Abstract
The MYC oncoprotein is a transcription factor that coordinates cell growth and division. MYC overexpression exacerbates genomic instability and sensitizes cells to apoptotic stimuli. Here we demonstrate that MYC directly stimulates transcription of the human Werner syndrome gene, WRN, which encodes a conserved RecQ helicase. Loss-of-function mutations in WRN lead to genomic instability, an elevated cancer risk, and premature cellular senescence. The overexpression of MYC in WRN syndrome fibroblasts or after WRN depletion from control fibroblasts led to rapid cellular senescence that could not be suppressed by hTERT expression. We propose that WRN up-regulation by MYC may promote MYC-driven tumorigenesis by preventing cellular senescence.
Keywords: Werner gene, Myc, transcription, senescence
Alterations in c-myc oncogene expression have been implicated in the pathogenesis of several human cancers, including Burkitt and diffuse large B-cell lymphomas, breast and prostate cancer, colon cancer, melanoma, and multiple myeloma (Nesbit et al. 1999). The MYC oncoprotein is a basic helix–loop–helix–leucine zipper (bHLHZIP) transcription factor that through dimerization with MAX protein binds to specific DNA elements (“E boxes”) and modulates transcription of a wide variety of genes (for review, see Dang 1999; Grandori et al. 2000; Oster et al. 2002). The proteins encoded by MYC transcriptional target genes appear to regulate cell-cycle progression and cell growth while sensitizing cells to apoptotic stimuli (Evan et al. 1992). MYC may also be able to promote tumorigenesis by up-regulating the expression of genes such as hTERT that play a role in cellular immortalization or the escape from senescence (Wang et al. 1998a; Greenberg et al. 1999; Wu et al. 1999). We reasoned that MYC might modulate the expression of other genes that control cellular senescence, and thus determined whether the gene encoding the Werner syndrome RecQ helicase protein is a MYC transcriptional target.
Werner syndrome (WRN) is an uncommon, autosomal recessive genetic instability syndrome that results from loss-of-function mutations in the chromosome 8p12-p11.2 WRN gene (Yu et al. 1996). The WRN phenotype resembles premature aging, and includes genomic instability, an elevated risk of malignancy, and accelerated cellular senescence. Genetic instability following loss of the 162-kD WRN RecQ helicase protein reflects the physiologic role of WRN in mitotic recombination and repair (Brosh and Bohr 2002; Saintigny et al. 2002). Conversely, the elevated levels of WRN observed in immortalized and human tumor cell lines may help insure continuous cell proliferation (Shiratori et al. 1999). In order to delineate potential interactions between MYC and WRN in tumorigenesis, we determined whether WRN expression is modulated by MYC, and monitored cellular responses to MYC overexpression in the absence of WRN. The results indicated that WRN expression appears to be required to avoid cellular senescence upon MYC up-regulation in hTERT-immortalized fibroblasts.
Results and Discussion
c-myc and WRN expression are tightly linked
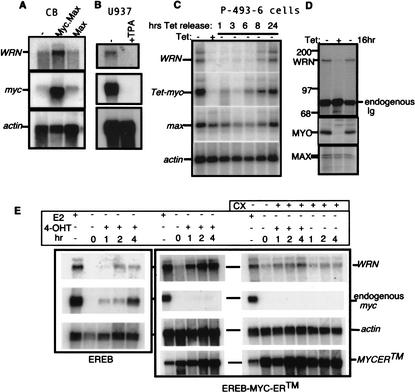
To determine whether MYC regulates WRN expression, we first correlated c-myc and WRN expression levels in different cell lines in which MYC expression was constitutively or could be conditionally altered. Analysis of WRN expression in EBV-immortalized B cells that constitutively expressed high levels of c-myc and max, max alone, or neither gene demonstrated the coordinate expression of c-myc and WRN:WRN mRNAwas expressed at high levels only in B cells that constitutively overexpressed myc and max (Fig. 1A; Gu et al. 1993). High constitutive c-myc and WRN mRNAlevels were also found in human U937 leukemia cells, and both myc and WRN mRNAlevels were coordinately down-regulated after TPA-induced differentiation (Fig. 1B). To determine the kinetics of induction of WRN by MYC, we used the B-cell line P-493-6, which expresses a Tet-myc repressible gene (Schuhmacher et al. 1999). Northern analysis indicated that c-myc induction was closely followed by an increase in steady-state WRN mRNAlevels (Fig. 1C). There was also a parallel increase in MYC and WRN protein levels in P-493-6 cells upon removal of tetracycline (Fig. 1D).
