Abstract
The gene encoding a denitrification enzyme, nitrous oxide reductase (EC 1.7.99.6), in Rhizobium meliloti and other gram-negative bacteria was detected by hybridization to an internal 1.2-kb PstI fragment of the structural gene (nosZ) cloned from Pseudomonas stutzeri Zobell (W.G. Zumft, A. Viebrock-Sambale, and C. Braun, Eur. J. Biochem. 192:591-599, 1990). Homology to the probe was detected in the DNAs of two N2-fixing strains of P. stutzeri, two denitrifying Pseudomonas species, one Alcaligenes eutrophus strain, and 36 of 56 R. meliloti isolates tested. Except for two isolates of R. meliloti, all showed nitrous oxide reduction activity (Nos+). Therefore, at least part of the nosZ sequence appears to be conserved and widely distributed among denitrifiers, which include free-living and symbiotic diazotrophs. By using Agrobacterium tumefaciens transconjugants harboring different megaplasmids of R. meliloti JJ1c10 and SU47, sequence homology with the nosZ probe was unequivocally located on the nod megaplasmid. A cosmid clone of JJ1c10 in which nosZ homology was mapped on a 4.2-kb BamHI fragment was selected. This cosmid, which conferred Nos+ activity to the R. meliloti wild-type strains ATCC 9930 and Balsac (Nos- and nondenitrifying, respectively) also restored Nos+ activity in the mutants of JJ1c10 and SU47 in which the 4.2-kb BamHI segment was deleted. Therefore, this segment contains sequences essential for nos gene expression in JJ1c10 and SU47 and thus confirms that the nod megaplasmid in JJ1c10 and SU47 which carries genes essential for symbiotic dinitrogen fixation also carries genes involved in the antagonistic process of denitrification.
Full text
PDF


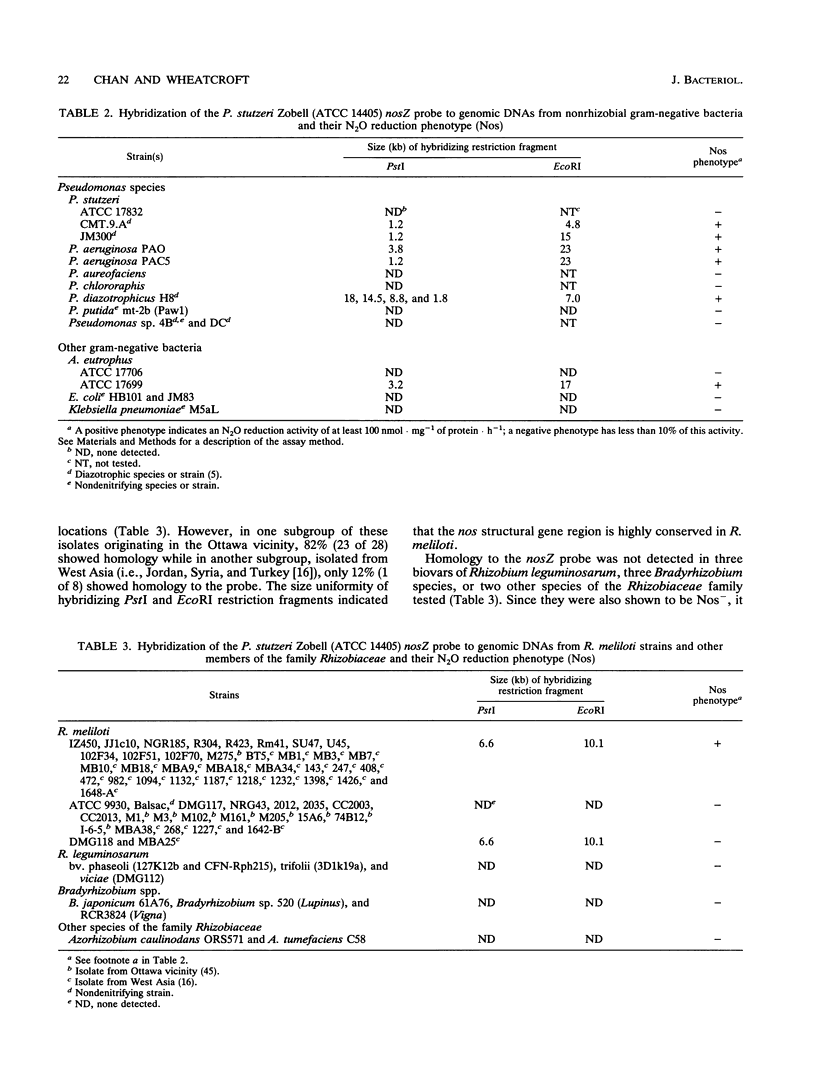
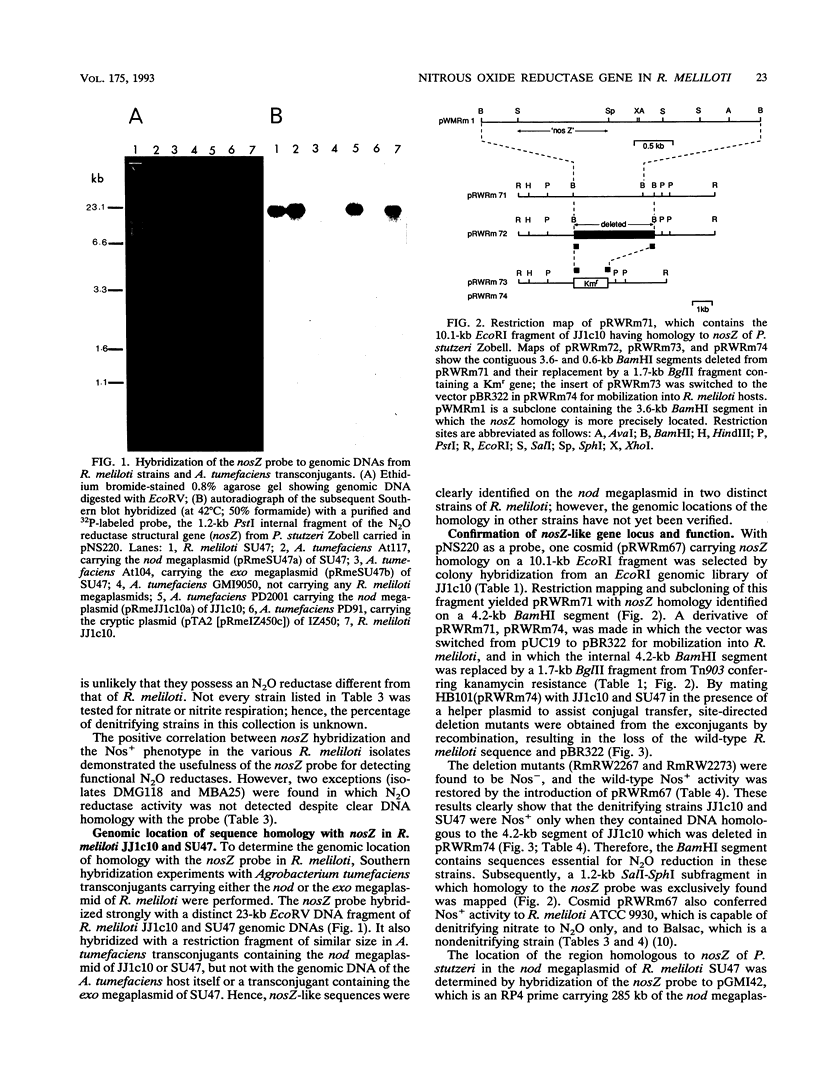



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barraquio W. L., Dumont A., Knowles R. Enumeration of free-living aerobic n(2)-fixing h(2)-oxidizing bacteria by using a heterotrophic semisolid medium and most-probable-number technique. Appl Environ Microbiol. 1988 Jun;54(6):1313–1317. doi: 10.1128/aem.54.6.1313-1317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bromfield E. S., Sinha I. B., Wolynetz M. S. Influence of Location, Host Cultivar, and Inoculation on the Composition of Naturalized Populations of Rhizobium meliloti in Medicago sativa Nodules. Appl Environ Microbiol. 1986 May;51(5):1077–1084. doi: 10.1128/aem.51.5.1077-1084.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Schilling-Cordaro C., Mergia A., Houck C. M. A new technique for genetic engineering of Agrobacterium Ti plasmid. Plasmid. 1983 Jul;10(1):21–30. doi: 10.1016/0147-619x(83)90054-9. [DOI] [PubMed] [Google Scholar]
