Abstract
Chromatin structure plays a critical role in the regulation of transcription. Drosophila GAGA factor directs chromatin remodeling to its binding sites. We show here that Drosophila FACT (facilitates chromatin transcription), a heterodimer of dSPT16 and dSSRP1, is associated with GAGA factor through its dSSRP1 subunit, binds to a nucleosome, and facilitates GAGA factor-directed chromatin remodeling. Moreover, genetic interactions between Trithorax-like encoding GAGA factor and spt16 implicate the GAGA factor–FACT complex in expression of Hox genes Ultrabithorax, Sex combs reduced, and Abdominal-B. Chromatin immunoprecipitation experiments indicated the presence of the GAGA factor–FACT complex in the regulatory regions of Ultrabithorax and Abdominal-B. These data illustrate a crucial role of FACT in the modulation of chromatin structure for the regulation of gene expression.
Keywords: FACT, GAGA factor, Hox gene, chromatin, Drosophila
The identity of the body segment of metazoans is determined by the expression patterns of Hox genes (McGinnis and Krumlauf 1992). Hox gene expression is initiated by the actions of many transcription factors during embryogenesis. Curiously, these expression patterns are maintained after disappearance of the transcription factors. The active or inactive state is memorized through chromatin structure and transmitted to daughter cells through cell division. Although the mechanism of this epigenetic gene expression is not clear, genetic studies have identified many genes involved in the process (Kennison 1995). These genes are classified into two groups. Pc group genes mainly govern the maintenance of the inactive state, and trx group genes are mainly responsible for the maintenance of the active state. These genes are conserved between Drosophila and vertebrates.
To elucidate the mechanism underlying the maintenance of the active state, we have analyzed the Drosophila Trithorax-like (Trl) gene product GAGA factor. GAGA factor has a BTB/POZ domain on its N terminus and a glutamine-rich domain on its C terminus. These domains are involved in protein–protein interactions. There is a Zn-finger domain in the middle of the molecule. Through this domain, the factor binds to a GAGAG sequence on DNA and hence, it is called GAGA factor (Wilkins and Lis 1997). GAGA factor-binding sites are present in the promoter regions of many Drosophila genes, including fushi tarazu (ftz), hsp70, and Ultrabithorax (Ubx; Soeller et al. 1993). Both GAGA factor and its binding sites are necessary for the proper expression of ftz and Ubx (Topol et al. 1991; Farkas et al. 1994; Bhat et al. 1996). GAGA factor can induce chromatin remodeling at the hsp70 promoter in an embryonic extract from Drosophila (Tsukiyama et al. 1994). The responsible chromatin remodeling factor NURF was purified, and its catalytic subunit was identified as ISWI (Tsukiyama and Wu 1995; Tsukiyama et al. 1995). We have shown that GAGA factor-induced and ISWI-mediated chromatin remodeling within the promoter region of ftz activates transcription on chromatin templates (Okada and Hirose 1998). On both promoters, the remodeling occurred only around the GAGA factor-binding sites.
To investigate how GAGA factor induces the site-specific chromatin remodeling, we searched for GAGA factor-associated proteins and obtained dSPT16 (Drosophila homolog of yeast SPT16) and dSSRP1 (Drosophila homolog of mammalian structure-specific recognition protein 1). SPT16 and SSRP1 are highly conserved among eukaryotes and present as a heterodimer. The human complex FACT (facilitates chromatin transcription) was purified as a factor that is required for transcription elongation through the barrier of nucleosomes (Orphanides et al. 1998). FACT has been suggested to change nucleosome structure through its binding to histones H2A and H2B (Orphanides et al. 1999). The heterodimer isolated from Xenopus, DUF1 (DNA unwinding factor 1), is necessary for DNA synthesis in an oocyte extract (Okuhara et al. 1999). In the yeast Saccharomyces cerevisiae, the SPN complex consists of the three proteins SPT16, POB3, and NHP6, in which the SSRP1 counterpart is split into two polypeptides. The yeast complex has been indicated to play an important role in regulating transcription and replication (Brewster et al. 1998; Formosa et al. 2001).
In this study, we analyzed physical and functional interactions between Drosophila FACT (dFACT) and GAGA factor. Our data demonstrate that, in addition to transcription elongation and DNA replication, FACT is involved in the modulating chromatin structure for regulation of gene expression.
Results
Isolation of GAGA factor-associated proteins
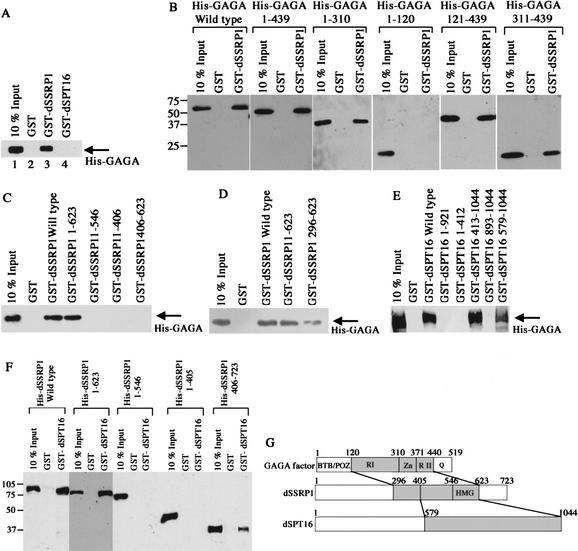
To isolate GAGA factor-associated proteins, we made a transgenic fly line capable of expressing Flag-tagged GAGA factor. Although two isoforms of GAGA factor have been reported (Benyajati et al. 1997), we chose a better characterized 519-amino acid isoform for the tagging. The expression level of Flag-GAGA factor was comparable to those of endogenous GAGA factor (data not shown). Nuclear extracts were prepared from embryos of the transgenic line. GAGA factor and its associated proteins were collected from the extracts using anti-Flag antibody beads, eluted with the Flag peptide after extensive washing, and resolved by SDS-PAGE. When we started from a nuclear extract of yw host line lacking the transgene, no protein bands were detectable (Fig. 1A, lane 1). In contrast, three proteins were recovered from a nuclear extract of the transgenic line (Fig. 1A, lane 2). The fastest migrating band appears to be Flag-GAGA factor, judging from its mobility and its detection with anti-Flag M2 antibody on a Western blot (Fig. 1A, lane 3). Peptide sequencing of a protein in the middle band (Fig. 1B) showed an unambiguous match with dSSRP1 (Hsu et al. 1993). Similar sequence analyses of peptides (Fig. 1B) revealed that the slowest migrating band is dSPT16 (AF023270). The identification of the two proteins was confirmed by detection of the bands with antibodies against dSSRP1 (Fig. 1A, lane 4) and dSPT16 (Fig. 1A, lane 5).