Figure 1.
c-myc and WRN expression are tightly linked. (A) EBV-immortalized CB B cells overexpressing c-myc and max (+MycMax; Gu et al. 1993) have elevated WRN mRNAlevels compared with cells that overexpress max alone (+Max). (B) U937 leukemia cells show decreased levels of WRN and c-myc mRNAs after TPA-induced differentiation (U937+TPA). (C) Kinetics of WRN mRNAinduction following MYC up-regulation upon tetracycline withdrawal in B-cell line P-493-6 expressing Tet-myc (Schuhmacher et al. 1999). (D) Parallel induction of WRN and MYC protein in P-493-6 B cells labeled with 35S-methionine for 4 h after MYC induction: myc-on (–Tet), after growth for 48 h in the presence of tetracycline (myc-off; +Tet); and after 16 h of MYC induction: (16hr). Normalized lysates were immunoprecipitated with anti-WRN (Moser et al. 2000), anti-MAX (Santa Cruz Biotechnology no. sc765), or anti-MYC antibody (Santa Cruz Biotechnology no. sc764). (E) WRN induction by MYC does not require new protein synthesis. EREB B cells (Kempkes et al. 1995) expressing a chimeric MYC-ER™ protein (Wu et al. 1999) were grown in the absence of estrogen (E2–) to inactivate EBNA2-ER, then exposed for the indicated times to 4-OHT to induce MYC-ER™ in the presence or absence of cycloheximide. PhosphorImager quantitation of data in E indicated an approximately fivefold induction of WRN mRNA relative to actin by MYC-ER™ and approximately fourfold with cycloheximide, compared with an ∼1.2-fold increase due to cycloheximide addition to control EREB cells not expressing MYC-ER™.
The possibility that MYC protein might directly induce transcription of WRN was tested using conditionally immortalized EREB (Kempkes et al. 1995) B cells expressing EBNA2-ER (EBNA2 fused to the estrogen receptor, ER) and MYC-ER™ chimeric proteins (Littlewood et al. 1995). The MYC-ER™ chimeric protein contains a mutant estrogen receptor domain (ER™) that is selectively activated by 4-hydroxytamoxifen (4-OHT). Upon estrogen removal, EBNA2-ER is inactivated and cells undergo cell-cycle arrest. Addition of 4-OHT preferentially activates the MYC-ER™ fusion protein, and leads to a rapid increase in WRN mRNA (Fig. 1E). WRN mRNAup-regulation by MYC-ER™ was found to be cycloheximide-resistant (Fig. 1E, right panel). This result is consistent with WRN being a direct target of the MYC oncoprotein.
The WRN promoter binds MYC in vitro and in vivo
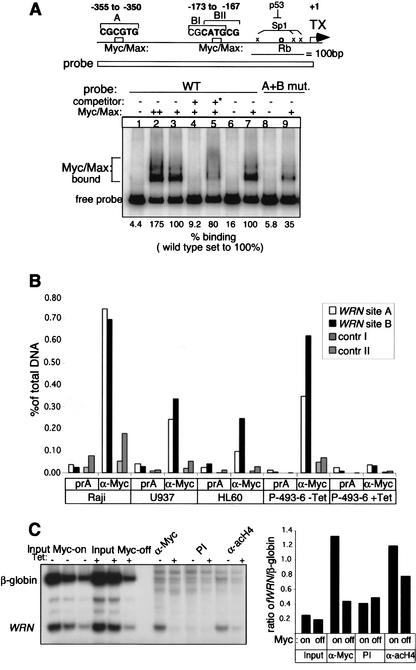
To establish whether MYC directly regulates WRN expression, we explored the WRN promoter region for the presence of MYC–MAX consensus elements. This search revealed several noncanonical MYC–MAX-binding sites (Grandori and Eisenman 1997) in close proximity to the transcriptional start (sites Aand B; Fig. 2A; Yamabe et al. 1998). The B site consists of two overlapping sites (BI and BII). Mobility shift assays demonstrated that these potential target sites could be bound in vitro by purified MYC–MAX heterodimers (Fig. 2A). Point mutations in sites Aand B reduced binding to ∼35% of wild type (Fig. 2A). Residual binding is likely due to several half-sites adjacent to the mutated site (C. Grandori, unpubl.). In vitro DNAseI footprinting demonstrated MYC–MAX heterodimer-dependent protection of the Aand B binding sites and immediate flanking nucleotides (see Supplementary Fig. 1). In vivo binding of MYC to the WRN promoter was documented by use of a chromatin immunoprecipitation (ChIP) assay (Frank et al. 2001). Comparable results demonstrating increased WRN A and B site binding by MYC were obtained in four different cell lines expressing deregulated c-myc (Fig. 2B). In vivo binding of MYC to the WRN promoter and a modest but consistent elevation in histone H4 acetylation, as observed for other MYC-target genes (Frank et al. 2001), was also shown by duplex PCR in the B-cell line P-493-6 that expresses a Tet-Myc-repressible gene (Fig. 2C). These results and the tight transcriptional correlation of WRN and c-myc indicate that WRN is a direct transcriptional target of the MYC oncoprotein.