- Eardly B. D., Materon L. A., Smith N. H., Johnson D. A., Rumbaugh M. D., Selander R. K. Genetic structure of natural populations of the nitrogen-fixing bacterium Rhizobium meliloti. Appl Environ Microbiol. 1990 Jan;56(1):187–194. doi: 10.1128/aem.56.1.187-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallik E., Chan Y. K., Robson R. L. Detection of alternative nitrogenases in aerobic gram-negative nitrogen-fixing bacteria. J Bacteriol. 1991 Jan;173(1):365–371. doi: 10.1128/jb.173.1.365-371.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M., Kunkel B., De Vos G. F., Signer E. R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986 Jul;167(1):66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Greenberg E. P., Becker G. E. Nitrous oxide as end product of denitrification by strains of fluorescent pseudomonads. Can J Microbiol. 1977 Jul;23(7):903–907. doi: 10.1139/m77-133. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüngst A., Braun C., Zumft W. G. Close linkage in Pseudomonas stutzeri of the structural genes for respiratory nitrite reductase and nitrous oxide reductase, and other essential genes for denitrification. Mol Gen Genet. 1991 Feb;225(2):241–248. doi: 10.1007/BF00269855. [DOI] [PubMed] [Google Scholar]
- Kahn D., David M., Domergue O., Daveran M. L., Ghai J., Hirsch P. R., Batut J. Rhizobium meliloti fixGHI sequence predicts involvement of a specific cation pump in symbiotic nitrogen fixation. J Bacteriol. 1989 Feb;171(2):929–939. doi: 10.1128/jb.171.2.929-939.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles R. Denitrification. Microbiol Rev. 1982 Mar;46(1):43–70. doi: 10.1128/mr.46.1.43-70.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner H., Frunzke K., Döhler K., Zumft W. G. Immunochemical patterns of distribution of nitrous oxide reductase and nitrite reductase (cytochrome cd1) among denitrifying pseudomonads. Arch Microbiol. 1987 Jun;148(1):20–24. doi: 10.1007/BF00429641. [DOI] [PubMed] [Google Scholar]