Figure 1.
Flag-GAGA factor-associated proteins. (A) Identification of Flag-GAGA factor-associated proteins. Proteins were purified by an anti-Flag M2 affinity column from a nuclear extract of a host line yw (lane 1) or a transgenic line expressing Flag-GAGA factor (lane 2), separated by 7.5% SDS-PAGE, transferred to a membrane, and stained with Coomassie Brilliant Blue. (Lanes 3–5) Proteins were blotted onto a membrane as in lane 2 and probed with the indicated antibodies. Positions of molecular weight markers are shown at the left of lane 1. (B) Peptide sequences from a protein in the middle or upper band. “X” represents an unidentified amino acid. Numbers in parentheses denote amino acid positions. (C) Deduced amino acid sequence of SPT16 from Drosophila embryos. Lines represent the stretches of acidic amino acids.
We then compared the cellular abundance of GAGA factor and dFACT. Western analyses showed that virtually all Flag-GAGA factors in the nuclear extract were captured on anti-Flag antibody beads and eluted with the Flag peptide, whereas ∼5% each of dSSRP1 and dSPT16 in the extract were recovered in the eluate (data not shown). Together with the presence of stoichiometric amounts of the three proteins in the eluate (Fig. 1A), these results suggest that there is a molar excess of dFACT over GAGA factor and that essentially all GAGA factors are associated with dFACT in embryonic nuclei. This is supported by coimmunoprecipitation of equimolar amounts of dSSRP1, dSPT16, and the natural form of GAGA factor from nuclear extracts of yw embryos using antibodies against GAGA factor (data not shown).
We cloned cDNAs encoding dSSRP1 and dSPT16 using a cDNA library from embryos. The deduced amino acid sequence of the dSSRP1 cDNA was exactly same as that reported by Hsu et al. (1993). However, amino acid sequence predicted from our dSPT16 cDNA (accession no. AB083008; Fig. 1C) was different from that deposited previously (accession no. AF023270): missing in our cDNA is the exon before the last one, encoding a 39-amino acid stretch near the C terminus. Analyses using reverse transcriptase PCR (RT–PCR) revealed that the mRNA corresponding to our cDNA is expressed throughout the life of Drosophila, whereas mRNA with the previously deposited sequence is expressed only in larvae and pupae (data not shown).
GAGA factor makes a direct contact with dFACT through dSSRP1
Protein–protein interactions among GAGA factor, dSPT16, and dSSRP1 were analyzed by using bacterially expressed and purified proteins. First we examined which subunit of dFACT binds directly to GAGA factor. GST pull-down assays showed that polyhistidine-tagged GAGA factor (His-GAGA) binds to GST-dSSRP1 fusion protein (GST-dSSRP1) but not to GST-dSPT16 (Fig. 2A). In a reciprocal test, His-dSSRP1 was pulled down with both GST-GAGA (data not shown) and GST-dSPT16 (Fig. 2E). These results demonstrate that GAGA factor makes a direct contact with dFACT through dSSRP1.
Figure 2.
GST pull-down assays. (A) His-GAGA was tested for binding to GST, GST-dSSRP1, or GST-dSPT16. (B) Indicated deletion derivatives of His-GAGA were examined for binding to GST or GST-dSSRP1. (C,D) His-GAGA factor was assayed for binding to GST or indicated deletion derivatives of GST-dSSRP1. (E) His-dSSRP1 was analyzed for binding to GST or indicated deletion derivatives of GST-dSPT16. (F) Indicated deletion derivatives of His-dSSRP1 were tested for binding to GST-dSPT16. (G) Summary of interactions among GAGA factor, dSSRP1, and dSPT16. In B–F, numbers in each deletion represent the remaining amino acid residues. In B and F, positions of molecular weight markers are shown at the left.
Then we prepared deletion derivatives of His-GAGA and GST-dSSRP1 to analyze regions responsible for interaction between GAGA factor and dSSRP1. GAGA factor was divided into BTB/POZ, Zn-finger and Q-rich domains, and two regions between these domains (RI and RII; Fig. 2G). GST pull-down assays showed that either RI or a region consisting of the Zn-finger domain and RII is sufficient for the binding to GST-dSSRP1 (Fig. 2B). Similar deletion analyses of dSSRP1 revealed that a region between amino acid positions 296 and 623 including an HMG box is enough for the binding of His-GAGA (Fig. 2C,D).
