Abstract
Anteroposterior patterning of neural tissue is thought to be directed by the axial mesoderm which is functionally divided into head and trunk organizer. The LIM class homeobox gene Xlim-1 is expressed in the entire axial mesoderm, whereas the distinct transcription factor Xbra is expressed in the notochord but not in the prechordal mesoderm. mRNA injection experiments showed that Xenopus animal explants (caps) expressing an activated form of Xlim-1 (a LIM domain mutant named 3m) induce anterior neural markers whereas caps coexpressing Xlim-1/3m and Xbra induce posterior neural markers. These data indicate that, in terms of neural inducing ability, Xlim-1/3m-expressing caps correspond to the head organizer and Xlim-1/3m plus Xbra-coexpressing caps to the trunk organizer. Thus the expression domains of Xlim-1 and Xbra correlate with, and possibly define, the functional domains of the organizer. In animal caps Xlim-1/3m initiates expression of a neuralizing factor, chordin, whereas Xbra activates embryonic fibroblast growth factor (eFGF) expression, as reported previously; these factors could mediate the neural inducing and patterning effects that were observed. A dominant-negative FGF receptor (XFD) inhibits posteriorization by Xbra in a dose-dependent manner, supporting the suggestion that eFGF or a related factor has posteriorizing influence.
In early development of vertebrates, the dorsal mesoderm, known as the Spemann organizer in amphibians, plays a central role in establishing the basic body plan. The axial mesoderm, formed by involuting mesodermal tissue at the dorsal midline during gastrula and neurula stages, can be divided morphologically and functionally into the prechordal (or head) mesoderm and the notochord (1–5). Several transcription factors are expressed in the axial mesoderm of Xenopus embryos (6). Among them, the LIM class homeobox gene Xlim-1 is expressed in the entire axial mesoderm (7), the homeobox genes goosecoid (8) and Otx2 (9, 10) are expressed in the prechordal mesoderm, and Xbra (the Xenopus ortholog of mouse T gene) is expressed in the notochord (11). Thus the expression domains of these genes seem to demarcate the morphological and functional domains of the axial mesoderm, but it is not known which transcription factors specify the anteroposterior prepattern of this tissue.
The functional division of the organizer into head and trunk organizer is based on their ability to induce brain or spinal cord, respectively, as shown in classical transplantation and explantation experiments. These studies indicated that the prechordal mesoderm has head organizer activity while the notochord has trunk organizer activity (1–5). An important feature of the organizer is posterior dominance, shown in combination of head and trunk organizers by Okada and Takaya (12) (see also the review in ref. 3). This observation, together with the fact that posteriorizing and neuralizing activities of axial mesoderm are inversely correlated, led to the proposal of models in which two signals (13, 14) or two gradients (15) are responsible for anteroposterior patterning of the early central nervous system (CNS); these models postulate the action of (at least) two secreted molecules, a neural inducer and a posteriorizing (or caudalizing) factor. According to the two-signal model, anterior type neural tissue is generated by initial neural induction while some of it is subsequently converted to posterior type by a factor which need not have neuralizing activity in itself.
Mouse embryos lacking the mouse ortholog of Xlim-1, Lim1 (also named Lhx1; see review in ref. 16), exhibit a headless phenotype, implying that Lim1 is involved in head organizer function, and suggesting that the head organizer can be separated genetically from the trunk organizer (17, 18). In a previous paper (19) we reported that an activated mutant form of Xlim-1, named 3m, can induce neural tissue in animal explants of Xenopus embryos. In the present work we analyze the nature of the induced neural tissue in further detail. Expression of Xlim-1/3m or Xlim-1/3m plus Xbra in animal explants reconstituted head or trunk organizer function, respectively, as defined by the ability to induce anterior or posterior neural tissue. Furthermore we found that Xlim-1/3m can induce chordin, a known neuralizing factor (20), and we provide evidence supporting a posteriorizing role for embryonic fibroblast growth factor (eFGF), previously shown to be activated by Xbra (21). Thus, the neuralizing and patterning effects of Xlim-1 and Xbra may be mediated by the secreted factors chordin and eFGF.
In addition to the role played by polypeptide factors, several studies have shown that retinoic acid (RA) can exert a posteriorizing influence on the early CNS (22, 23), in part mediated by modification of the differentiation of the mesoderm (24, 25). Therefore we tested the effect of RA on Xlim-1/3m-induced anterior neural tissue. The results suggest that RA can modify the anteroposterior nature of this tissue, albeit in a more limited way than Xbra-expressing animal caps.