Figure 2.
In vitro and in vivo binding of MYC to the WRN promoter. (A) Purified MYC–MAX heterodimers bind and retard the mobility of a WRN promoter fragment encompassing nucleotides –412 to +8 in EMSAs. Where indicated, the A and B sites were mutated (see Materials and Methods) to determine the specificity of binding. In lane 2, 25 ng of MYC and 7.5 ng of MAX were used; In lanes 3–5, 7, and 9, 12.5 ng of MYC and 3.75 ng of MAX were used. Controls include a specific competitor oligonucleotide containing a MYC CACGTG consensus site added at 1 pM (∼10-fold molar excess over the probe; +), as well as a methylated CACGTG that is not recognized by MYC (*). (B) ChIP detects MYC binding to WRN promoter elements in vivo. Cross-linked chromatin from Raji, U937 and HL60 cell lines, and the EBV-immortalized P-493-6 cell expressing a Tet-myc was immunoprecipitated with protein Aonly (prA) or with anti-MYC antibodies. Bound DNAwas monitored by real-time PCR with WRN promoter-specific primers. Two negative controls were used as nontarget E-box-containing genes (see Materials and Methods). The percent of total DNAbound was calculated as described (Frank et al. 2001). (C) MYC induction increases MYC residence on the WRN promoter and correlates with increased histone H4 acetylation. ChIP assay using chromatin from P-493-6 cells expressing Tet-myc (–Tet; MYC-on), or from cells grown in the presence of tetracycline for 48 h (+Tet; MYC-off). Chromatin was immunoprecipitated with anti-MYC or anti-acetylated H4 (acH4; Upstate Biotechnology) antibodies or with rabbit preimmune serum. Bound DNAwas quantified by duplex PCR using primers flanking site B of the WRN promoter and the β-globin gene as internal control (Schubeler et al. 2000). The right panel shows the quantitation of the ratio of WRN promoter to β-globin. Similar results were obtained in three independent experiments.
MYC overexpression in the absence ofWRN promotes cell senescence
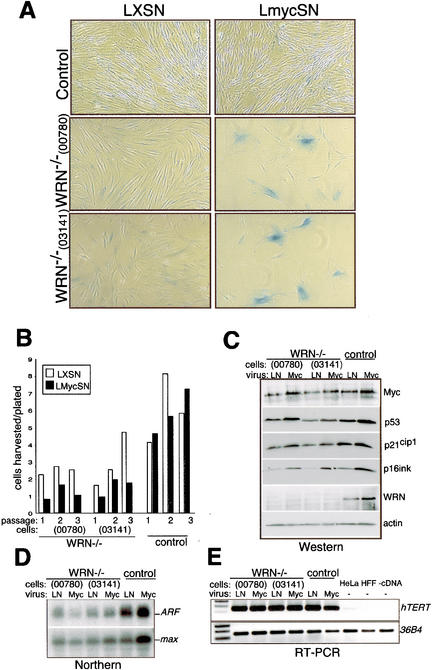
To determine whether the proliferation and survival of cells lacking WRN were altered upon MYC overexpression, we compared the proliferative effects of c-myc overexpression in normal and in two different WRN syndrome fibroblasts (WRN–/– strains AG03141 and AG00780) that had been immortalized by hTERT as previously described (Hirata et al. 2002). Abrief burst of cell proliferation was observed 48 h following retroviral transduction of c-myc in all cell strains. However, within 2–3 passages, ∼30%–70% of the WRN–/– cells expressed senescence-associated (SA-) β-galactosidase (β-gal; Fig. 3A) and lost proliferative capacity (Fig. 3B). The senescent phenotype in c-myc-transduced WRN–/– cells was also confirmed at the gene expression level by microarray analysis, which demonstrated elevated expression of several genes characteristic of replicative senescence, such as the matrix proteases (Shelton et al. 1999; see Supplementary Table 1). In contrast, only a small percentage of the hTERT+ WRN–/– cells transduced with a control retroviral vector expressed SA-β-gal (∼1%), similarly to hTERT-immortalized normal fibroblasts (hTERT+, from two independent donors) upon MYC overexpression, (Figs. 3A, 4C). These results indicate that WRN counteracts senescence in the presence of elevated MYC.
