Abstract
A recessive, fully penetrant mutation (cm1OR) at the mouse albino locus that results in coat-color mottling has been characterized at the molecular level. Restriction mapping and DNA sequencing analyses provide evidence that mutants carry a 5.4-kb intracisternal A particle (IAP) element insertion upstream of the tyrosinase (Tyr) promoter. Northern blot analysis and reverse transcription–PCR results show that the tyrosinase gene is expressed at much lower levels in mutant than in wild-type mice. The mutant Tyr gene still retains the tissue-specific expression pattern, and the Tyr transcript is not initiated from the IAP long terminal repeat promoter. We propose that the IAP insertion isolates the promoter of the tyrosinase gene from upstream cis-acting regulatory elements, leading to a substantially decreased level of Tyr gene expression in mutants.
Tyrosinase (monophenol, l-dopa: oxygen oxidoreductase, EC 1.14.18.1), a key enzyme for melanin biosynthesis in pigment cells, catalyzes the hydroxylation of tyrosine to dopa and its subsequent oxidation to dopaquinone (1). The structural gene for tyrosinase (Tyr) has been mapped to the mouse albino (c) locus in chromosome 7 (2, 3), and full-length tyrosinase cDNAs have been cloned and sequenced (3–5). A cysteine-to-serine mutation in tyrosinase leads to albinism in the original c mutation found in a number of laboratory mouse strains (6). Alternatively, spliced tyrosinase gene transcripts have been identified, along with two promoters from which transcripts are initiated; the major transcription start site is at position +1, and the minor transcription start site is at position −56 (3). The roles for these multiple transcripts are unclear, but the functional tyrosinase transcript is believed to be initiated from the minor transcription start site (3).
We report here a spontaneous c-locus mutation, cm1OR, which causes coat-color mottling in homozygous mutant mice. Molecular analysis of the Tyr gene in this mutation shows a genomic rearrangement resulting from the insertion of an intracisternal A particle (IAP) sequence at the 5′ end of the gene. IAPs are retrovirus-like elements present in approximately 1000 copies per haploid genome in Mus musculus (7). Functionally, IAP elements are known to act as retrotransposons in the mouse germ line. An IAP can affect expression of an adjacent gene either by acting as a regulatory element for such a gene or by initiating transcription of the adjacent gene from within the IAP’s long terminal repeats (LTRs) (8).
MATERIALS AND METHODS
Mice.
The cm1OR mutation occurred spontaneously in the C3Hf/Rl inbred strain in 1988 at the Oak Ridge National Laboratory (Oak Ridge, TN).
Southern and Northern Blot Analyses and DNA Probes.
Procedures for the preparation of DNA and RNA and for Southern and Northern blot analyses are described elsewhere (9). Isolation of poly(A+) RNA was performed essentially as described (10). Probe PTY-1H is a 650-bp HindIII–HincII restriction fragment containing the 5′ end of the tyrosinase coding sequences. Probe MTY811 is a tyrosinase cDNA probe containing 75% of the full-length tyrosinase transcript (5).
Cloning of Tyr DNA Fragments.
Using MTY811 as probe, a mutant 3-kb EcoRI fragment from the 5′ Tyr sequence was cloned from cm1OR/cm1OR DNA into EcoRI-digested pBSSK+ to yield plasmid pRN209. The corresponding wild-type 4.7-kb EcoRI fragment was subcloned from λFix2–1, a 5′ Tyr gene λ clone obtained from B. S. Kwon (Indiana University), into EcoRI-digested pBSSK+ to yield plasmid pRN210. cm1OR/cm1OR genomic DNA was also partially digested with Sau3A, size selected, and ligated to λ2001 arms to construct a cm1OR/cm1OR genomic λ library. PTY-1H was used to select a mutant 15-kb 5′ Tyr λ clone, designated λcm9, from this λ library. A mutant 5.4-kb BamHI fragment was cloned from λcm9 into BamHI-digested pBSSK+ vector to yield plasmid 5cm9.
Reverse Transcription (RT)–PCR and DNA Sequencing.