MATERIALS AND METHODS
Injection of embryos with synthetic mRNA, dissection, and culture of single or combined animal caps has been described, as has RNA extraction and analysis by Northern blot analysis (19, 26). The homeodomain mutant of Xlim-1/3m, named 3mHDm, was constructed using an in vitro site-directed mutagenesis kit (CLONTECH) with a mutated antisense oligonucleotide (5′-CCAGACCTGGggTACTCGCATG) to replace isoleucine with proline at amino acid 223, which is the fourth position of the DNA recognition helix in the homeodomain (27). The following RNAs for embryo injection were used: Xlim-1/3m (19), Xbra (28), noggin (29), chordin (30), β-globin (31), dominant-negative FGF receptor (XFD), and d50 (32). The following plasmids were used as probes: Otx2 (9), neural cell adhesion molecule (NCAM) (33), β2-tubulin/N-tubulin/clone 24–10 (34, 35), cpl-1 (36), XCG7 (37), en-2 (38), Krox-20 (39), HoxB9/XlHbox6 (40), chordin (30), eFGF (21), and Sonic hedgehog (Shh) (41). Antibody staining on sections was performed as described (42) with anti-HoxB9 antibodies (43). Dissected animal caps were treated with 10 μM RA/0.5% dimethyl sulfoxide (DMSO) or 0.5% DMSO (control) for 3 hr, washed with 1× Marc’s modified Ringer’s solution (MMR) (44) twice, then transferred to 0.5× MMR/50 μg/ml gentamicin.
RESULTS
Neural Patterning Elicited by Xlim-1/3m and by Xlim-1/3m Plus Xbra.
Combined animal cap assays demonstrated that injection of synthetic mRNA encoding the LIM domain mutant of Xlim-1, named 3m, initiates neural differentiation in injected animal caps as well as in adjacent uninjected caps, suggesting that a neural inducing signal emanates from the Xlim-1/3m-expressing cells (19). We proceeded to study the nature of the neural cells induced by Xlim-1/3m, and its modulation by Xbra. To study the modulation of neural induction by Xbra, combined animal cap assays provide an important advantage: while Xbra inhibits neural differentiation in the cells in which it is expressed by converting them from ectoderm to mesoderm (28), the influence of Xbra still can be studied in adjacent uninjected caps.
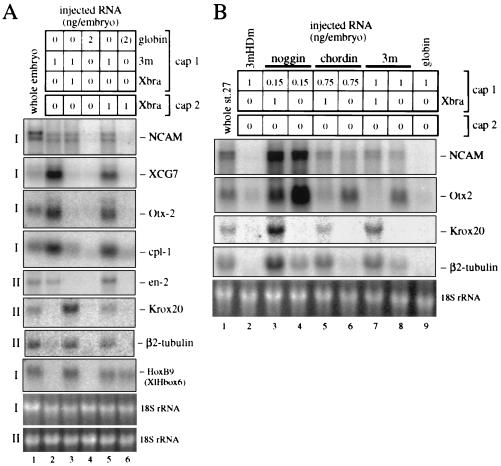
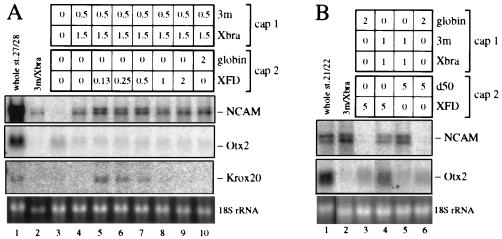
Fig. 1 presents results of combined animal cap experiments as assayed by the expression of marker genes characteristic for different regions in the CNS. Recombinates between uninjected caps and Xlim-1/3m expressing or Xlim-1/3m plus Xbra-expressing caps generated similar levels of total neural tissue, as assayed by the expression of the pan-neural marker NCAM (Fig. 1A, lanes 2 and 3). However, regionally specific markers revealed major differences. Recombinates with a Xlim-1/3m-injected partner expressed the anterior neural markers Otx2 and cpl-1, the midbrain–hindbrain marker en-2, and the cement gland marker XCG7, but not the posterior markers Krox-20 (hindbrain), HoxB9 (spinal cord; previously known as XlHbox6), and β2-tubulin [hindbrain and spinal cord (45)] (Fig. 1A, lane 2). Thus, Xlim-1/3m generates a signal that results in the induction of anterior neural tissue. In contrast, recombinates containing caps coexpressing Xlim-1/3m plus Xbra were converted from anterior to posterior marker expression (lane 3). These data suggest that Xbra elicits signals that inhibit the expression of anterior and induce the expression of posterior marker genes in Xenopus animal explants.
Figure 1.