Figure 3.
Cells lacking WRN undergo rapid proliferative arrest and senescence upon c-myc overexpression. (A) hTERT-immortalized WRN–/– fibroblast strains AG00780 and AG03141 and control normal fibroblasts were transduced with LmycSN or LXSN (control) retrovirus; (Miller and Rosman 1989), then selected in G418. Approximately 30%–70% of WRN–/– infected with LmycSN showed morphological changes typical of senescent cells after 2–3 passages, and expressed SA-β-gal activity. (B) Proliferation of the cells as in A is indicated by the ratio of cells harvested/plated. (C) Western analysis of WRN and control cells at passage 3 indicates that c-myc overexpression leads to modest overexpression of p53 (Santa Cruz Biotechnology no. sc-126), p21Cip (BD Transduction Laboratories no. 610233), and p16ink4A (Pharmingen no. 13251A) in all cell types. WRN (BD Transduction Laboratories no. 611168) is undetectable in the WRN–/– lines as reported (Moser et al. 2000), but induced by MYC in control cells. (D) ARF mRNAexpression in WRN–/– and control hTERT+ fibroblasts was analyzed by Northern blot using an INK4a exon 1-specific probe. max was used as loading control. (E) hTERT mRNAexpression at passage 3 was quantitated by RT–PCR primers, and the ribosomal protein 36B4 was used as a control. HeLa and primary human foreskin fibroblasts (HFF) cDNAs were used as controls. The apparently higher levels of ARF mRNAin control cultures are due to RNAloading differences.
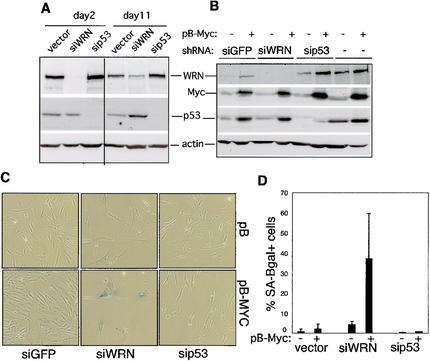
Figure 4.
WRN depletion promotes MYC-induced senescence in normal hTERT+ fibroblasts. (A) Cells transduced with a control retroviral vector (LXSH) or with pBH-siWRN or pBH-sip53 were collected for Western analysis after 2 or 11 d of hygromycin selection (cells were passaged at day +7). Antibodies are as described in Figure 3B. Western analysis of simultaneous transduction of hTERT+ fibroblasts with pBH-siWRN and pB-MYC (+) or pBabe (–) retroviruses. Controls were pBH-siGFP or pBH-sip53. Cell lysates were collected after 3 d growth in the presence of hygromycin. (C) Representative fields of cells as in B, stained for SA-β-gal. (D) Percentage of SA-β-gal+ cells as in B after 7–9 d in hygromycin. Data are from three independent experiments, counting at least 5 different fields and/or 500 cells. As controls, the data obtained with pBH empty vector were pooled with pBH-siGFP.
To better characterize the senescent phenotype of WRN–/–, hTERT+ cells following MYC overexpression, we examined the cell-cycle distribution, the expression of cell-cycle regulatory proteins, and the apoptotic response of transduced cells. WRN–/– cells that were senescent by virtue of MYC overexpression were largely G1-arrested with a small G2 fraction (data not shown). c-myc overexpression induced p53 and its target gene p21Cip (Brown et al. 1997), as well as p16ink4A (Alcorta et al. 1996), the major inhibitor of Rb phosphorylation, in WRN–/– and control cells (Fig. 3C). No significant difference in the magnitude of these responses was observed, with the exception of the predicted higher levels of WRN protein in control cells transduced with a c-myc-containing retrovirus (Fig. 3C). ARF, a prominent response to elevated MYC in rodent cells, was not significantly induced by MYC (Fig. 3D), whereas hTERT expression, as expected, was consistently elevated in all cells (Fig. 3E). The limited proliferation of WRN–/– cells following c-myc transduction might reflect increased apoptosis. However, we did not detect appreciable cell death early after infection or during later passages, by FACS or by TUNEL assay (data not shown).
