Abstract
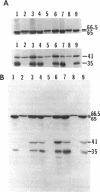
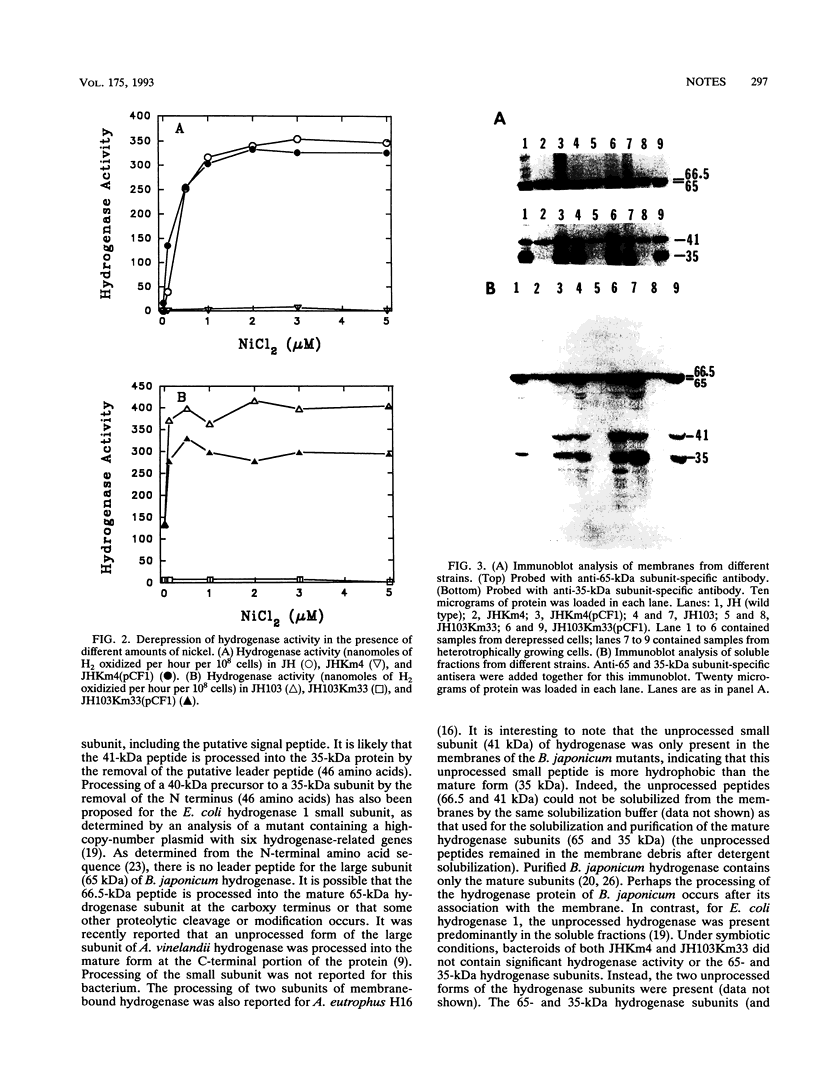
Deletion of a 2.9-kb chromosomal EcoRI fragment of DNA located 2.2 kb downstream from the end of the hydrogenase structural genes resulted in the complete loss of hydrogenase activity. The normal 65- and 35-kDa hydrogenase subunits were absent in the deletion mutants. Instead, two peptides of 66.5 and 41 kDa were identified in the mutants by use of anti-hydrogenase subunit-specific antibody. A hydrogenase structural gene mutant did not synthesize either the normal hydrogenase subunits or the larger peptides. Hydrogenase activity in the deletion mutants was complemented to near wild-type levels by plasmid pCF1, containing a 6.5-kb BglII fragment, and the 65- and 35-kDa hydrogenase subunits were also recovered in the mutants containing pCF1.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arp D. J. Rhizobium japonicum hydrogenase: purification to homogeneity from soybean nodules, and molecular characterization. Arch Biochem Biophys. 1985 Mar;237(2):504–512. doi: 10.1016/0003-9861(85)90303-0. [DOI] [PubMed] [Google Scholar]
- Barany F. Single-stranded hexameric linkers: a system for in-phase insertion mutagenesis and protein engineering. Gene. 1985;37(1-3):111–123. doi: 10.1016/0378-1119(85)90263-x. [DOI] [PubMed] [Google Scholar]
- Cauvin B., Colbeau A., Vignais P. M. The hydrogenase structural operon in Rhodobacter capsulatus contains a third gene, hupM, necessary for the formation of a physiologically competent hydrogenase. Mol Microbiol. 1991 Oct;5(10):2519–2527. doi: 10.1111/j.1365-2958.1991.tb02098.x. [DOI] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Du L., Stejskal F., Tibelius K. H. Characterization of two genes (hupD and hupE) required for hydrogenase activity in Azotobacter chroococcum. FEMS Microbiol Lett. 1992 Sep 1;75(1):93–101. doi: 10.1016/0378-1097(92)90462-w. [DOI] [PubMed] [Google Scholar]
- Fu C. L., Maier R. J. Identification of a locus within the hydrogenase gene cluster involved in intracellular nickel metabolism in Bradyrhizobium japonicum. Appl Environ Microbiol. 1991 Dec;57(12):3502–3510. doi: 10.1128/aem.57.12.3502-3510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Maier R. J. Nickel-dependent reconstitution of hydrogenase apoprotein in Bradyrhizobium japonicum Hupc mutants and direct evidence for a nickel metabolism locus involved in nickel incorporation into the enzyme. Arch Microbiol. 1992;157(6):493–498. doi: 10.1007/BF00276768. [DOI] [PubMed] [Google Scholar]
- Gollin D. J., Mortenson L. E., Robson R. L. Carboxyl-terminal processing may be essential for production of active NiFe hydrogenase in Azotobacter vinelandii. FEBS Lett. 1992 Sep 14;309(3):371–375. doi: 10.1016/0014-5793(92)80809-u. [DOI] [PubMed] [Google Scholar]