(A) Expression of anteroposterior neural marker genes in combined animal caps expressing activated Xlim-1/3m and Xbra. An animal cap from a embryo injected with mRNAs (cap 1) in the indicated amounts (ng/embryo) was combined with another cap (cap 2) as indicated. Two experiments of identical design, identified as I and II, are presented in one panel. Six combined caps were collected at equivalent stage 22/23 (blot I) or 27/28 (blot II), and the RNA was assayed by Northern blot analysis. NCAM is a pan-neural marker, and XCG7 identifies the cement gland, an anterior ectodermal derivative; the following neural markers from Otx2 through HoxB9 are arranged in anteroposterior order of their expression. (B) Xbra-induced posteriorization of neural tissue generated by noggin and chordin. The injected RNAs are indicated above the blot; 3mHDm is a homeodomain mutant derivative of Xlim-1/3m that is expected to be inactive and is used as control, as is globin mRNA.
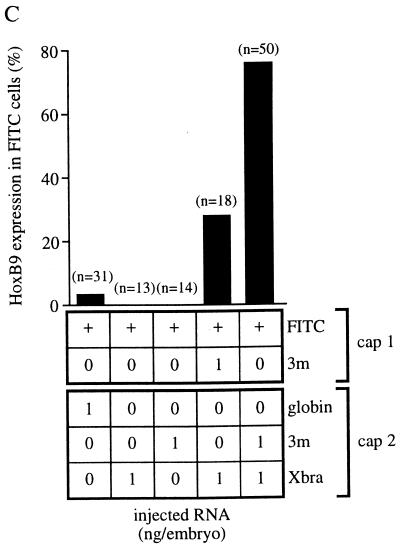
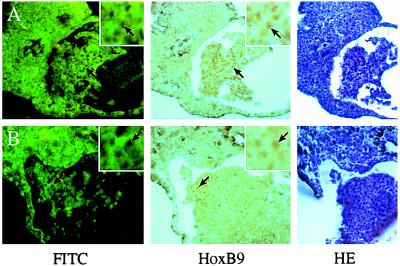
Injection of Xbra mRNA alone also activated the HoxB9 gene but not NCAM or any other neural marker in animal explants (Fig. 1A, lane 6), suggesting that in this case we are observing the mesodermal component of the HoxB9 expression pattern (43). To ascertain the germ layer in which HoxB9 is expressed in combined caps with one Xlim-1/3m plus Xbra-injected partner we carried out immunostaining of lineage-labeled recombinates. Fluorescein isothiocyanate (FITC)–dextran-labeled animal caps not injected with any RNA were apposed to caps that expressed Xlim-1/3m plus Xbra. After culture and sectioning we observed staining with HoxB9 antibody in the nuclei of many FITC-labeled cells (Fig. 2A) in a high proportion of the recombinates (Fig. 2C). These results indicate that HoxB9 expression in this preparation occurred in the ectodermal (i.e., neural) compartment and can thus be considered a consequence of posteriorization of the induced neural tissue.
Figure 2.
Immunostaining of HoxB9 in combined animal caps. (A and B) FITC fluorescence, staining for HoxB9, and hematoxylin/eosine (HE) staining, as indicated. HoxB9-positive nuclei (arrows) are seen as black holes in the FITC panel because of fluorescence quenching by the immunoprecipitate. (A) An FITC–dextran-injected cap was combined with a cap coinjected with Xlim-1/3m and Xbra mRNAs. (B) A cap coinjected with FITC–dextran and Xlim-1/3m was combined with an Xbra mRNA-injected cap. (C) Frequency of HoxB9 expression in FITC lineage-labeled caps, injected with RNAs as indicated in the figure. n, Number of combined caps examined.
Xbra-Expressing Cells Exert a Posteriorizing Influence.
The results shown above indicate that Xlim-1/3m-expressing caps exert an anterior neural inducing influence whereas Xlim-1/3m plus Xbra-expressing caps exert a posterior neural inducing influence. Because classical experiments have suggested the existence of posteriorizing factor(s) that do not possess neuralizing activity, we tested whether caps that express Xbra alone can provide a posteriorizing signal. Xlim-1/3m-injected caps combined with Xbra-injected caps express all anterior and posterior marker genes examined (Fig. 1A, lane 5), and immunostaining showed that HoxB9 is induced in the Xlim-1/3m-injected (FITC-labeled) caps (Fig. 2 B and C). Thus, expression of Xbra can posteriorize cells that have been induced to a neural fate by the expression of Xlim-1/3m. We next tested whether Xbra is also able to posteriorize anterior neural tissue induced by the secreted neuralizing factors noggin or chordin. In all cases, coexpression of Xbra activated the posterior neural markers Krox-20 and β2-tubulin and substantially reduced expression of the anterior marker Otx2 (Fig. 1B). Thus, Xbra-expressing cells generate a general posteriorizing signal.