Depletion ofWRN protein in normal hTERT+ fibroblasts by RNA interference (RNAi)
The results above suggest that the coordinate up-regulation of WRN by MYC may be important to insure MYC-driven cell proliferation by attenuating the nonspecific suppressive effect of cell senescence. To determine whether the senescent phenotype of WRN–/– fibroblasts results directly from the loss of WRN, we examined the effects of MYC overexpression in normal hTERT+ fibroblasts following the acute depletion of WRN by RNAi. Initial experiments indicated that WRN protein could be almost completely depleted from WRN+ hTERT+ fibroblasts after transduction with a retroviral vector (pBH-siWRN) expressing a short hairpin silencing RNAspecific for the WRN gene and 2 d of antibiotic selection (Fig. 4A). WRN protein could be detected at later time points, so we simultaneously transduced cells with pBH-siWRN and a retrovirus expressing MYC (pB-MYC) or an empty virus (pB). Hygromycin selection was used to recover cells that expressed siWRN. WRN was again undetectable after 3 d of selection in hygromycin, whereas MYC was overexpressed (Fig. 4B). WRN-depleted cells proliferated poorly compared to empty vector or cells expressing a control RNAi (sip53; data not shown). MYC overexpression failed to rescue the proliferative capacity of WRN-depleted cells and led to a significant increase in the percentage of SA-β-gal-positive cells (Fig. 4C,D).
Conclusions
The results detailed above indicate that WRN is able to modulate cell proliferation and senescence upon MYC overexpression in human hTERT+ fibroblasts. Amodel to explain this interdependence of MYC and WRN is shown in Figure 5: MYC overexpression can promote cell growth and proliferation, as well as genomic instability (Felsher and Bishop 1999; Soucek and Evan 2002; Vafa et al. 2002). WRN suppresses genomic instability and insures cell survival after DNAdamage by virtue of its role(s) in DNArecombination and repair (Prince et al. 2001; Brosh and Bohr 2002; Saintigny et al. 2002). Cells lacking WRN function display increased genetic instability and have limited growth potential. These defects can in part be compensated for by the re-expression of telomerase activity (Wyllie et al. 2000; Choi et al. 2001). However, hTERT+ cells lacking WRN cannot overcome the additional demands imposed by MYC-driven proliferation, and thus exhibit impaired cell proliferation and enter senescence.
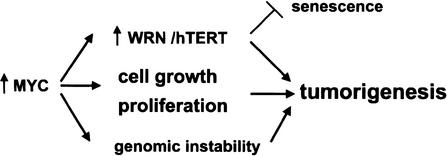
Figure 5.
Role of WRN induction in MYC-driven tumorigenesis. WRN appears to suppress genomic instability and cellular senescence by insuring the error-free repair of DNAfollowing damage or replication arrest. MYC overexpression promotes tumorigenesis by influencing cell growth, cell proliferation (for review, see Grandori et al. 2000), and genomic instability (Felsher and Bishop 1999; Vafa et al. 2002). MYC induction of WRN and hTERT (Wang et al. 1998a; Wu et al. 1999) may further promote tumorigenesis by suppressing cellular senescence while insuring a high probability of continued cell division.
One prediction of this model is that WRN patients should not be susceptible to tumors such as Burkitt or diffuse large B-cell lymphomas or to breast or prostate carcinomas, where MYC overexpression is commonly observed (Nesbit et al. 1999). This is indeed the case: the most common neoplasms observed in WRN patients are thyroid carcinomas, osteosarcomas, soft tissue sarcomas, and meningiomas, and the leukemias and lymphomas that are observed in WRN patients are largely of histopathologic subtypes other than those associated with MYC abnormalities (Goto et al. 1996). It should also be possible to further define the functional interdependence of MYC and WRN in transgenic systems, such as Eμ-c-myc mice (Adams et al. 1985), which rapidly develop MYC-dependent B-cell lymphomas. The model in Figure 5 predicts that lymphoma development in these mice will be suppressed by cellular senescence in the absence of WRN (Lebel and Leder 1998; Lombard et al. 2000). This new link between MYC and WRN may also identify new opportunities for the treatment of MYC-overexpressing human tumors.