We also analyzed regions responsible for interaction between dSSRP1 and dSPT16. Binding of His-dSSRP1 to various deletion derivatives of GST-dSPT16 demonstrated that a C-terminal region of dSPT16 between amino acid positions 579 and 1044 is enough for the interaction with dSSRP1 (Fig. 2E). Similar deletion analyses of dSSRP1 narrowed down the binding region to a sequence between amino acid positions 405 and 623 including the HMG box (Fig. 2F). Protein–protein interactions among GAGA factor, dSSRP1, and dSPT16 are summarized in Figure 2G.
dFACT can bind to a nucleosome
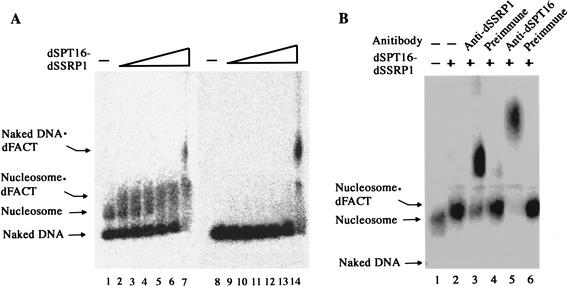
Because human FACT and the yeast counterpart SPN complex bind to nucleosomes (Orphanides et al. 1999; Formosa et al. 2001), we anticipated that dFACT also interacts with chromatin. To test this possibility, we carried out gel electrophoresis mobility shift assays. When His-dSPT16 and His-dSSRP1 were incubated with mononucleosomes, we observed retardation of a mononucleosome band (Fig. 3A, cf. lanes 2–6 and 1; Fig. 3B, cf. lanes 2 and 1). The shifted band appears to be a dFACT–mononucleosome complex, because further addition of anti-dSSRP1 (Fig. 3B, lane 3) or anti-dSPT16 (Fig. 3B, lane 5) antibodies resulted in supershift of the band, whereas addition of each preimmune serum did not change the mobility (Fig. 3B, lanes 4,6). Under these conditions, dFACT did not bind to naked DNA from which the mononucleosomes were assembled (Fig. 3A, lanes 9–13; see also behavior of naked DNA contained in the preparation of mononucleosomes in Fig. 3A, lanes 2–6). When we added larger amounts of His-dSPT16 and His-dSSRP1, they formed a complex with the naked DNA (Fig. 3A, lane 14). These results show that dFACT binds to mononucleosome with a higher affinity than for naked DNA.
Figure 3.
EMSA for binding of dFACT to nucleosome. (A) Binding of dFACT to nucleosome and naked DNA. Four femtomoles of 32P-labeled naked DNA fragments (lanes 8–14) or mononucleosomes assembled on them (lanes 1–7) were incubated with buffer (lanes 1,8) or 0.24fmoles (lanes 2,9), 0.73 fmoles (lanes 3,10), 2.2 fmoles (lanes 4,11), 6.6 fmoles (lanes 5,12), 20 fmoles (lanes 6,13), and 60 fmoles (lanes 7,14) each of His-dSPT16 and His-dSSRP1. The samples were then analyzed by agarose gel electrophoresis. (B) Supershift by antibodies. Mononucleosomes were incubated with buffer (lane 1) or 12 fmoles each of His-dSPT16 and His-dSSRP1 as in A. In lanes 3–6, the mixtures also contained 1.2 μg of indicated serum. The preparation of mononucleosomes contained naked DNA in A but not in B.
dFACT facilitates GAGA factor-directed chromatin remodeling
To examine whether dFACT can affect GAGA factor-directed chromatin remodeling, we reconstituted chromatin on a plasmid DNA carrying the ftz promoter in an embryonic extract from Drosophila (S150), and analyzed GAGA factor-induced disruption of the chromatin structure in the presence or absence of dFACT. Because the S150 extract was derived from embryonic cytoplasm, it did not contain detectable levels of nuclear proteins dSPT16 and dSSRP1 (data not shown), and hence we expected a response to exogenously added proteins.
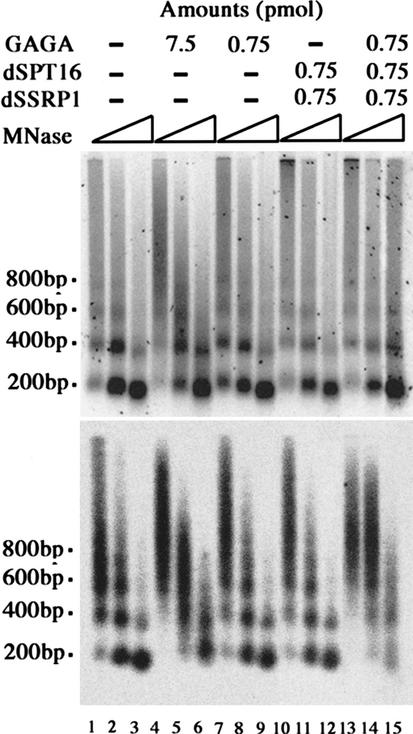
After digestion of the chromatin with micrococcal nuclease, DNA was purified and electrophoresed on an agarose gel and then stained with SYBR green I dye. No change in the pattern of nucleosome ladders was detectable upon incubation with GAGA factor in the presence or absence of dFACT (Fig. 4, top panel), indicating that GAGA factor and/or dFACT did not affect the bulk chromatin structure under these conditions. The DNA in the gel was then transferred to a membrane, and probed with an oligonucleotide carrying the GAGA factor-binding sites in the ftz promoter. As reported (Okada and Hirose 1998), GAGA factor induced smearing of nucleosome ladders around its binding sites in the S150 extract (Fig. 4, bottom panel, cf. lanes 4–6 and 1–3). Such smearing was barely detectable when the amount of GAGA factor was reduced to one-tenth (Fig. 4, bottom panel, lanes 7–9). However, addition of His-dSPT16 and His-dSSRP1 induced the smearing even in the low concentration of GAGA factor (Fig. 4, bottom panel, lanes 13–15). In the absence of GAGA factor, addition of the two proteins did not induce the smearing (Fig. 4, bottom panel, lanes 10–12). Essentially the same results were obtained by using natural dFACT purified from the embryonic nuclear extract (data not shown). These results indicate that dFACT stimulates GAGA factor-directed chromatin remodeling.