Five micrograms of total RNA were reverse-transcribed in a 20-μl reaction volume (11). PCR amplification was performed in volumes of 50 μl containing 1 μl of the RT reaction (12). Primers for the RT-PCR analysis are as follows: primer a, 5′-GACGGCGAATGTGGGGGCGG-3′; primer b, 5′-CCTCGAGCCTGTGCCTCCTC-3′; primer c, 5′-GGGAGCCTGGGGGTTTTGGC-3′. The cycling parameters were: 94°C for 30 sec, 64°C for 1 min, 72°C for 3 min for 35 cycles, followed by 72°C for 5 min.
All plasmid clones were sequenced by the dideoxynucleotide method using the Sequenase 2.0 kit (United States Biochemical) (13).
Histology.
Dorsal skin samples were taken from 4-day-old cm1OR/cm1OR or control isogenic C3Hf mice. These samples were cleared and fixed in 2.5% glutaraldehyde. Following postfixation in 1% osmium tetraoxide, samples were embedded in plastic. Ultrathin sections were stained with uranyl acetate and lead citrate and examined under a Phillip 300 electron microscope (14).
Immunohistochemical Studies.
Dorsal skin samples were taken from 4-day-old mice and frozen in embedding media (O.C.T.) without prior fixation. Immunohistochemical experiments were performed on 8-micron skin sections as described (15) using rabbit antiPEP8 as a marker for melanocytes. AntiPEP8, provided by Vincent J. Hearing (National Institutes of Health, Bethesda), is a rabbit polyclonal antibody against the synthetic peptide PEP8, and recognizes specifically DOPAchrome tautomerase (TRP-2) in melanocytes (16). The sections were subsequently stained with hematoxylin/eosin.
RESULTS
Description of the cm1OR Phenotype.
A recessive, fully penetrant mutation that arose spontaneously in the C3Hf/Rl inbred strain results in pink-eyed homozygotes exhibiting a mottled appearance, with interspersed dark hairs on an otherwise unpigmented background (Fig. 1). Genetic tests confirmed this mutation, designated cm1OR, to be an allele of albino (c). The mutant phenotype is fully recessive to wild type, i.e., +/cm1OR mice are wild-type in pigmentation; however, the mottling phenotype is dominant to both c and cch (chinchilla), so that mottling is evident in cm1OR/c or cm1OR/cch heterozygotes.
Figure 1.
Two C3Hf-cm1OR/cm1OR mutant mice and one wild-type C3Hf mouse.
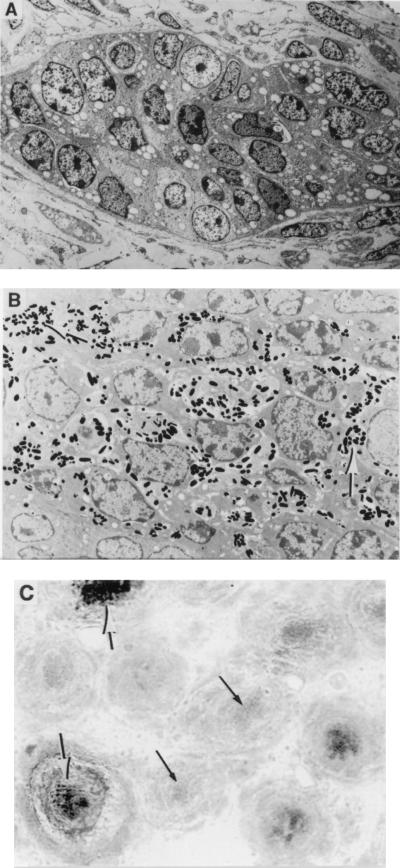
A comparison of skin sections from 4-day-old cm1OR/cm1OR mice and C3Hf wild-type controls revealed many fewer pigmented follicles in the former (data not shown). Electron micrographs were also taken from cm1OR/cm1OR and C3Hf skin sections (Fig. 2). Fig. 2A shows a typical hair follicle from cm1OR/cm1OR skin, in which no melanosomes are visible. Fig. 2B shows a typical hair follicle from C3Hf skin, with melanosomes scattered throughout. In the pigmented follicles from cm1OR/cm1OR skin, however, the pattern of melanosome distribution is similiar to that in C3Hf hair follicles (data not shown).
Figure 2.
(A) Electron micrograph of a typical hair follicle from cm1OR/cm1OR skin (×1500). Note the complete absence of melanin. (B) Electron micrograph of a typical hair follicle from C3Hf skin (×3000). Note the presence of melanin, indicated by thick arrows. (C) Detection of melanocytes in cm1OR/cm1OR hair follicles by antiPEP8 (×200). Thick arrows point to some of the pigmented hair follicles. Thin arrows point to some of the unpigmented hair follicles, which still react with antiPEP8 to yield the faint yellow staining in the original color picture.