Xlim-1/3m Activates Chordin While Xbra Activates eFGF Expression.
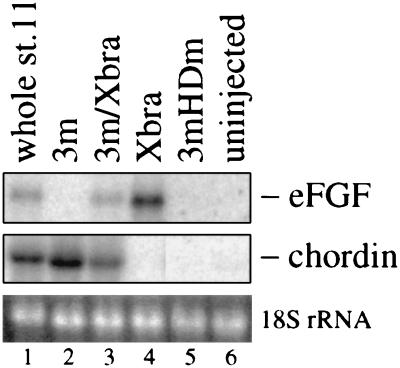
Because Xlim-1 and Xbra are transcription factors displaying nuclear localization (46, 47), their non-cell autonomous effects in the experiments reported previously (19) and above are likely due to secreted factors produced under their control. Among known neural inducers we already excluded noggin and follistatin as candidates because their expression is not activated by Xlim-1/3m (19). In contrast, the neuralizing and dorsalizing factor chordin (20) is induced by Xlim-1/3m (Fig. 3, lane 2), as also noted by Sasai et al. (20); a homeodomain mutant of Xlim-1/3m (3mHDm), used as control, does not induce chordin (lane 5). Xbra does not elicit chordin expression (Fig. 3, lane 4), but strongly activates eFGF as already reported by Isaacs et al. (21). Importantly in the context of these experiments, the combined injection of Xlim-1/3m and Xbra mRNAs into animal caps led to the expression of both eFGF and chordin (Fig. 3, lane 3). These observations may provide an explanation for the types of neural markers that are activated in our combined animal cap experiments (Fig. 1), because basic FGF (bFGF) was shown to posteriorize anterior neural plate explants and noggin or follistatin-induced anterior neural tissue (48–50), and the biological activities of bFGF and eFGF in the embryo are similar (21). Further, bFGF has been reported to exert neuralizing activity in animal cap cells that had been dissociated and reaggregated, or in explants that were cultured in low Ca2+ and Mg2+ solutions, conditions that may lead to reduced cell adhesion (50, 51). No neuralizing activity of Xbra-expressing caps was observed under our experimental conditions that involve close association between signaling and receiving cells within an intact tissue.
Figure 3.
Early gene expression in animal caps injected with Xlim-1/3m, Xbra, and 3m plus Xbra mRNAs. 3mHDm was used as a negative control. The amount of mRNA was 1 ng/embryo for each species. Twelve animal caps were collected at equivalent stage 11 and assayed by Northern blot analysis.
A Dominant-Negative FGF Receptor Inhibits Posteriorization by Xbra.
Additional evidence for a dose-dependent involvement of FGF in posteriorization was obtained in experiments that used the dominant-negative FGF receptor, XFD (32). Recombinates between uninjected animal caps and caps injected with Xlim-1/3m plus Xbra in a 1:3 ratio, expressed NCAM but not Krox-20, possibly because they are too strongly posteriorized (Fig. 4A, lane 4). Injection of XFD into the second cap (recipient of the signal) led to Krox-20 expression when low levels of RNA were injected, and to inhibition at higher levels (Fig. 4A, lanes 5–9). In this experiment no recovery of Otx2 expression was observed. However, in a separate experiment with a different ratio of Xlim-1/3m and Xbra and at a high level of XFD, recovery of Otx2 expression in the recombinates could be seen (Fig. 4B, lane 4). This recovery is important because it indicates that increased levels of XFD do not suppress Krox-20 due to general toxicity. In this experiment the inactive mutant FGF receptor d50 was used as control and showed no effect (Fig. 4B, lane 5). These results indicate that Xlim-1/3m plus Xbra-expressing caps emit a signal whose effect can be titrated by XFD, indicating that the signaling molecule is a member of the FGF family.
Figure 4.
(A) Dose dependence of the effect of dominant-negative FGF receptor (XFD) on Krox-20 expression induced by Xlim-1/3m plus Xbra. At the 3m-to-Xbra ratio used, Otx2 is suppressed but Krox-20 is not activated, possibly because the tissue is posteriorized too extensively. Low levels of XFD in the recipient cap (cap 2) allow Krox-20 expression, but high levels inhibit. (B) Restoration of Otx2 expression by XFD in combined caps. Caps injected with a high level of XFD mRNA were combined with caps coinjected with Xlim-1/3m and Xbra mRNAs. Eight combined caps were collected at equivalent stage 27/28 and assayed by Northern blot analysis. d50 is an inactive form of the FGF receptor that was used as control.
