Abstract
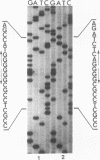
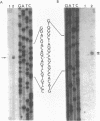
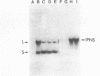
The complete nucleotide sequence of a small linear plasmid (pSCL1) from Streptomyces clavuligerus has been determined. This plasmid is 11,696 bp in length, has a 72% G+C content, and has approximately 900-bp inverted terminal repeat sequences. A comparison of the inverted terminal repeats of pSCL1 with those of a linear plasmid from S. rochei shows that the two terminal sequences have a high degree of similarity (approximately 70%). Several small inverted repeats found in the long terminal sequences of both plasmids are also conserved. An analysis of the sequence and codon preferences indicates that pSCL1 has seven or eight highly probable protein-coding open reading frames (ORFs). However, only two RNA species encoded by pSCL1 were detected in S. clavuligerus grown in liquid culture. The larger of these transcripts (900 nucleotides) corresponds to an ORF and is likely to be an mRNA for a protein similar to the KorA protein of pIJ101. The smaller transcript (460 nucleotides) does not correspond to any ORF; however, its 5' end is complementary to the 5' end of a predicted mRNA, suggesting that it may function as an antisense RNA. The larger of the two RNA species was present at a high level during the early stage of growth in liquid medium, and then its apparent rate of transcription decreased and remained at a lower level through the later stages; the level of the smaller RNA species remained relatively constant through all stages of growth.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison S. L., Phillips A. T. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Pseudomonas putida. J Bacteriol. 1990 Sep;172(9):5470–5476. doi: 10.1128/jb.172.9.5470-5476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour A. G., Garon C. F. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987 Jul 24;237(4813):409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990 Apr 25;265(12):6513–6516. [PubMed] [Google Scholar]
- Bernad A., Lázaro J. M., Salas M., Blanco L. The highly conserved amino acid sequence motif Tyr-Gly-Asp-Thr-Asp-Ser in alpha-like DNA polymerases is required by phage phi 29 DNA polymerase for protein-primed initiation and polymerization. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4610–4614. doi: 10.1073/pnas.87.12.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Buck D., Guest J. R. Overexpression and site-directed mutagenesis of the succinyl-CoA synthetase of Escherichia coli and nucleotide sequence of a gene (g30) that is adjacent to the suc operon. Biochem J. 1989 Jun 15;260(3):737–747. doi: 10.1042/bj2600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone F. J., Britten R. J., Davidson E. H. Mapping of gene transcripts by nuclease protection assays and cDNA primer extension. Methods Enzymol. 1987;152:611–632. doi: 10.1016/0076-6879(87)52069-9. [DOI] [PubMed] [Google Scholar]
- Chen C. M., Ye Q. Z., Zhu Z. M., Wanner B. L., Walsh C. T. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J Biol Chem. 1990 Mar 15;265(8):4461–4471. [PubMed] [Google Scholar]
- Epp J. K., Burgett S. G., Schoner B. E. Cloning and nucleotide sequence of a carbomycin-resistance gene from Streptomyces thermotolerans. Gene. 1987;53(1):73–83. doi: 10.1016/0378-1119(87)90094-1. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Hirochika H., Nakamura K., Sakaguchi K. A linear DNA plasmid from Streptomyces rochei with an inverted terminal repetition of 614 base pairs. EMBO J. 1984 Apr;3(4):761–766. doi: 10.1002/j.1460-2075.1984.tb01881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G. H., Leavitt M. C., Hsieh J. C., Ito J. Bacteriophage PRD1 DNA polymerase: evolution of DNA polymerases. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8287–8291. doi: 10.1073/pnas.84.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkus J., Reh M., Schlegel H. G. Hydrogen autotrophy of Nocardia opaca strains is encoded by linear megaplasmids. J Gen Microbiol. 1990 Jun;136(6):1145–1151. doi: 10.1099/00221287-136-6-1145. [DOI] [PubMed] [Google Scholar]