Figure 4.
Effect of dFACT on GAGA factor-directed chromatin remodeling. (Top) Nucleosome ladders derived from bulk chromatin. Chromatin was reconstituted on a plasmid DNA carrying the ftz promoter in S150 extract and then treated with indicated amounts of His-GAGA and/or a mixture of His-dSPT16 and His-dSSRP1. After incubation, chromatin was digested with micrococcal nuclease, and nucleosome ladders were visualized by staining with SYBR green I dye. (Bottom) Chromatin structure around GAGA factor-binding sites. DNA was then transferred to a nylon membrane and detected by Southern hybridization with 32P-labeled oligonucleotide GAWT harboring two tandemly repeated GAGA factor-binding sites in the ftz promoter (Okada and Hirose 1998).
GAGA factor–dFACT complex is involved in expression of Hox genes
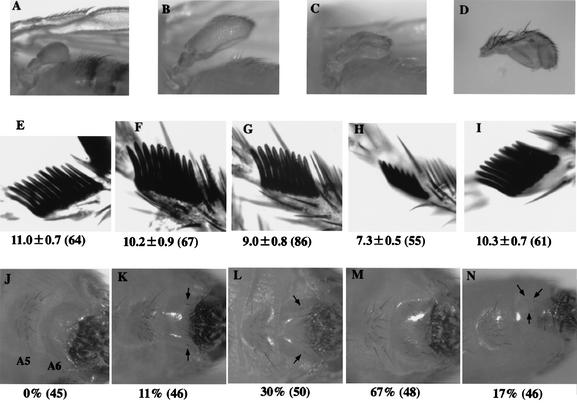
To address the biological significance of the GAGA factor–dFACT complex, we carried out genetic studies. First we focused on a phenotype of Ubx function. Most insects have two pairs of wings; in dipterans such as Drosophila, the posterior wing becomes a tiny structure termed haltere. This is due to suppression of the wing formation by Ubx in the body segment T3 (Galant and Carroll 2002). Ubx expression is initiated during embryogenesis and maintained epigenetically in cells of imaginal discs. The imaginal cells proliferate until differentiation starts in late third-instar larvae. Therefore, when the maintenance of Ubx expression is abolished in earlier stages, the haltere becomes larger and more wing-like in structure. We analyzed the genetic interaction between Ubx and Trl encoding GAGA factor. Because these genes are essential for development, the phenotypes were observed in heterozygous states. In Ubx130 heterozygotes, halteres were homogeneous in size and shape (Fig. 5A), being slightly larger than those in wild type (data not shown). In 56% of flies doubly heterozygous for Trl13C and Ubx130, halteres were quite heterogeneous in size but at least larger than those in Ubx130 single heterozygotes (e.g., see Fig. 5, cf. B and A). This confirmed the previous conclusion that Trl, a member of trx group, is involved in the maintenance of Ubx expression (Farkas et al. 1994). We then analyzed the genetic interaction between Ubx and spt16. Whereas reduction of a single dose of spt16+ in the wild-type background showed no obvious phenotype (data not shown), we observed similar large halteres with heterogeneous sizes as in Trl13C, Ubx130 double heterozygotes in 63% of files doubly heterozygous for Δspt16 and Ubx130 (e.g., see Fig. 5C). The frequency of flies with the large halteres was increased to 78% in Δspt16, Trl13C, Ubx130 triple heterozygotes. Moreover, the triple heterozygotes showed a stronger phenotype than each double heterozygote in qualitative aspects (Fig. 5, cf. D and B,C); the haltere became a wing-like structure with wing veins and hairs on the wing margin. These effects were reversed by expression of dSPT16 from a transgene in the same background (data not shown). Similar results were obtained using another Ubx allele, Ubx1 (data not shown). These results indicate that the maintenance of Ubx expression is stochastically compromised at various stages of development in Trl, Ubx and spt16, Ubx double heterozygotes and the timing is earlier in the spt16, Trl, Ubx triple heterozygotes than in the double heterozygotes.
Figure 5.
Genetic interactions between Trl and spt16. (A–D) Haltere phenotypes. (A) Ubx130/+. (B) Trl13C+/+ Ubx130. (C) Δspt16+/+ Ubx130. (D) Δspt16 Trl13C+/++ Ubx130. These were photographed at the same magnification so that the size of haltere in each panel can be compared directly. (E–I) Sex comb phenotypes. (E) +/+. (F) Trl13C/+. (G) Δspt16/+. (H) Δspt16 Trl13C/+. (I) P[SPT16+];Δspt16 Trl13C/+. Average number of teeth per sex comb with standard deviation is shown at the bottom of each figure. Numerals in parentheses represent total numbers of combs examined. (J–N) A6-to-A5 transformation. (J) +/+. (K) Trl13C/+. (L) Δspt16/+. (M) Δspt16 Trl13C/+. (N) P[SPT16+]; Δspt16 Trl13C/+. Arrows indicate bristles on A6. Also shown are percentages of males with bristles on A6. Numerals in parentheses represent total numbers of males observed.