RA Can Posteriorize Neural Tissue Generated by Xlim-1/3m.
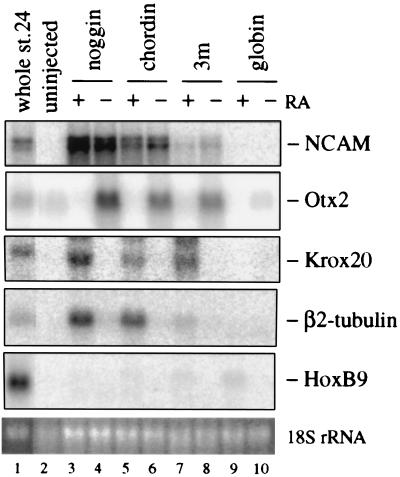
RA has been postulated to be a posteriorizing factor based on the observations that RA treatment of embryos leads to truncation of anterior structures in Xenopus (52), and that RA affects both ectoderm and mesoderm explants, initiating the expression of posterior neural markers in ectoderm–mesoderm recombinates (23–25). We therefore tested whether RA can posteriorize neural tissue generated by Xlim-1/3m, chordin, or noggin. As shown in Fig. 5, RA strongly inhibits Otx2 expression and induces Krox-20 and β2-tubulin expression in all cases, indicating that RA can act as a posteriorizing factor for neural tissue in the absence of mesoderm. However, HoxB9 expression was barely induced and Krox-20 was not repressed even at the high RA concentration of 10 μM, suggesting that RA cannot fully posteriorize neural tissue in contrast to Xbra-expressing caps.
Figure 5.
Posteriorization of anterior-type neural tissue by RA. The amounts of injected RNAs (ng/embryo) were 1 for globin and Xlim-1/3m, 0.75 for chordin, and 0.1 for noggin. Animal caps were treated with or without 10 μM RA. Eight caps were collected at equivalent stage 23/24 and assayed by Northern blot analysis.
DISCUSSION
Earlier studies using transplantation and explant experiments in amphibian embryos suggested a two-signal model (13, 14) or two-gradient model (15) for anteroposterior patterning of the CNS in which neuralization and posteriorization (or caudalization) were thought of as distinct phenomena; these two phenomena have also been referred to as activation and transformation (see reviews in refs. 3–5). Further, while head mesoderm induces anterior neural tissues such as forebrain and eyes, and posterior axial mesoderm (notochord) induces posterior neural tissue such as hindbrain and spinal cord, the combination of both inducers generates posterior characteristics, leading to the proposal of posterior dominance in neural patterning (12, 15).
In this paper we suggest a role for two transcription factors, Xlim-1 and Xbra, in anteroposterior patterning of neural tissue in the Xenopus embryo. As previously reported (19), mRNA injection of an activated mutant of Xlim-1, named 3m, into the animal region followed by explant culture led to the expression of neural and cement gland markers; we further showed that this effect was non-cell autonomous— i.e., was mediated by secreted factor(s). In the present paper we report an analysis of the nature of the induced neural tissue. Combined explants containing Xlim-1/3m-injected and uninjected animal caps express the anterior neural markers Otx2, cpl-1, and en-2, but not the posterior markers Krox-20 and HoxB9. Although explants expressed several anterior neural markers under the influence of Xlim-1/3m they did not differentiate into forebrain or eyes, suggesting that additional inducers are required for anterior CNS morphogenesis. Such factors might include Shh (53, 54) and cerberus (55), which are involved in CNS patterning and head/eye formation, respectively; we have observed that Xlim-1/3m does not initiate Shh expression in animal caps (data not shown).
In contrast to the anterior differentiation elicited by Xlim-1/3m, animal caps cultured with explants expressing Xlim-1/3m and Xbra activate the posterior neural markers Krox-20 and HoxB9 while repressing the expression of the anterior markers, Otx2 and cpl-1. Like the neural inducing effect of Xlim-1/3m, the posteriorizing effect of Xbra is non-cell autonomous—i.e., is mediated by a secreted factor(s) which does not, in itself, have neural-inducing activity. We suggest that the activities of Xlim-1/3m and Xbra are mediated by chordin and eFGF, possibly in combination with additional factors, because Xlim-1/3m activates the expression of chordin but not of noggin or follistatin in animal caps (Fig. 3; refs. 19 and 20), whereas Xbra is known to elicit eFGF expression (Fig. 3; ref. 21). Several recent studies have provided evidence for a neural inducing function for chordin (20) and noggin (56) as mediated by inhibition of BMP4 action (57, 58), and a posteriorizing function for bFGF (48–50). We have also shown that RA can posteriorize anterior neural tissue generated by Xlim-1/3m, chordin, and noggin; however, RA did not strongly induce the posterior marker HoxB9 (Fig. 5), suggesting that RA could be involved in incomplete posteriorization of Xlim-1 expressing caps.