To determine if melanocytes are present in unpigmented regions of the follicles of cm1OR/cm1OR mice, skin sections from 4-day-old mice were reacted with the melanocyte marker antiPEP8. AntiPEP8 is a polyclonal antibody recognizing specifically DOPAchrome tautomerase (TRP-2) (16). As shown in Fig. 2C, melanocytes are indeed present in cm1OR/cm1OR hair follicles, including those in which there is no pigment.
Insertion of an IAP Element into the 5′ Flanking Region of the Tyrosinase Gene in cm1OR.
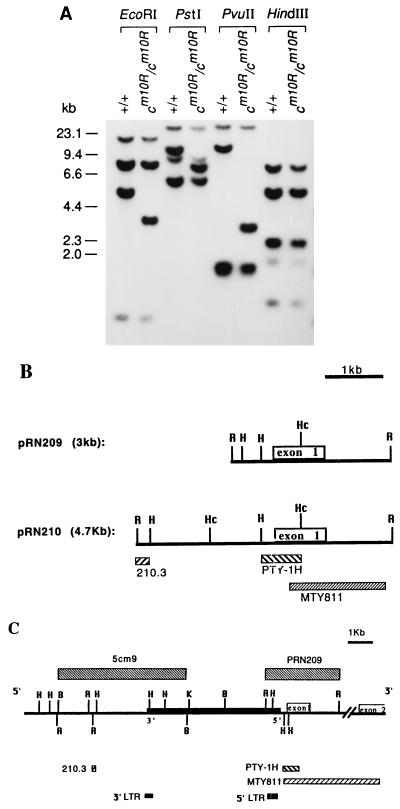
Restriction fragment length polymorphisms for a number of enzymes were detected between cm1OR/cm1OR and C3Hf wild-type genomic DNAs probed with the tyrosinase cDNA probe MTY811 (Fig. 3A). For example, the wild-type 4.7-kb EcoRI fragment, which is the most 5′ exon-containing EcoRI fragment (17), is absent in mutant DNA, and is replaced by a rearranged 3-kb EcoRI fragment. This mutant 3-kb EcoRI fragment (pRN209) was then cloned from genomic DNA of cm1OR/cm1OR mice and compared with the wild-type 4.7-kb fragment (pRN210) by restriction mapping. Fig. 3B shows that these fragments diverge at a point upstream of the HindIII site nearest the Tyr coding sequences, suggesting that DNA rearrangement occurred in that region. The ends of the mutant 3-kb EcoRI fragment in pRN209 were sequenced, revealing approximately 700 bp of IAP sequence in one end of pRN209; the rest of the pRN209 sequence is identical to the published sequence for the wild-type Tyr upstream region.
Figure 3.
Structure of the mutant tyrosinase gene. (A) Southern blot analysis. Genomic DNA were prepared from C3Hf-cm1OR/cm1OR and C3Hf mice, digested with the enzymes indicated, blotted, and hybridized with the tyrosinase cDNA probe, MTY811. (B) Restriction map of pRN209 and pRN210. R, EcoRI; H, HindIII; Hc, HincII. (C) Restriction map of the mutant tyrosinase gene. The solid box on the horizontal line represents the 5.4-kb IAP element, which is inserted head-to-head into the upstream region of the Tyr gene. Boxes below the horizontal line represent the positions of individual probes used in Southern blot and Northern blot analyses. R, EcoRI; H, HindIII; Hc, HincII; B, BamHI; K, KpnI; X, XhoI.