We also examined the number of teeth per sex comb on the male anterior leg, which reflects the expression level of the Sex combs reduced (Scr) gene. Whereas on average ∼11 teeth per sex comb were seen in the wild type (Fig. 5E), we observed the average numbers of ∼10 in Trl13C/+ (Fig. 5F), ∼9 in Δspt16/+ (Fig. 5G), and ∼7 in Trl13C Δspt16/+ flies (Fig. 5H). This reduction in the sex comb teeth was restored by expression of dSPT16 from the transgene (Fig. 5I). These data suggest that the GAGA factor–dFACT complex is involved in Scr expression.
Finally, we searched for bristles on the male abdominal segment 6(A6). This is a sign of homeotic transformation from A6 to A5 due to a defect in the function of a cis-regulatory element iab6 of the Abdominal-B (Abd-B) gene (Celniker et al. 1990). No bristle was detectable on the wild-type A6 (Fig. 5J). In contrast, we observed bristles on A6 in 11% of Trl13C/+ (Fig. 5K) and 30% of Δspt16/+ flies (Fig. 5L). The frequency was increased to 67% in Trl13C, Δspt16 double heterozygotes (Fig. 5M). Moreover, we observed a significant increase in the number of bristles on A6 in the double heterozygotes compared to each single heterozygote. The effects were reversed by expression of dSPT16 from the transgene (Fig. 5N). These data indicate that the GAGA factor–dFACT complex affects Abd-B expression through the cis-regulatory element iab-6.
GAGA factor–dFACT complex is present in regulatory regions of Ubx and Abd-B
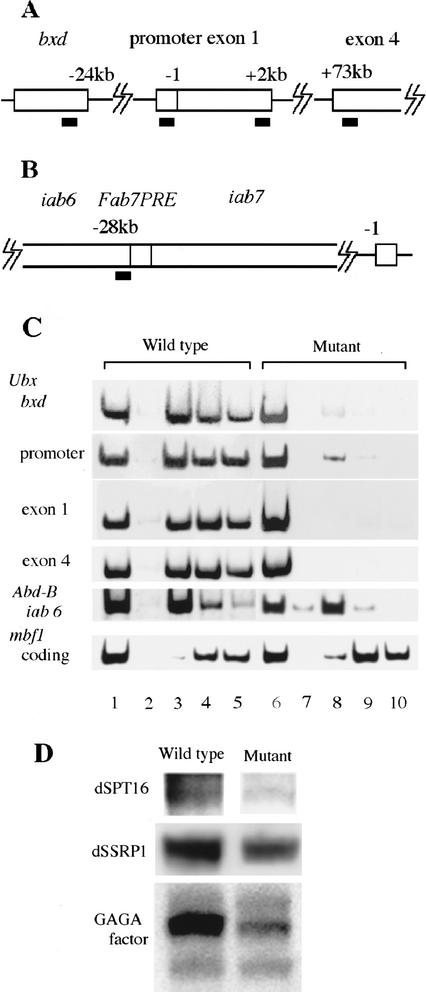
If the GAGA factor–dFACT complex is directly required for regulation of Ubx and Abd-B expression, the complex should be present in the regulatory regions of the genes. At least two regulatory regions are known for expression of Ubx (Fig. 6A). One is the bxd region that is present ∼24kb upstream of the transcription start site. The region serves as both a Pc response element and a Trx response element (Poux et al. 2002). The other is the promoter region just upstream of the transcription start site (Biggin and Tjian 1988). Expression of Abd-B on A6 is regulated through the cis-element iab-6 (Fig. 6B; Celniker et al. 1990). These regions carry a cluster of GAGA factor-binding sites. We examined whether the GAGA factor–dFACT complexes occupy these sites in late embryos, where the maintenance of Hox gene expression has been established. This was done by chromatin immunoprecipitation assays. Formaldehyde-fixed chromatin was fragmented by sonication, and protein–DNA complexes were immunoprecipitated using antibodies against GAGA factor, dSSRP1, and dSPT16. We analyzed the distribution of these proteins within the Ubx and Abd-B loci by PCR amplification of the immunoprecipitated DNA fragments. As shown in Figure 6C, all three proteins, GAGA factor, dSSRP1, and dSPT16 were present in the bxd region, the promoter region, and the coding regions of Ubx, and the iab-6 element of Abd-B in wild-type embryos.
Figure 6.
Binding of GAGA factor, dSSRP1, and dSPT16 to regulatory regions of Ubx and Abd-B in vivo. (A,B) Schematic presentation of the regions in Ubx (A) and Abd-B (B) used for ChIP assays. Positions are relative to the transcription start site. Thick bars represent the PCR amplified regions. (C) PCR amplification of DNA isolated from immunoprecipitated GAGA factor (lanes 3,8), dSSRP1 (lanes 4,9), and dSPT16 (lanes 5,10) chromatin in wild-type or the mutant embryos. Lanes 1 and 6 represent amplification of 0.1% of input DNA. Lanes 2 and 7 show amplification of DNA from mock immunoprecipitated chromatin. (D) Western analyses of GAGA factor, dSSRP1, and dSPT16 in wild-type or the mutant embryos.