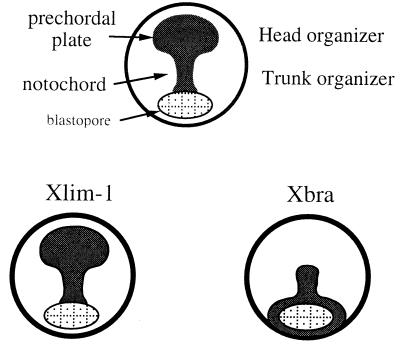
A model of the dorsal mesoderm at the late gastrula stage (Fig. 6) shows the traditional separation into notochord and prechordal plate (future head mesoderm) and the correspondence of these two regions to the head and trunk organizer (5). The figure illustrates the correlation between the expression domains of Xlim-1 and Xbra and these functional domains of the dorsal mesoderm. Xlim-1 and chordin are found in both head mesoderm and notochord (7, 30), whereas Xbra and eFGF are limited to the notochord (11, 59); the expression domain of Xbra may thus define the boundary between head and trunk organizer. These considerations (Fig. 6) are in broad agreement with the classical two-signal and two-gradient models of neural patterning (13–15). As in the original models, the posteriorizing activity of Xbra-injected caps cannot by itself neuralize apposed animal explants, but does posteriorize anterior neural tissue generated by Xlim-1/3m, chordin, and noggin.
Figure 6.
Schematic dorsal views of the Xenopus embryo at late gastrula stage. (Upper) Drawing shows the dorsal mesoderm divided into the developing notochord and the prechordal plate (future head mesoderm); these two regions correspond to the head and trunk organizer identified in classical transplantation experiments. The expression domain of Xlim-1 encompasses the entire dorsal mesoderm at this stage whereas Xbra is expressed only in the notochord (Lower). These expression domains may define the head and trunk organizer, as further discussed in the text.
The results described here are consistent with the phenotype of Lim1 knockout mice in which the head region anterior to the otic vesicles fails to develop (17). Neural tissue generated by Xlim-1/3m expressed those anterior markers (Otx2 and en-2) that are missing in Lim1-null mice, whereas Krox-20 is expressed in Lim1-null mice but not in Xlim-1/3m-injected caps. An apparent inconsistency with the headless phenotype of Lim1 knockout mice is the fact that ventral expression of Xlim-1/3m in Xenopus embryos initiates only partial secondary axes that do not contain forebrain or eyes (19). We believe that in these experiments, Xlim-1/3m-generated anterior neural tissue may be posteriorized by the surrounding Xbra-expressing cells in the ventral equatorial zone in a situation akin to posterior dominance seen in earlier transplantation experiments (12), or in cell mixing experiments with anterior neural plate and posterior axial mesoderm (60).
In conclusion, we suggest that the expression domains of Xlim-1 and Xbra specify the classically defined subdomains of the dorsal mesoderm, the head and trunk organizer (Fig. 6). Further we suggest that Xlim-1 and Xbra exert their inducing influence by eliciting the production of the secreted signaling factors chordin and eFGF that, in interaction with each other, with BMP4 (57, 58), and possibly additional factors, pattern the early CNS along its anteroposterior axis.
Acknowledgments
We thank J. Smith, K. Cho, E. Boncinelli, C. Kintner, M. Jamrich, R. Harland, D. Wilkinson, E. De Robertis, H. Isaacs, J. Slack, Y.-B. Shi, and C. Wright for plasmids and anti-HoxB9 antibodies. We also thank T. Sargent for critical reading of the manuscript.
Footnotes
Abbreviations: CNS, central nervous system; FGF, fibroblast growth factor; eFGF, embryonic FGF; bFGF, basic FGF; FITC, fluorescein isothiocyanate; RA, retinoic acid; NCAM, neural cell adhesion molecule; XFD, dominant-negative FGF receptor.