- Renalier M. H., Batut J., Ghai J., Terzaghi B., Gherardi M., David M., Garnerone A. M., Vasse J., Truchet G., Huguet T. A new symbiotic cluster on the pSym megaplasmid of Rhizobium meliloti 2011 carries a functional fix gene repeat and a nod locus. J Bacteriol. 1987 May;169(5):2231–2238. doi: 10.1128/jb.169.5.2231-2238.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C., Boistard P., Dénarié J., Casse-Delbart F. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet. 1981;184(2):326–333. doi: 10.1007/BF00272926. [DOI] [PubMed] [Google Scholar]
- Stouthamer A. H. Metabolic regulation including anaerobic metabolism in Paracoccus denitrificans. J Bioenerg Biomembr. 1991 Apr;23(2):163–185. doi: 10.1007/BF00762216. [DOI] [PubMed] [Google Scholar]
- Sundqvist A., Berg M., Moreno-López J., Linné T. The haemagglutinin-neuraminidase glycoprotein of the porcine paramyxovirus LPMV: comparison with other paramyxoviruses revealed the closest relationship to simian virus 5 and mumps virus. Arch Virol. 1992;122(3-4):331–340. doi: 10.1007/BF01317194. [DOI] [PubMed] [Google Scholar]
- Viebrock A., Zumft W. G. Molecular cloning, heterologous expression, and primary structure of the structural gene for the copper enzyme nitrous oxide reductase from denitrifying Pseudomonas stutzeri. J Bacteriol. 1988 Oct;170(10):4658–4668. doi: 10.1128/jb.170.10.4658-4668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wheatcroft R., Laberge S. Identification and nucleotide sequence of Rhizobium meliloti insertion sequence ISRm3: similarity between the putative transposase encoded by ISRm3 and those encoded by Staphylococcus aureus IS256 and Thiobacillus ferrooxidans IST2. J Bacteriol. 1991 Apr;173(8):2530–2538. doi: 10.1128/jb.173.8.2530-2538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatcroft R., Watson R. J. A Positive Strain Identification Method for Rhizobium meliloti. Appl Environ Microbiol. 1988 Feb;54(2):574–576. doi: 10.1128/aem.54.2.574-576.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yoshinari T. N2O reduction by Vibrio succinogenes. Appl Environ Microbiol. 1980 Jan;39(1):81–84. doi: 10.1128/aem.39.1.81-84.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W. G., Viebrock-Sambale A., Braun C. Nitrous oxide reductase from denitrifying Pseudomonas stutzeri. Genes for copper-processing and properties of the deduced products, including a new member of the family of ATP/GTP-binding proteins. Eur J Biochem. 1990 Sep 24;192(3):591–599. doi: 10.1111/j.1432-1033.1990.tb19265.x. [DOI] [PubMed] [Google Scholar]