To test the effect of Δspt16, Trl13C double mutation on the presence of GAGA factor–dFACT complex in the regulatory regions of Ubx and Abd-B, chromatin immunoprecipitation assays were carried out using late embryos of a Δspt16 Trl13C/TM3y+ line. Actually, the population consisted of embryos heterozygous for Δspt16 Trl13C (50%), those homozygous for Δspt16 Trl13C (25%), and dead embryos homozygous for TM3y+ (25%), but we call the population simply “mutant embryos” here. We observed significantly reduced binding of GAGA factor, dSSRP1, and dSPT16 to the bxd, promoter, and coding regions of Ubx, and iab-6 element of Abd-B in the mutant embryos (Fig. 6C). As a control, the binding of dSSRP1 and dSPT16 to the coding region of a GAGA factor-independent gene mbf1 (Liu et al. 2003) was barely affected by these mutations (Fig. 6C). The observed large impact of the double mutation on the cross-linking of GAGA factor and dFACT to the regulatory regions of Ubx and Abd-B could be due to an unexpectedly large decrease in the expression of GAGA factor or dFACT in the mutant embryos. However, this possibility was excluded by our Western analyses of GAGA factor, dSSRP1, and dSPT16. As shown in Figure 6D, the amounts of these proteins in the mutant embryos were not reduced by more than a factor of two. Collectively these data indicate that the GAGA factor–dFACT complex binds to the regulatory regions of Ubx and Abd-B, and contributes to expression of these genes through chromatin remodeling.
Discussion
Role of GAGA factor–dFACT complex in chromatin remodeling
We anticipated that we would identify a chromatin remodeling factor among the GAGA factor-associated proteins. However, to our surprise, we identified GAGA factor–dFACT complex in the nuclei of Drosophila embryos. GST pull-down assays showed that GAGA factor makes a direct contact with dFACT through its dSSRP1 subunit. Gel electrophoresis mobility shift assays revealed that dFACT binds to nucleosome. Furthermore, we found that dFACT stimulates GAGA factor-directed chromatin remodeling in the embryonic extract of Drosophila. Based on these data, we propose the following model for GAGA factor-directed site-specific chromatin remodeling. The GAGA factor–dFACT complex binds to a GAGAG sequence on DNA. dFACT binds to nucleosome and stimulates chromatin remodeling. This allows remodeling in a GAGA factor binding site-dependent manner. Because human FACT binds to histones H2A and H2B (Orphanides et al. 1999), and the yeast SPN complex enhances DNase I sensitivity of nucleosome in a region where H2A and H2B contact the DNA (Formosa et al. 2001), it is most likely that FACT binds to DNA at the entry and exit site of the nucleosome through its HMG subunit SSRP1, and then acts to destabilize and remove the H2A/H2B dimers to facilitate chromatin remodeling. However, the H2A/H2B dimers remain associated with the FACT–nucleosome complex through SPT16 such that they can quickly rebind to the H3/H4 tetramer when required. In support of this model, an acidic amino acid stretch found in histone-interacting proteins such as nucleoplasmin and NAP1 is conserved in the C-terminal tail of SPT16 (Fig. 1C; Evans et al. 1998; Okuhara et al. 1999; Orphanides et al. 1999). Furthermore, H2B (and probably H2A) has been shown to turn over more rapidly than H3 and H4during transcription (Kimura and Cook 2001).
There are many ATP-dependent chromatin remodeling factors (Vignali et al. 2000; Narlikar et al. 2002). Which factor is responsible for the GAGA factor–dFACT complex-induced chromatin remodeling? Because an antibody against ISWI abolished the GAGA factor-induced chromatin remodeling in the embryonic extract (Okada and Hirose 1998), remodeling factors containing ISWI as the catalytic subunit must play a role. Among them, at least NURF appears to be involved in the remodeling, because GAGA factor interacts directly with NURF (Xiao et al. 2001). NURF subunits seem to have escaped from our survey of GAGA factor-associated proteins due to our severe washing conditions for Flag-antibody beads after the binding of proteins. The GAGA factor–dFACT complex tolerated this, but the GAGA factor–NURF complex dissociated in the washing buffer containing 0.34M NaCl.
Although ISWI is essential for the expression of en and Ubx in imaginal discs, Deuring et al. (2000) suggested that ISWI is mainly involved in transcription repression in vivo. Specific acetylation of histone H4at Lys 16 by MOF counteracts the action of ISWI (Corona et al. 2002) and leads to derepression of chromatin transcription (Akhtar and Becker 2000). Interestingly, the yeast SPT16–Pob3 complex interacts with Sas3, a yeast homolog of MOF (John et al. 2001). It is possible that dFACT may also recruit MOF to shut out ISWI and induce a change from repression to activation. Under such conditions, remodeling factor(s) other than NURF may cooperate with the GAGA factor–dFACT complex. Brahma (BRM) remodeling complex may be a candidate for the replacer, but no genetic interaction between Trl and brm has been demonstrated (Papoulas et al. 1998). CHD1 may be another candidate because mouse, Drosophila, and yeast CHD1 have been reported to interact with SSRP1 (Kelley et al. 1999) or its yeast counterpart POB3 (Simic et al. 2003).
Protein–protein interactions among GAGA factor, dSPT16,and dSSRP1
We have defined the regions of GAGA factor, dSPT16, and dSSRP1 that are responsible for interactions of these proteins, as summarized in Figure 2G. GAGA factor interacts with dSSRP1 through the region containing the Zn-finger domain and its flanking sequences. This finding is consistent with the previous report that both BTB/POZ and Q-rich domains are not required for the GAGA factor-induced chromatin remodeling in the embryonic extract (Agianian et al. 1999). It also suggests that GAGA factor can form an oligomer through its BTB/POZ and/or Q-rich domains and bind cooperatively to clusters of its binding sites (Espinas et al. 1999; Katsani et al. 1999) even as the complex with dFACT. The cooperative and stable binding of GAGA factor–dFACT complex to chromatin may be important for the epigenetic maintenance of the active state.