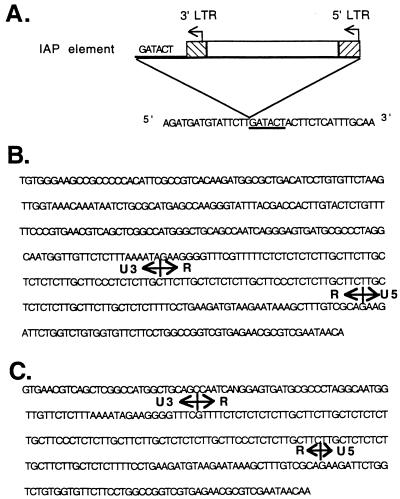
To characterize further the nature of the 5′ Tyr rearrangement, PTY-1H, a HindIII–HincII restriction fragment upstream of the Tyr gene, was used as a probe to isolate a 15-kb clone, λcm9, from a cm1OR/cm1OR genomic λ library. A restriction map was then constructed for λcm9 (Fig. 3C). An IAP LTR probe hybridizes to separate fragments from λcm9, suggesting that LTR sequences are present on both ends of the IAP insertion. To define the other end of the Tyr–IAP junction, the 5.4-kb BamHI fragment from λcm9 was subcloned into pBSSK+ to yield plasmid 5cm9 (Fig. 3C). Wild-type Tyr sequences around the point of divergence were obtained by sequencing the corresponding region of pRN210. A primer was generated from the wild-type Tyr sequence near the divergence point and used to sequence toward the IAP in 5cm9. As is typical of IAP transpositions, there is a 6-bp duplication of endogenous gene sequences at the boundary of the Tyr–IAP junction, which is at the −225 bp position relative to the tyrosinase major transcription start site. Overall, restriction mapping of λcm9 and limited DNA sequence analysis of λcm9 subclones revealed that there is an IAP element of approximately 5.4 kb inserted upstream of the tyrosinase promoter sequences (Fig. 3C). This IAP insertion appears to be intact and of the IΔ1 type, which contains a diagnostic 4-kb HindIII restriction fragment (8). Further DNA sequencing analysis of the LTR on both ends of the IAP insertion showed that the IAP element is inserted “head-to-head” with the Tyr gene (Fig. 3C). Fig. 4 shows the sequences near the IAP integration site, as well as the sequences for both the 5′ LTR and the 3′ LTR.
Figure 4.
Partial DNA sequences of the cm1OR Tyr gene covering both ends of the IAP element. (A) Wild-type tyrosinase sequences at the IAP insertion site. The underlined 6-bp sequences are the duplicated endogenous sequences at the divergent site. (B) 5′ LTR sequences of the IAP insertion. (C) 3′ LTR sequences of the IAP insertion.
Tissue-Specific, but Lower, Expression of Tyrosinase Transcripts in cm1OR/cm1OR Skin.
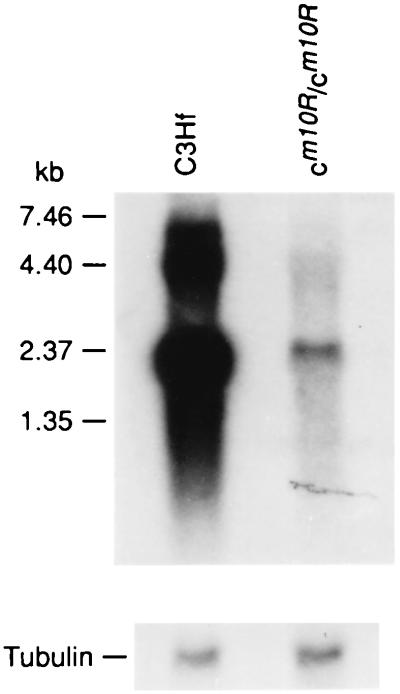
Poly(A)+ RNAs were prepared from the dorsal skin of 4-day-old mice of C3Hf and C3Hf-cm1OR/cm1OR inbred strains, blotted, and hybridized with MTY811 (Fig. 5). After a 1-week exposure of the autoradiogram, weak signals appeared in cm1OR/cm1OR skin RNA, whereas strong signals appeared in the C3Hf control skin RNA. These results show that the total expression of Tyr gene in the mutant skin is substantially decreased.
Figure 5.
Northern blot of poly(A)+ skin RNA, prepared from 4-day-old C3Hf-cm1OR/cm1OR and C3Hf mice, hybridized to probe MTY811 (Upper). The blot was rehybridized to a chicken tubulin probe for loading control (Lower).
To detect whether tyrosinase transcripts are aberrantly expressed in other tissues, RNAs from brain, heart, liver, spleen, testis, thymus, and kidney of wild-type and mutant mice were extracted, Northern blotted, and probed with MTY811. No hybridization was detected in either mutant or wild-type mice (data not shown). Therefore, as in wild-type mice, tyrosinase gene expression in cm1OR/cm1OR mutant mice is restricted to skin.