Materials and methods
Electromobility gel shift assays (EMSAs)
EMSAs were performed with bacterially expressed C92-MYC and with full-length MAX as described (Wu et al. 1999). Approximately 3 ng of a 32-P-labeled WRN promoter fragment (nucleotides –412 to +8; GenBank accession no. AB003173; Wang et al. 1998b; Yamabe et al. 1998) was used with different amounts of purified MYC–MAX protein. Where specified, the WRN promoter Aand B sites were mutated to CGCCTG or CATTCT, respectively. MYC–MAX–DNA complexes were resolved on 4% polyacrylamide-HEPES gels prior to PhosphorImager quantitation.
Chromatin cross-linking and immunoprecipitations (ChIP assay)
The detailed ChIP protocol and quantification has been described (Frank et al. 2001). For additional details, see Supplemental Materials and Methods. In the experiment shown in Figure 2C, cross-linked chromatin from P-493-6 cells expressing Tet-myc gene (–Tet; MYC on) or from cells exposed to tetracycline for 48 h (+Tet; MYC off) was prepared as described (Schubeler et al. 2000). Identical chromatin fractions were precipitated with anti-MYC, anti-acetylated H4 (acH4; Upstate Biotechnology), or rabbit preimmune serum. Bound DNAwas quantified by duplex PCR using primers flanking site B of the WRN promoter and the β-globin gene as internal control as described (Schubeler et al. 2000).
Northern analyses and reverse transcriptase PCR (RT–PCR)
See Supplemental Materials and Methods.
Cell culture
Primary WRN fibroblast strains AG00780 and AG03141 (Coriell Cell Repository) and normal primary human fibroblasts were immortalized by infection with a human hTERT-expressing retrovirus, MLV-LTSP (Hirata et al. 2002). Polyclonal populations of cells were then transduced with the MYC expressing LmycSN retrovirus or the control retrovirus LXSN (Miller and Rosman 1989) prior to selection in G418 (700 μg/mL). G418-resistant cells were plated at the same density for proliferation assays and for counting at each passage. Cell staining for the senescent marker SA-β-gal activity was performed as described (Dimri et al. 1995). For the experiments shown in Figure 4, hTERT+ fibroblasts obtained with MLV-LTSP or LXSN expressing hTERT were simultaneously infected with pBH silencing vectors (see below) and pBabe-MYC or empty vector, pBabe (Morgenstern and Land 1990). After 3 d in hygromycin, cells were replated at 0.5 × 105 – 105 in 10-cm plates and stained for SA-β-gal within 6–9 d.
U937, Raji, HL60, and P-493-6 cells were grown in RPMI1640 medium supplemented with 10% fetal calf serum (FCS). For the P-493-6 cells ChIP assay, 2 L of exponentially growing cells were diluted to 3 × 105 cells/mL, and 0.1 μg/mL tetracycline was added 72 h prior to harvest. To induce expression of Tet-myc, P-493-6 cells were washed 3 times in warm RPMI medium containing 10% FCS in order to remove tetracycline before culturing, as indicated in Figure 2B.
RNAi depletion
Amodified pBabeHygro retroviral vector (pBH) was constructed to stably express siRNAs in primary fibroblasts. An expression cassette containing the human H1 promoter followed by gene-specific short hairpin sequences (as described by Brummelkamp et al. 2002) was inserted into the unique NheI site in the downstream U3 region of the pBH backbone to create pBH-siWRN. The anti-WRN hairpin corresponds to human WRN cDNAbps 391–415 (GenBank accession no. L76937). This WRN region does not share significant homology with the other human RecQ helicases or to other expressed genes. pBH-sip53 contains a similar H1-siRNAcassette containing an anti-p53 hairpin (Brummelkamp et al. 2002). An empty pBH vector and pBH-siGFP, expressing a siRNAagainst green fluorescent protein, were used as negative controls.
Western analyses
See Supplemental Material and Methods.