The GAGA factor-interacting region of dSSRP1 contains the HMG box and its N-terminal flanking sequence that overlaps with the dSPT16-binding region of dSSRP1. However, the presence of equimolar complex of GAGA factor, dSSRP1, and dSPT16 in the embryonic nuclear extract indicates that the overlapped region in dSSRP1 does not interfere with the simultaneous binding of GAGA factor and dSPT16. Interestingly, dSSRP1 bound to naked DNA, but addition of increasing amounts of dSPT16 decreased the DNA binding of dSSRP1 in a dose-dependent manner (data not shown). This observation suggests that dSPT16 suppresses the binding of dSSRP1 to naked DNA through its interaction with the HMG box region without affecting the affinity for nucleosome.
We defined the dSSRP1-binding sequence of dSPT16 as the C-terminal highly conserved region. This is in good agreement with the previous observation that expression of the corresponding region of yeast SPT16 is required to rescue yeast temperature-sensitive mutants of spt16 (Evans et al. 1998).
Role of GAGA factor–dFACT complex in epigenetic expression of Hox genes
The most interesting finding in this study is the involvement of the GAGA factor–dFACT complex in the regulation of gene expression. The anterior transformation of T3 and A6 in Δspt16 Trl double heterozygotes and the binding of the GAGA factor–dFACT complex to the bxd region of Ubx and the iab-6 element of Abd-B in vivo indicate that the complex contributes to the epigenetic maintenance of Hox gene expression. Based on our data, we envision the following scheme for the maintenance of the active state. The GAGA factor–dFACT complex induces chromatin remodeling in the regulatory regions of various GAGA factor-dependent genes and potentiates transcription. Whereas the expression of ftz and hsp70 is transient, the active state is maintained in Hox genes such as Ubx, Scr, and Abd-B with the aid of other trx group gene products.
What is the mechanism underlying the maintenance? Among trx group proteins, BRM constitutes an SWI/SNF-type chromatin remodeling complex (Papoulas et al. 1998). This type of chromatin remodelers possesses a unique ability to act on condensed mitotic chromatin (Krebs et al. 2000). A sequence-specific regulator, Zeste, has been shown to recruit the BRM complex to its target sites (Kal et al. 2000). Functionally distinct chromatin remodeling induced by the GAGA–dFACT and Zeste–BRM complexes may be important to keep the active state through many rounds of cell cycle. In addition to the GAGA factor–dFACT and the BRM complexes, three trx group protein complexes have been identified to date. One is TAC1 consisting of Trx, dCBP, and Sbf1, which acetylates core histones in nucleosomes (Petruck et al. 2001). Mutations in trx or nej encoding dCBP have been shown to reduce the expression of Ubx. The others are ASH1 and ASH2 complexes (Papoulas et al. 1998). ASH1 also has been known to interact directly with dCBP (Bantignies et al. 2000). These data suggest that acetylation of core histones or other proteins plays a crucial role in the maintenance of the active state. In support of this hypothesis, Cavalli and Paro (1999) showed that hyper-acetylation of H4is a heritable epigenetic mark of the active state. The finding that a counteracting Pc group complex ESC/E(Z) contains histone deacetylase RPD3 (Tie et al. 2001) is also consistent with this hypothesis. Chromatin remodeling induced by the GAGA factor–d-FACT and the Zeste–BRM complexes might be essential for maintenance of the hyperacetylated state of H4.
Materials and methods
Fly stocks
To obtain a transgenic line capable of expressing a Flag-GAGA factor, we inserted an entire open reading frame (ORF) of the 519-amino acid isoform of GAGA factor (Benyajati et al. 1997) with a Flag peptide at its C terminus into a CaSpeR–Hsp83 vector (Horabin and Schedl 1993) and used it for germline transformation. Trl13C is a P-element insertion line (Farkas et al. 1994). The Δspt16 line used in this study is Df(3L)Aprt21, which deletes a small chromosomal region including spt16, CG13937, and Aprt (Wang et al. 1994). For the rescue experiments, we inserted an entire ORF of dSTP16 with a polyhistidine tag at its N terminus into the CaSpeR–Hsp83 vector and established a transgenic line, P[w+,hsp83–spt16+] in the second chromosome. Other stocks have been described in Lindsley and Zimm (1992).
Identification of GAGA factor-associated proteins
Embryos 0–12 h after egg laying (AEL) were collected from a population of the hsp83–Flag-GAGA transgenic line reared at 25°C. Nuclear extracts were prepared from the embryos as described previously (Ueda et al. 1990). For isolation of Flag-GAGA factor-associated proteins, the nuclear extract (15 mL) was added to anti-Flag M2 affinity gel (2 mL packed volume; Sigma), and the mixture was rotated at 4°C for 1 h. The gel was collected by centrifugation and washed with a buffer (20 mM HEPES-KOH at pH 7.9, 0.34M NaCl, 3 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF, 5% glycerol) three times. The bound proteins were then eluted with the Flag peptide (50 μg/mL in the wash buffer; Sigma).
To identify Flag-GAGA factor-associated proteins, the eluted proteins were resolved by SDS-PAGE and transferred to a PVDF membrane (Boehringer Mannheim) by a Trans-Blot Semi-Dry apparatus (Bio-Rad). The membranes were subsequently stained with Coomassie Brilliant Blue, and protein in each band was subjected to sequence analysis after digestion with lysylendopeptidase.
Preparation of recombinant proteins
cDNAs encoding dSPT16 and dSSRP1 were obtained by PCR with primers designed according to the deposited sequences (Hsu et al. 1993; AF023270). A Drosophila cDNA library from 10–12-h AEL embryos in λEXlox (Novagen) was used as PCR templates. For expression of polyhistidine-tagged and GST-fusion proteins, the cDNAs encoding intact GAGA factor, dSPT16, and dSSRP1 or their deletion derivatives were cloned into pET14b (Novagen) and pGEX4T-3 NdeI, respectively. pGEX4T-3NdeI was prepared by generating an NdeI site in the multicloning site of pGEX4T-3 (Amersham Biosciences) to transfer the DNA insert directly from the pET vector to the pGEX vector. Because we were unable to express dSPT16 with the previously deposited sequence in both bacterial and baculovirus systems, we used our cDNA for this study.