No IAP-Tyrosinase Fusion Transcript Is Detected in the Mutant Mice.
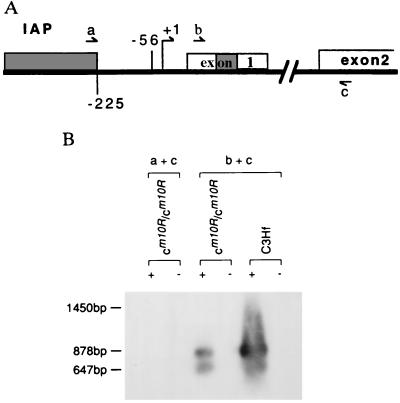
Some IAP LTRs have promoter function in both sense and antisense orientations (18). To detect whether the transcription of the tyrosinase gene is initiated from the IAP LTR promoter, RT-PCR analysis was performed on C3Hf-cm1OR/cm1OR skin RNAs by using primers from the 5′ end of the IAP LTR (primer a) and from the 5′ end of the second exon of the tyrosinase gene (primer c) (Fig. 6A). As a control, RT-PCR analysis was also done on both C3Hf and C3Hf-cm1OR/cm1OR 4-day skin RNAs with primers derived from the first (primer b) and second (primer c) exons of the tyrosinase gene (Fig. 6A). Whereas the primer b + c combination amplified the correct sized Tyr cDNA fragments from both C3Hf-cm1OR/cm1OR and C3Hf skin sample, no PCR product was detected using the primer a + c combination (Fig. 6B). Therefore, abnormal tyrosinase expression is not associated with an aberrant transcript originating from the IAP LTR promoter.
Figure 6.
RT-PCR analysis. (A) Schematic representation of the positions for the primers used in RT-PCR analysis. The +1 represents the tyrosinase major transcription start site. The −56 represents the tyrosinase minor transcription start site (3). The IAP insertion is presented as a solid box. There are two alternatively spliced Tyr transcripts that are different in exon 1 (19). tyr1 contains the full-length of exon 1, tyr2 deletes the shaded box in exon 1. The distance between primers b and c is 878 bp in tyr1 and 647 bp in tyr2. If the tyr transcripts are initiated from IAP promoter in cm1OR/cm1OR mice, the expected distance between primers a and c is 1.45 kb in tyr1 and 1.24 kb in tyr2. (B) Total skin RNA, prepared from 4-day-old C3Hf mice and cm1OR/cm1OR, respectively, was reverse transcribed in the presence (+) or absence (−) of reverse transcriptase; the resulting cDNA was then amplified by PCR using different primer combinations. The amplified PCR products were electrophoresed, blotted, and hybridized with MTY811.
DISCUSSION
Mosaicism for coat pigmentation has been observed in many naturally occurring mutations at various coat-color loci. For example, chinchilla-mottled mice (cm/cm) have a DNA rearrangement starting 5.4 kb upstream of the tyrosinase gene transcription start site (20), whereas the mottled yellow and pseudoagouti coat color of Aiapy/− mice is caused by a 5.2-kb IAP insertion immediately 5′ of the first coding exon of the agouti gene (21). Mosaic coat pigmentation is also frequently observed in transgenic mice carrying tyrosinase minigenes with various upstream sequences linked to the tyrosinase promoter and coding sequences (22–24).
cm1OR results in a mottled coat-color appearance with interspersed dark hairs on an albino background. Many fewer pigmented hairs are present in cm1OR/cm1OR skin than in C3Hf wild-type controls, which correlates with the observations from electron micrographs that some cm1OR/cm1OR hair follicles produce melanin, whereas others do not; we have shown that melanocytes are present even in cm1OR/cm1OR follicles not producing melanin.
We have characterized cm1OR at the molecular level, and have shown this mutation to be caused by an IAP element insertion 225 bp upstream from the major initiation site for tyrosinase transcription. Northern blot analysis and RT-PCR data demonstrate that tyrosinase expression is significantly decreased in C3Hf-cm1OR/cm1OR skin compared with wild-type controls.