- Haugland R. A., Cantrell M. A., Beaty J. S., Hanus F. J., Russell S. A., Evans H. J. Characterization of Rhizobium japonicum hydrogen uptake genes. J Bacteriol. 1984 Sep;159(3):1006–1012. doi: 10.1128/jb.159.3.1006-1012.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo E., Palacios J. M., Murillo J., Ruiz-Argüeso T. Nucleotide sequence and characterization of four additional genes of the hydrogenase structural operon from Rhizobium leguminosarum bv. viciae. J Bacteriol. 1992 Jun;174(12):4130–4139. doi: 10.1128/jb.174.12.4130-4139.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom S. S., Graham L. A., Maier R. J. Isolation of genes (nif/hup cosmids) involved in hydrogenase and nitrogenase activities in Rhizobium japonicum. J Bacteriol. 1985 Mar;161(3):882–887. doi: 10.1128/jb.161.3.882-887.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Yu C., Maier R. J. Common cis-acting region responsible for transcriptional regulation of Bradyrhizobium japonicum hydrogenase by nickel, oxygen, and hydrogen. J Bacteriol. 1991 Jul;173(13):3993–3999. doi: 10.1128/jb.173.13.3993-3999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlüke C., Friedrich B. Maturation of membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992 Oct;174(19):6290–6293. doi: 10.1128/jb.174.19.6290-6293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978 Jun 15;87(1):206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Menon A. L., Mortenson L. E., Robson R. L. Nucleotide sequences and genetic analysis of hydrogen oxidation (hox) genes in Azotobacter vinelandii. J Bacteriol. 1992 Jul;174(14):4549–4557. doi: 10.1128/jb.174.14.4549-4557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon N. K., Robbins J., Wendt J. C., Shanmugam K. T., Przybyla A. E. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J Bacteriol. 1991 Aug;173(15):4851–4861. doi: 10.1128/jb.173.15.4851-4861.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri F., Maier R. J. Conformational changes in the membrane-bound hydrogenase of Bradyrhizobium japonicum. Evidence that the redox state of the enzyme affects its accessibility to protease and membrane-impermeant reagents. J Biol Chem. 1988 Nov 25;263(33):17809–17816. [PubMed] [Google Scholar]
- Sauter M., Böhm R., Böck A. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol. 1992 Jun;6(11):1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
- Sayavedra-Soto L. A., Arp D. J. The hoxZ gene of the Azotobacter vinelandii hydrogenase operon is required for activation of hydrogenase. J Bacteriol. 1992 Aug;174(16):5295–5301. doi: 10.1128/jb.174.16.5295-5301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayavedra-Soto L. A., Powell G. K., Evans H. J., Morris R. O. Nucleotide sequence of the genetic loci encoding subunits of Bradyrhizobium japonicum uptake hydrogenase. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8395–8399. doi: 10.1073/pnas.85.22.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults L. W., Moshiri F., Maier R. J. Aerobic purification of hydrogenase from Rhizobium japonicum by affinity chromatography. J Bacteriol. 1986 Jun;166(3):795–800. doi: 10.1128/jb.166.3.795-800.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults L. W., O'Hara E. B., Maier R. J. Nickel is a component of hydrogenase in Rhizobium japonicum. J Bacteriol. 1984 Jul;159(1):153–158. doi: 10.1128/jb.159.1.153-158.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. W., Wall J. D. Clustering of genes necessary for hydrogen oxidation in Rhodobacter capsulatus. J Bacteriol. 1991 Apr;173(7):2401–2405. doi: 10.1128/jb.173.7.2401-2405.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]