- Keen C. L., Mendelovitz S., Cohen G., Aharonowitz Y., Roy K. L. Isolation and characterization of a linear DNA plasmid from Streptomyces clavuligerus. Mol Gen Genet. 1988 Apr;212(1):172–176. doi: 10.1007/BF00322461. [DOI] [PubMed] [Google Scholar]
- Kemble R. J., Mans R. J. Examination of the mitochondrial genome of revertant progeny from S cms maize with cloned S-1 and S-2 hybridization probes. J Mol Appl Genet. 1983;2(2):161–171. [PubMed] [Google Scholar]
- Kendall K. J., Cohen S. N. Complete nucleotide sequence of the Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988 Oct;170(10):4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinashi H., Shimaji-Murayama M., Hanafusa T. Nucleotide sequence analysis of the unusually long terminal inverted repeats of a giant linear plasmid, SCP1. Plasmid. 1991 Sep;26(2):123–130. doi: 10.1016/0147-619x(91)90052-x. [DOI] [PubMed] [Google Scholar]
- Kinashi H., Shimaji M. Detection of giant linear plasmids in antibiotic producing strains of Streptomyces by the OFAGE technique. J Antibiot (Tokyo) 1987 Jun;40(6):913–916. doi: 10.7164/antibiotics.40.913. [DOI] [PubMed] [Google Scholar]
- Kinashi H., Shimaji M., Sakai A. Giant linear plasmids in Streptomyces which code for antibiotic biosynthesis genes. 1987 Jul 30-Aug 5Nature. 328(6129):454–456. doi: 10.1038/328454a0. [DOI] [PubMed] [Google Scholar]
- Kirby R., Hopwood D. A. Genetic determination of methylenomycin synthesis by the SCP1 plasmid of Streptomyces coelicolor A3(2). J Gen Microbiol. 1977 Jan;98(1):239–252. doi: 10.1099/00221287-98-1-239. [DOI] [PubMed] [Google Scholar]
- Kuzmin E. V., Levchenko I. V. S1 plasmid from cms-S-maize mitochondria encodes a viral type DNA-polymerase. Nucleic Acids Res. 1987 Aug 25;15(16):6758–6758. doi: 10.1093/nar/15.16.6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskiw B. K., Aharonowitz Y., Mevarech M., Wolfe S., Vining L. C., Westlake D. W., Jensen S. E. Cloning and nucleotide sequence determination of the isopenicillin N synthetase gene from Streptomyces clavuligerus. Gene. 1988;62(2):187–196. doi: 10.1016/0378-1119(88)90557-4. [DOI] [PubMed] [Google Scholar]
- Lis J. T. Fractionation of DNA fragments by polyethylene glycol induced precipitation. Methods Enzymol. 1980;65(1):347–353. doi: 10.1016/s0076-6879(80)65044-7. [DOI] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988 Mar 11;16(5):1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miyashita S., Hirochika H., Ikeda J. E., Hashiba T. Linear plasmid DNAs of the plant pathogenic fungus Rhizoctonia solani with unique terminal structures. Mol Gen Genet. 1990 Jan;220(2):165–171. doi: 10.1007/BF00260476. [DOI] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H., Kondo A., Ohshima A., Ogura T., Hiraga S. Structure and function of the F plasmid genes essential for partitioning. J Mol Biol. 1986 Nov 5;192(1):1–15. doi: 10.1016/0022-2836(86)90459-6. [DOI] [PubMed] [Google Scholar]
- Morrison D. A. Transformation and preservation of competent bacterial cells by freezing. Methods Enzymol. 1979;68:326–331. doi: 10.1016/0076-6879(79)68023-0. [DOI] [PubMed] [Google Scholar]
- Motallebi-Veshareh M., Rouch D. A., Thomas C. M. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol Microbiol. 1990 Sep;4(9):1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- Neal R. J., Chater K. F. Nucleotide sequence analysis reveals similarities between proteins determining methylenomycin A resistance in Streptomyces and tetracycline resistance in eubacteria. Gene. 1987;58(2-3):229–241. doi: 10.1016/0378-1119(87)90378-7. [DOI] [PubMed] [Google Scholar]