One possible explanation for this decrease in expression is that the 5.4-kb IAP insertion may separate an upstream cis-acting element(s) from the tyrosinase-coding sequences. Wild-type tyrosinase minigenes containing 5.5 or 2.6 kb of DNA upstream from coding sequences fail to restore coat pigmentation to wild-type levels in the resulting transgenic mice (25, 26). In contrast, a 250-kb yeast artificial chromosome transgene containing the full-length tyrosinase gene plus many kilobases of surrounding genomic sequences can restore wild-type pigmentation (27). These findings suggest the existence of an upstream regulatory element(s) that is important for full expression of the tyrosinase gene in melanocytes. A melanocyte-specific DNase I hypersensitive site has been identified 15 kb upstream of the mouse Tyr gene by analysis of transgenic mice and by transient transfection assays (24, 28). The hypersensitive site region is believed to contain a strong cell-specific enhancer that significantly increases the expression of tyrosinase in melanocytes. In cm1OR, the 5.4-kb IAP element is inserted 225 bp upstream of the tyrosinase promoter, thereby increasing the distance between this enhancer element and the tyrosinase promoter. Alternatively, the IAP insertion itself may confer certain suppressor effects on tyrosinase gene expression.
Several possibilities have been proposed for the cis-acting mechanisms that lead to mosaicism. Mosaicism of gene expression may be caused by the failure to form the appropriate chromatin structure required for full expression of that gene. For example, in cm/cm mice, light-melanocyte clones have reduced DNase I hypersensitivity in tyrosinase chromatin when compared with dark-melanocyte clones (24). This variable DNase I hypersensitivity may reflect variable ability within individual cells to form appropriate chromatin configurations required for Tyr gene transcription, leading to the mosaic coat-color appearance. In cm1OR/cm1OR mice, the 5.4-kb IAP insertion upstream of the tyrosinase promoter may have changed the chromatin structure, preventing the formation of a stable initiation complex, and causing different levels of tyrosinase gene expression in individual melanocytes.
Variable methylation may also play a role in the mosaic expression pattern of genes. In Aiapy/- mice, increased expression of the Aiapy allele, which leads to higher production of phaeomelanin, is correlated with a lower methylation status of the the HpaII and HhaI restriction enzyme sites in the 5′ LTR of the inserted IAP (21). A similiar experiment was performed in cm1OR/cm1OR mice to compare, with respect to dark and light patches of skin, the methylation status of the HhaI and HpaII sites in the 5′ LTR of the IAP insertion in genomic DNA. The HhaI and HpaII sites were found to be methylated in both samples (data not shown). Therefore, the methylation status of the cm1OR allele does not seem to be directly associated with the variation of tyrosinase expression in melanocytes. However, because the methylation study was done with DNA extracted from total skin, and melanocytes are a minor component of cells represented in the sample, the analysis of melanocyte-specific methylation differences at the LTR sites will require the cloning of individual lines of melanocytes for in vitro studies of methylation patterns. Such clones would also allow the analysis of local chromatin structure in cells expressing differing levels of tyrosinase.
Because pigmentation patterns can be easily visualized, coat-color gene mutations are excellent models for the study of mosaicism of gene expression. It is clear from this study that even among cells of identical genotype in an inbred strain of mice, epigenetic events impose differences on the regulation of tyrosinase expression in individual skin melanocytes. Clarifying the mechanisms of cm1OR tyrosinase gene expression will help us to understand the heterogeneity of gene expression.
Acknowledgments
We gratefully acknowledge Kay Houser, Richard Machanoff, and P. R. Hunsicker for expert technical assistance, and Liane Russell and E. J. Michaud for critical reading of the manuscript. This manuscript has been authored by a contractor of the United States Government under contract No. DE-AC05–96OR22464.
Footnotes
Abbreviations: IAP, intracisternal A particle; LTR, long terminal repeat; RT, reverse transcription.