Acknowledgments
We thank Robert Eisenman for his continued support and advice throughout this work; Michela Mattioli for control Northern analysis of EREB cells without Myc-ER™; Jeffrey Delrow for microarray analysis; Jenny Benanti, Lindy Gewin, and Denise Galloway for reagents and advice; Susan Mendrysa and Shaun Cowley for critically reviewing this manuscript; and Amir Oryan for stimulating discussions. This work was supported by grants from the U.S. NIH to C.G. (CA7515), Robert Eisenman (CA20525), R.J.M., and D.W.R., and from the Nippon Boehringer-Ingelheim Virtual Research Institute on Aging to R.J.M.
This paper is dedicated to the memory of Carlo Grandori.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1100303.
Corresponding author.
Footnotes
Supplemental material is available at http://parma.fhcrc.org/CGrandori.
References
- Adams J.M., Harris, A.W., Pinkert, C.A., Corcoran, L.M., Alexander, W.S., Cory, S., Palmiter, R.D., and Brinster, R.L. 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318: 533–538. [DOI] [PubMed] [Google Scholar]
- Alcorta D.A., Xiong, Y., Phelps, D., Hannon, G., Beach, D., and Barrett, J.C. 1996. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc. Natl. Acad. Sci. 93: 13742–13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R.M. and Bohr, V.A. 2002. Roles of the Werner syndrome protein in pathways required for maintenance of genome stability. Exp. Gerontol. 37: 491–506. [DOI] [PubMed] [Google Scholar]
- Brown J.P., Wei, W., and Sedivy, J.M. 1997. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science 277: 831–834. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards, R., and Agami, R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. [DOI] [PubMed] [Google Scholar]
- Choi D., Whittier, P.S., Oshima, J., and Funk, W.D. 2001. Telomerase expression prevents replicative senescence but does not fully reset mRNAexpression patterns in Werner syndrome cell strains. FASEB J. 15: 1014–1020. [DOI] [PubMed] [Google Scholar]
- Dang C.V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P., Lee, X., Basile, G., Acosta, M., Scott, G., Roskelley, C., Medrano, E.E., Linskens, M., Rubelj, I., Pereira-Smith, O., et al. 1995. Abiomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. 92: 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G.I., Wyllie, A.H., Gilbert, C.S., Littlewood, T.D., Land, H., Brooks, M., Waters, C.M., Penn, L.Z., and Hancock, D.C. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69: 119–128. [DOI] [PubMed] [Google Scholar]
- Felsher D.W. and Bishop, J.M. 1999. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc. Natl. Acad. Sci. 96: 3940–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.R., Schroeder, M., Fernandez, P., Taubert, S., and Amati, B. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes & Dev. 15: 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M., Miller, R.W., Ishikawa, Y., and Sugano, H. 1996. Excess of rare cancers in Werner syndrome (adult progeria). Cancer Epidemiol Biomark. Prev. 5: 239–246. [PubMed] [Google Scholar]
- Grandori C. and Eisenman, R.N. 1997. Myc target genes. Trends Biochem. Sci. 22: 177–181. [DOI] [PubMed] [Google Scholar]
- Grandori C., Cowley, S.M., James, L.P., and Eisenman, R.N. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16: 653–699. [DOI] [PubMed] [Google Scholar]
- Greenberg R.A., O'Hagan, R.C., Deng, H., Xiao, Q., Hann, S.R., Adams, R.R., Lichtsteiner, S., Chin, L., Morin, G.B., and DePinho, R.A. 1999. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene 18: 1219–1226. [DOI] [PubMed] [Google Scholar]
- Gu W., Cechova, K., Tassi, V., and Dalla-Favera, R. 1993. Opposite regulation of gene transcription and cell proliferation by c-Myc and Max. Proc. Natl. Acad. Sci. 90: 2935–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata R., Chamberlain, J., Dong, R., and Russell, D.W. 2002. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol. 20: 735–738. [DOI] [PubMed] [Google Scholar]