- Oeser B., Tudzynski P. The linear mitochondrial plasmid pClK1 of the phytopathogenic fungus Claviceps purpurea may code for a DNA polymerase and an RNA polymerase. Mol Gen Genet. 1989 May;217(1):132–140. doi: 10.1007/BF00330952. [DOI] [PubMed] [Google Scholar]
- Paillard M., Sederoff R. R., Levings C. S. Nucleotide sequence of the S-1 mitochondrial DNA from the S cytoplasm of maize. EMBO J. 1985 May;4(5):1125–1128. doi: 10.1002/j.1460-2075.1985.tb03749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrich A. K., Wu X., Roy K. L., Jensen S. E. Transcriptional analysis of the isopenicillin N synthase-encoding gene of Streptomyces clavuligerus. Gene. 1992 Feb 1;111(1):77–84. doi: 10.1016/0378-1119(92)90605-o. [DOI] [PubMed] [Google Scholar]
- Rekosh D. M., Russell W. C., Bellet A. J., Robinson A. J. Identification of a protein linked to the ends of adenovirus DNA. Cell. 1977 Jun;11(2):283–295. doi: 10.1016/0092-8674(77)90045-9. [DOI] [PubMed] [Google Scholar]
- Rigaud G., Grange T., Pictet R. The use of NaOH as transfer solution of DNA onto nylon membrane decreases the hybridization efficiency. Nucleic Acids Res. 1987 Jan 26;15(2):857–857. doi: 10.1093/nar/15.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K. Invertrons, a class of structurally and functionally related genetic elements that includes linear DNA plasmids, transposable elements, and genomes of adeno-type viruses. Microbiol Rev. 1990 Mar;54(1):66–74. doi: 10.1128/mr.54.1.66-74.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas M., Mellado R. P., Viñuela E. Characterization of a protein covalently linked to the 5' termini of the DNA of Bacillus subtilis phage phi29. J Mol Biol. 1978 Feb 25;119(2):269–291. doi: 10.1016/0022-2836(78)90438-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Bender R. A. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Klebsiella aerogenes. J Bacteriol. 1990 Sep;172(9):5477–5481. doi: 10.1128/jb.172.9.5477-5481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiffman D., Cohen S. N. Reconstruction of a Streptomyces linear replicon from separately cloned DNA fragments: existence of a cryptic origin of circular replication within the linear plasmid. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):6129–6133. doi: 10.1073/pnas.89.13.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Stark M. J., Mileham A. J., Romanos M. A., Boyd A. Nucleotide sequence and transcription analysis of a linear DNA plasmid associated with the killer character of the yeast Kluyveromyces lactis. Nucleic Acids Res. 1984 Aug 10;12(15):6011–6030. doi: 10.1093/nar/12.15.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein D. S., Kendall K. J., Cohen S. N. Identification and analysis of transcriptional regulatory signals for the kil and kor loci of Streptomyces plasmid pIJ101. J Bacteriol. 1989 Nov;171(11):5768–5775. doi: 10.1128/jb.171.11.5768-5775.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the pyruvate dehydrogenase component. Eur J Biochem. 1983 Jun 1;133(1):155–162. doi: 10.1111/j.1432-1033.1983.tb07441.x. [DOI] [PubMed] [Google Scholar]
- Strohl W. R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992 Mar 11;20(5):961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Smith C. A. The trfB region of broad host range plasmid RK2: the nucleotide sequence reveals incC and key regulatory gene trfB/korA/korD as overlapping genes. Nucleic Acids Res. 1986 Jun 11;14(11):4453–4469. doi: 10.1093/nar/14.11.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasino M., Ricci S., Galeotti C. L. Genome organization of the killer plasmid pGK12 from Kluyveromyces lactis. Nucleic Acids Res. 1988 Jul 11;16(13):5863–5878. doi: 10.1093/nar/16.13.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Winship P. R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989 Feb 11;17(3):1266–1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]