References
- 1.Lerner A B, Fitzpatrick T B, Calkins E, Summerson W H. J Biol Chem. 1949;178:185–195. [Google Scholar]
- 2.Kwon B S, Haq A K, Pomerantz S H, Halaban R. Proc Natl Acad Sci USA. 1987;84:7473–7477. doi: 10.1073/pnas.84.21.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müller G, Ruppert S, Schmid E, Schütz G. EMBO J. 1988;7:2723–2730. doi: 10.1002/j.1460-2075.1988.tb03126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto H, Takeuchi S, Kudo T, Makino K, Nakata A, Shinoda T, Takeuchi T. Jpn J Genet. 1987;62:271–274. [Google Scholar]
- 5.Kwon B S, Wakulckik M, Haq A K, Halaban R, Kestler D. Biochem Biophys Res Commun. 1988;153:1301–1309. doi: 10.1016/s0006-291x(88)81370-6. [DOI] [PubMed] [Google Scholar]
- 6.Yokoyama T, Silversides D, Waymire K G, Known B S, Takeuchi T, Overbeek P A. Nucleic Acids Res. 1990;18:7293–7298. doi: 10.1093/nar/18.24.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono M, Cole M D, White A T, Huang R C C. Cell. 1980;21:465–473. doi: 10.1016/0092-8674(80)90483-3. [DOI] [PubMed] [Google Scholar]
- 8.Kuff E L, Lueders K K. Adv Cancer Res. 1988;51:183–276. doi: 10.1016/s0065-230x(08)60223-7. [DOI] [PubMed] [Google Scholar]
- 9.Nicholls R D, Gottlieb W, Russell L B, Dauda M, Horsthemke B, Rinchik E M. Proc Natl Acad Sci USA. 1993;90:2050–2054. doi: 10.1073/pnas.90.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothstein J R, Johnson D, DeLoia J A, Sknowronsk J, Solter D, Knowles B. Genes Dev. 1992;6:1190–1201. doi: 10.1101/gad.6.7.1190. [DOI] [PubMed] [Google Scholar]
- 11.Kawasaki E S. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 21–27. [Google Scholar]
- 12.Pieretti M, Zhang F, Fu Y H, Warren S T, Oostra B A, Caskey C T, Nelson D L. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 13.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson J E, Lillie J H, Suter M M, Smith C A. Am J Vet Res. 1989;50:1161–1165. [PubMed] [Google Scholar]
- 15.Suter M M, Greenberg L J, Wilkinson J E, Lewis R M. J Histochem Cytochem. 1990;38:541–549. doi: 10.1177/38.4.2319124. [DOI] [PubMed] [Google Scholar]
- 16.Tsukamoto K, Jackson I J, Urabe K, Montague P M, Hearing V J. EMBO J. 1992;11:519–526. doi: 10.1002/j.1460-2075.1992.tb05082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rüppert S, Müller G, Kwon B, Schütz G. EMBO J. 1988;7:2715–2722. doi: 10.1002/j.1460-2075.1988.tb03125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christy R J, Huang C C. Mol Cell Biol. 1988;8:1093–1102. doi: 10.1128/mcb.8.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terao M, Tabe L, Garattini E, Sartori D, Studer M, Mintz B. Biochem Biophys Res Commun. 1989;159:848–853. doi: 10.1016/0006-291x(89)90072-7. [DOI] [PubMed] [Google Scholar]
- 20.Porter S, Larue L, Mintz B. Dev Genet. 1991;12:393–402. doi: 10.1002/dvg.1020120604. [DOI] [PubMed] [Google Scholar]
- 21.Michaud E, Vugt M J V, Bultman S J, Sweet H O, Davisson M T, Woychik R P. Genes Dev. 1994;8:1463–1472. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- 22.Mintz B, Bradl M. Proc Natl Acad Sci USA. 1991;88:9643–9647. doi: 10.1073/pnas.88.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi, T., Tanaka, S. & Tanaka, M. (1993) J. Invest. Dermatol. 100, Suppl., 141S–145S. [PubMed]
- 24.Porter S, Meyer C J. Development (Cambridge, UK) 1994;120:2103–2111. doi: 10.1242/dev.120.8.2103. [DOI] [PubMed] [Google Scholar]
- 25.Beermann F, Ruppert S, Hummler E M, Bosch F X, Muller G, Ruther U, Schutz G. EMBO J. 1990;9:2819–2826. doi: 10.1002/j.1460-2075.1990.tb07470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka S, Yamamoto H, Takeuchi S, Takeuchi T. Development (Cambridge, UK) 1990;108:223–227. doi: 10.1242/dev.108.2.223. [DOI] [PubMed] [Google Scholar]
- 27.Schedl A, Montoliu L, Kelsey G, Schutz G. Nature (London) 1993;362:258–261. doi: 10.1038/362258a0. [DOI] [PubMed] [Google Scholar]
- 28.Ganss R, Montoliu L, Monaghan A P, Schütz G. EMBO J. 1994;13:3083–3093. doi: 10.1002/j.1460-2075.1994.tb06607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]