- Kempkes B., Spitkovsky, D., Jansen-Durr, P., Ellwart, J.W., Kremmer, E., Delecluse, H.J., Rottenberger, C., Bornkamm, G.W., and Hammerschmidt, W. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14: 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M. and Leder, P. 1998. Adeletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc. Natl. Acad. Sci. 95: 13097–13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood T.D., Hancock, D.C., Danielian, P.S., Parker, M.G., and Evan, G.I. 1995. Amodified estrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23: 1686–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard D.B., Beard, C., Johnson, B., Marciniak, R.A., Dausman, J., Bronson, R., Buhlmann, J.E., Lipman, R., Curry, R., Sharpe, A., et al. 2000. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol. Cell Biol. 20: 3286–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.D. and Rosman, G.J. 1989. Improved retroviral vectors for gene transfer and expression. Biotechniques 7: 980–982. [PMC free article] [PubMed] [Google Scholar]
- Morgenstern J.P. and Land, H. 1990. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18: 3587–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M.J., Kamath-Loeb, A.S., Jacob, J.E., Bennett, S.E., Oshima, J., and Monnat Jr., R.J. 2000. WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res. 28: 648–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbit C.E., Tersak, J.M., and Prochownik, E.V. 1999. MYC oncogenes and human neoplastic disease. Oncogene 18: 3004–3016. [DOI] [PubMed] [Google Scholar]
- Oster S.K., Ho, C.S., Soucie, E.L., and Penn, L.Z. 2002. The myc oncogene: MarvelouslY complex. Adv. Cancer Res. 84: 81–154. [DOI] [PubMed] [Google Scholar]
- Prince P.R., Emond, M.J., and Monnat Jr., R.J. 2001. Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes & Dev. 15: 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny Y., Makienko, K., Swanson, C., Emond, M.J., and Monnat Jr., R.J. 2002. Homologous recombination resolution defect in Werner syndrome. Mol. Cell Biol. 22: 6971–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., Francastel, C., Cimbora, D.M., Reik, A., Martin, D.I., and Groudine, M. 2000. Nuclear localization and histone acetylation: A pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes & Dev. 14: 940–950. [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M., Staege, M.S., Pajic, A., Polack, A., Weidle, U.H., Bornkamm, G.W., Eick, D., and Kohlhuber, F. 1999. Control of cell growth by c-Myc in the absence of cell division. Curr. Biol. 9: 1255–1258. [DOI] [PubMed] [Google Scholar]
- Shelton D.N., Chang, E., Whittier, P.S., Choi, D., and Funk, W.D. 1999. Microarray analysis of replicative senescence. Curr. Biol. 9: 939–945. [DOI] [PubMed] [Google Scholar]
- Shiratori M., Sakamoto, S., Suzuki, N., Tokutake, Y., Kawabe, Y., Enomoto, T., Sugimoto, M., Goto, M., Matsumoto, T., and Furuichi, Y. 1999. Detection by epitope-defined monoclonal antibodies of Werner DNAhelicases in the nucleoplasm and their up-regulation by cell transformation and immortalization. J. Cell Biol. 144: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L. and Evan, G. 2002. Myc—Is this the oncogene from Hell? Cancer Cell 1: 406–408. [DOI] [PubMed] [Google Scholar]
- Vafa O., Wade, M., Kern, S., Beeche, M., Pandita, T.K., Hampton, G.M., and Wahl, G.M. 2002. c-Myc can induce DNAdamage, increase reactive oxygen species, and mitigate p53 function. Amechanism for oncogene-induced genetic instability. Mol. Cell 9: 1031–1044. [DOI] [PubMed] [Google Scholar]
- Wang J., Xie, L.Y., Allan, S., Beach, D., and Hannon, G.J. 1998a. Myc activates telomerase. Genes & Dev. 12: 1769–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hunt, K.E., Martin, G.M., and Oshima, J. 1998b. Structure and function of the human Werner syndrome gene promoter: Evidence for transcriptional modulation. Nucleic Acids Res. 26: 3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.J., Grandori, C., Amacker, M., Simon-Vermot, N., Polack, A., Lingner, J., and Dalla-Favera, R. 1999. Direct activation of TERT transcription by c-MYC. Nat. Genet. 21: 220–224. [DOI] [PubMed] [Google Scholar]
- Wyllie F.S., Jones, C.J., Skinner, J.W., Haughton, M.F., Wallis, C., Wynford-Thomas, D., Faragher, R.G., and Kipling, D. 2000. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat. Genet. 24: 16–17. [DOI] [PubMed] [Google Scholar]
- Yamabe Y., Shimamoto, A., Goto, M., Yokota, J., Sugawara, M., and Furuichi, Y. 1998. Sp1-mediated transcription of the Werner helicase gene is modulated by Rb and p53. Mol. Cell Biol. 18: 6191–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.E., Oshima, J., Fu, Y.H., Wijsman, E.M., Hisama, F., Alisch, R., Matthews, S., Nakura, J., Miki, T., Ouais, S., et al. 1996. Positional cloning of the Werner's syndrome gene. Science 272: 258–262. [DOI] [PubMed] [Google Scholar]