialization of B-type cyclins for mitosis or meiosis in S. cerevisiae. Genetics 140: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L., Goetsch, L., Ammerer, G., and Byers, B. 1998. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in 281: 1854–1857. Saccharomyces cerevisiae. Science [DOI] [PubMed] [Google Scholar]
- Foiani M., Nadjar-Boger, E., Capone, R., Sagee, S., Hashimshoni, T., and Kassir, Y. 1996. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol. Gen. Genet. 253: 278–288. [DOI] [PubMed] [Google Scholar]
- Grandin N. and Reed, S.I. 1993. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol. Cell. Biol. 13: 2113–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether M.E. and Herskowitz, I. 1999. Genetic and biochemical characterization of the yeast Spo12 protein. Mol. Biol. Cell 10: 3689–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttmann-Raviv N., Boger-Nadjar, E., Edri, I., and Kassir, Y. 2001. Cdc28 and Ime2 possess redundant functions in promoting entry into premeiotic DNA replication in Saccharomyces cerevisiae. Genetics 159: 1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttmann-Raviv N., Martin, S., and Kassir, Y. 2002. Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1. Mol. Cell. Biol. 22: 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S.R., Ebisuzaki, L.K., and Segall, J. 1995. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S.R., Friesen, H., and Segall, J. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 5750–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horecka J. and Jigami, Y. 1999. The trp1-Δ FA designer deletion for PCR-based gene functional analysis in Saccharomyces cerevisiae. Yeast 15: 1769–1774. [DOI] [PubMed] [Google Scholar]
- Jinno A., Tanaka, K., Matsushime, H., Haneji, T., and Shibuya, M. 1993. Testis-specific mak protein kinase is expressed specifically in the meiotic phase in spermatogenesis and is associated with a 210-kilodalton cellular phosphoprotein. Mol. Cell. Biol. 13: 4146–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K., Yamaguchi, E., and Arisawa, M. 1995. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene 165: 203–206. [DOI] [PubMed] [Google Scholar]
- Kominami K., Sakata, Y., Sakai, M., and Yamashita, I. 1993. Protein kinase activity associated with the IME2 gene product, a meiotic inducer in the yeast Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 57: 1731–1735. [DOI] [PubMed] [Google Scholar]
- Kupiec M., Byers, B., Esposito, R.E., and Mitchell, A.P. 1997. Meiosis and sporulation in Saccharomyces cerevisiae. In The molecular and cellular biology of the yeast Saccaromyces (eds. J.R. Pringle et al.), pp. 889–1036. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Longtine M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lydall D., Nikolsky, Y., Bishop, D.K., and Weinert, T. 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383: 840–843. [DOI] [PubMed] [Google Scholar]
- Mendenhall M.D. and Hodge, A.E. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62: 1191–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A.P., Driscoll, S.E., and Smith, H.E. 1990. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 2104–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller, R., and Funk, M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. [DOI] [PubMed] [Google Scholar]
- Nebreda A.R. and Ferby, I. 2000. Regulation of the meiotic cell cycle in oocytes. Curr. Opin. Cell Biol. 12: 666–675. [DOI] [PubMed] [Google Scholar]
- O ourke, S.M. and Herskowitz, I. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes & Dev. 12: 2874–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R., Cao, L., and Kleckner, N. 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256. [DOI] [PubMed] [Google Scholar]
- Pak J. and Segall, J. 2002a. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 6417–6429. —;—;—; [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002b. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 6430–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Lengauer, C., and Nasmyth, K. 1995. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a `reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14: 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D. 1999. Regulation of heterologous proteins by fusion to a hormone binding domain. In Nuclear receptors: a practical approach (ed. D. Picard), pp. 261–274. Oxford University Press, Oxford.
- Primig M., Williams, R.M., Winzeler, E.A., Tevzadze, G.G., Conway, A.R., Hwang, S.Y., Davis, R.W., and Esposito, R.E. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26: 415–423. [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Satoh, H., Takeda, N., Fukuda, M., Chiba, E., Kato, T., Kuramochi, T., and Araki, Y. 2002. A testicular germ cell-associated serine–threonine kinase, MAK, is dispensable for sperm formation. Mol. Cell. Biol. 22: 3276–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster E.O. and Byers, B. 1989. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces ce-revisiae. Genetics 123: 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia R.A. and Mitchell, A.P. 1995. Stimulation of later functions of the yeast meiotic protein kinase Ime2p by the IDS2 gene product. Mol. Cell. Biol. 15: 5279–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209–219. [DOI] [PubMed] [Google Scholar]
- Smith K.N., Penkner, A., Ohta, K., Klein, F., and Nicolas, A. 2001. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr. Biol. 11: 88–97. [DOI] [PubMed] [Google Scholar]
- Sopko R., Raithatha, S., and Stuart, D. 2002. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 22: 7024–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., and Futcher, B. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D. and Wittenberg, C. 1998. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes & Dev. 12: 2698–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone W.M., Aerne, B.L., Morgan, B.A., and Johnston, L.H. 1997. Getting started: Regulating the initiation of DNA replication in yeast. Annu. Rev. Microbiol. 51: 125–149. [DOI] [PubMed] [Google Scholar]
- Tung K.S., Hong, E.J., and Roeder, G.S. 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. 97: 12187–12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon A.K. and Pierce, M. 2000. Transcriptional regulation of meiosis in yeast. Curr. Opin. Cell Biol. 12: 334–339. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat, A., Pohlmann, R., and Philippsen, P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Xu L., Ajimura, M., Padmore, R., Klein, C., and Kleckner, N. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]