References
- 1.Mangold O. Naturwissenschaften. 1933;43:761–766. [Google Scholar]
- 2.Saxén L. Int J Dev Biol. 1989;33:21–48. [PubMed] [Google Scholar]
- 3.Slack J M W, Tannahill D. Development (Cambridge, UK) 1992;114:285–302. doi: 10.1242/dev.114.2.285. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert S F, Saxén L. Mech Dev. 1993;41:73–89. doi: 10.1016/0925-4773(93)90039-z. [DOI] [PubMed] [Google Scholar]
- 5.Yamada T. Development (Cambridge, UK) 1994;120:3051–3062. doi: 10.1242/dev.120.11.3051. [DOI] [PubMed] [Google Scholar]
- 6.Dawid I B. J Biol Chem. 1994;269:6259–6262. [PubMed] [Google Scholar]
- 7.Taira M, Otani H, Jamrich M, Dawid I B. Development (Cambridge, UK) 1994;120:1525–1536. doi: 10.1242/dev.120.6.1525. [DOI] [PubMed] [Google Scholar]
- 8.Steinbeisser H, De Robertis E M. CR Acad Sci III. 1993;316:959–971. [PubMed] [Google Scholar]
- 9.Pannese M, Polo C, Andreazzoli M, Vignali R, Kablar B, Barsacchi G, Boncinelli E. Development (Cambridge, UK) 1995;121:707–720. doi: 10.1242/dev.121.3.707. [DOI] [PubMed] [Google Scholar]
- 10.Blitz I L, Cho K W. Development (Cambridge, UK) 1995;121:993–1004. doi: 10.1242/dev.121.4.993. [DOI] [PubMed] [Google Scholar]
- 11.Green J B, New H V, Smith J C. Cell. 1992;71:731–739. doi: 10.1016/0092-8674(92)90550-v. [DOI] [PubMed] [Google Scholar]
- 12.Okada Y K, Takaya H. Proc Imp Acad (Tokyo) 1942;18:514–519. [Google Scholar]
- 13.Yamada T. Embryologia. 1950;1:1–20. [Google Scholar]
- 14.Nieuwkoop P D. J Exp Zool. 1952;120:83–108. [Google Scholar]
- 15.Saxén L, Toivonen S. J Embryol Exp Morphol. 1961;9:514–533. [PubMed] [Google Scholar]
- 16.Dawid I B, Toyama R, Taira M. CR Acad Sci III. 1995;318:295–306. [PubMed] [Google Scholar]
- 17.Shawlot W, Behringer R R. Nature (London) 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- 18.De Robertis E M. Nature (London) 1995;374:407–408. doi: 10.1038/374407a0. [DOI] [PubMed] [Google Scholar]
- 19.Taira M, Otani H, Saint-Jeannet J P, Dawid I B. Nature (London) 1994;372:677–679. doi: 10.1038/372677a0. [DOI] [PubMed] [Google Scholar]
- 20.Sasai Y, Lu B, Steinbeisser H, De Robertis E M. Nature (London) 1995;376:333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- 21.Isaacs H V, Pownall M E, Slack J M. EMBO J. 1994;13:4469–4481. doi: 10.1002/j.1460-2075.1994.tb06769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz i Altaba A, Jessell T M. Development (Cambridge, UK) 1991;112:945–958. doi: 10.1242/dev.112.4.945. [DOI] [PubMed] [Google Scholar]
- 23.Sharpe C R. Neuron. 1991;7:239–247. doi: 10.1016/0896-6273(91)90262-x. [DOI] [PubMed] [Google Scholar]
- 24.Sive H L, Draper B W, Harland R M, Weintraub H. Genes Dev. 1990;4:932–942. doi: 10.1101/gad.4.6.932. [DOI] [PubMed] [Google Scholar]
- 25.Ruiz i Altaba A, Jessell T. Genes Dev. 1991;5:175–187. doi: 10.1101/gad.5.2.175. [DOI] [PubMed] [Google Scholar]
- 26.Taira M, Jamrich M, Good P J, Dawid I B. Genes Dev. 1992;6:356–366. doi: 10.1101/gad.6.3.356. [DOI] [PubMed] [Google Scholar]
- 27.Wright C V, Cho K W, Hardwicke J, Collins R H, De Robertis E M. Cell. 1989;59:81–93. doi: 10.1016/0092-8674(89)90871-4. [DOI] [PubMed] [Google Scholar]
- 28.Cunliffe V, Smith J C. Nature (London) 1992;358:427–430. doi: 10.1038/358427a0. [DOI] [PubMed] [Google Scholar]
- 29.Smith W C, Harland R M. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- 30.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont L K, De Robertis E M. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krieg P A, Melton D A. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amaya E, Musci T J, Kirschner M W. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- 33.Kintner C R, Melton D A. Development (Cambridge, UK) 1987;99:311–325. doi: 10.1242/dev.99.3.311. [DOI] [PubMed] [Google Scholar]
- 34.Richter K, Grunz H, Dawid I B. Proc Natl Acad Sci USA. 1988;85:8086–8090. doi: 10.1073/pnas.85.21.8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Good P J, Richter K, Dawid I B. Nucleic Acids Res. 1989;17:8000. doi: 10.1093/nar/17.19.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knecht A K, Good P J, Dawid I B, Harland R M. Development (Cambridge, UK) 1995;121:1927–1935. doi: 10.1242/dev.121.6.1927. [DOI] [PubMed] [Google Scholar]
- 37.Jamrich M, Sato S. Development (Cambridge, UK) 1989;105:779–786. doi: 10.1242/dev.105.4.779. [DOI] [PubMed] [Google Scholar]
- 38.Hemmati-Brivanlou A, de la Torre J R, Holt C, Harland R M. Development (Cambridge, UK) 1991;111:715–724. doi: 10.1242/dev.111.3.715. [DOI] [PubMed] [Google Scholar]
- 39.Bradley L C, Snape A, Bhatt S, Wilkinson D G. Mech Dev. 1993;40:73–84. doi: 10.1016/0925-4773(93)90089-g. [DOI] [PubMed] [Google Scholar]
- 40.Sharpe C R, Fritz A, De Robertis E M, Gurdon J B. Cell. 1987;50:749–758. doi: 10.1016/0092-8674(87)90333-3. [DOI] [PubMed] [Google Scholar]
- 41.Stolow M A, Shi Y B. Nucleic Acids Res. 1995;23:2555–2562. doi: 10.1093/nar/23.13.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saint-Jeannet J P, Dawid I B. Proc Natl Acad Sci USA. 1994;91:3049–3053. doi: 10.1073/pnas.91.8.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright C V, Morita E A, Wilkin D J, De Robertis E M. Development (Cambridge, UK) 1990;109:225–234. doi: 10.1242/dev.109.1.225. [DOI] [PubMed] [Google Scholar]
- 44.Peng H B. Methods Cell Biol. 1991;36:657–662. [PubMed] [Google Scholar]
- 45.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Nature (London) 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- 46.Karavanov A A, Saint-Jeannet J-P, Karavanova I, Taira M, Dawid I B. Int J Dev Biol. 1996;40:453–461. [PubMed] [Google Scholar]
- 47.Conlon F L, Sedgwick S G, Weston K M, Smith J C. Development (Cambridge, UK) 1996;122:2427–2435. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
- 48.Cox W G, Hemmati-Brivanlou A. Development (Cambridge, UK) 1995;121:4349–4358. doi: 10.1242/dev.121.12.4349. [DOI] [PubMed] [Google Scholar]
- 49.Kengaku M, Okamoto H. Development (Cambridge, UK) 1995;121:3121–3130. doi: 10.1242/dev.121.9.3121. [DOI] [PubMed] [Google Scholar]
- 50.Lamb T M, Harland R M. Development (Cambridge, UK) 1995;121:3627–3636. doi: 10.1242/dev.121.11.3627. [DOI] [PubMed] [Google Scholar]
- 51.Kengaku M, Okamoto H. Development (Cambridge, UK) 1993;119:1067–1078. doi: 10.1242/dev.119.4.1067. [DOI] [PubMed] [Google Scholar]
- 52.Durston A J, Timmermans J P, Hage W J, Hendriks H F, de Vries N J, Heideveld M, Nieuwkoop P D. Nature (London) 1989;340:140–144. doi: 10.1038/340140a0. [DOI] [PubMed] [Google Scholar]
- 53.Echelard Y, Epstein D J, St-Jacques B, Shen L, Mohler J, MacMahon J A, MacMahon A P. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 54.Chiang C, Litingtung Y, Lee E, Young K, Corden J L, Westphal H, Beachy P. Nature (London) 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 55.Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis E M. Nature (London) 1996;382:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- 56.Lamb T M, Knecht A K, Smith W C, Stachel S E, Economides A N, Stahl N, Yancopolous G D, Harland R M. Science. 1993;262:713–718. doi: 10.1126/science.8235591. [DOI] [PubMed] [Google Scholar]
- 57.Piccolo S, Sasai Y, Lu B, De Robertis E M. Cell. 1996;86:589–598. doi: 10.1016/s0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimmerman L B, De Jesus-Escobar J M, Harland R M. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 59.Isaacs H V, Pownall M E, Slack J M. Int J Dev Biol. 1995;39:575–579. [PubMed] [Google Scholar]
- 60.Toivonen S, Saxén L. Science. 1968;159:539–540. doi: 10.1126/science.159.3814.539. [DOI] [PubMed] [Google Scholar]