His-GAGA and its deletion derivatives were expressed in Escherichia coli BL21(DE3)/pLysS at 25°C and purified by Ni-NTA (QIAGEN) chromatography as described by Agianian et al. (1999). His-dSPT16 and His-dSSRP1 were expressed in E. coli BL21 (DE3)/codon+ at 18°C. Cells were disrupted by sonication in buffer A (20 mM Tris-HCl at pH 7.9, 5 mM 2-mercaptoethanol, 10% glycerol, 0.1% NP-40, 1 mM PMSF) containing 0.4 M NaCl and centrifuged at 15,000g for 30 min. His-dSSRP1 was purified from the supernatant by Ni-NTA chromatography and dialyzed against buffer A containing 0.4M NaCl except that 0.1% NP-40 was replaced by 0.5% Triton X-100. His-dSPT16 was purified from the pellet. The pellet was washed twice by resuspending in buffer A containing 0.4M NaCl with brief sonication followed by centrifugation. The washed pellet was resuspended in buffer A containing 1 M NaCl by brief sonication and centrifuged as above. The supernatant was ∼90% pure as His-dSPT16. Because further fractionation by Ni-NTA chromatography did not improve the purity, we used the supernatant as His-dSTP16.
GST fusion proteins were expressed in E. coli BL21 (DE3)/pLysS (for GST or GST-GAGA) or in E. coli BL21(DE3)/codon+ (for GST-dSPT16, GST-dSSRP1, and their derivatives) at 25°C. Cells were disrupted by sonication in buffer A containing 0.4M NaCl and centrifuged at 15,000g for 30 min. Proteins were purified from the supernatant using glutathion Sepharose 4B (Amersham Biosciences) as described below (GST pull-down assays).
Antibodies
For production of anti-dSPT16 or anti-dSSRP1 serum, a cDNA fragment encoding amino acid residues 1–309 of dSPT16 or that encoding residues 1–420 of dSSRP1 was inserted into pET14b. Protein fragments were expressed in E. coli BL21/groEL at 25°C, purified by preparative disk SDS-PAGE (NA-1800, Nippon EIDO) and used to generate polyclonal antibodies in rabbits. Serum was used directly for Western analyses at a 10,000-fold dilution. Other antibodies include mouse anti-Flag M2 monoclonal antibody (Kodak), rabbit anti-GAGA factor antibodies (a gift from C. Wu, National Cancer Institute, NIH, Bethesda, MD) and rabbit anti-polyhistidine antibodies (Sigma).
GST pull-down assays
Bacterial lysate containing 1 μg of GST or GST-fusion protein was mixed with glutathione Sepharose 4B beads in buffer A containing 0.4M NaCl at 4°C for 60 min. The beads were collected by centrifugation and washed three times with the same buffer. The beads were then incubated with target protein (1 μg) in 50 μL of binding buffer (20 mM Tris-HCl at pH 7.9, 200 mM NaCl, 4mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.1% NR-40, 20% glycerol, and 100 μg/ml BSA) at 4°C for 60 min. The beads were collected by centrifugation and washed three times with the binding buffer. The bound proteins were resolved by SDS-PAGE and detected on immunoblots using anti-His tag or anti-dSSRP1 antibodies.
Electrophoresis mobility shift assays (EMSAs)
Mononucleosomes were assembled by a salt dialysis method (Stein 1989) using purified core histones from HeLa cells (Simon and Felsenfeld 1979) and a 216-bp EcoRI-DdeI DNA fragment from pXP10 (Wolffe and Brown 1986), which had been 5′-terminally labeled with 32P. The mononucleosomes were purified by sucrose gradient centrifugation (5%–20% in 20 mM Tris-HCl at pH 7.9, 1 mM EDTA, 10 mM 2-mercapto ethanol) in a Beckman SW50Ti rotor at 46,000 rpm for 8.5 h at 4°C.
Purified mononucleosomes or the 216-bp DNA fragments were incubated with His-dSPT16 and His-dSSRP1 in reaction mixtures (12.5 μL) containing 20 mM Tris-HCl at pH 7.9, 100 mM NaCl, 0.2 mM EDTA, 10 mM 2-mercaptoethanol, 0.4mM PMSF, 0.5 mg/mL BSA at 30°C for 60 min. The samples were then electrophoresed at 4°C on a 0.7% agarose gel in 0.5XTBE.
Chromatin remodeling assays
Chromatin was reconstituted on a mixture of pE(Hu)5-N carrying the ftz promoter (20 ng; Li et al. 1994) and φX174RFI DNA (180 ng) in reaction mixtures (20 μL) containing 10 mM HEPES-KOH at pH 7.6, 0.5 mM EGTA, 60 mM KCl, 4mM MgCl2, 10% glycerol, 30 mM creatine phosphate, 3 mM ATP, 1 mM DTT, 1 μg/mL creatine phosphokinase, 160 ng purified core histones from HeLa cells, and 6 μL S150 extract (Becker and Wu 1992) from Drosophila 0–6-h AEL embryos. After incubation at 26°C for 2.5 h, His-GAGA, His-dSPT16, and His-dSSRP1 were added and the incubation was continued for an additional 2.5 h. The samples were then treated with micrococcal nuclease, and nucleosome ladders were analyzed as described by Okada and Hirose (1998).
Chromatin immunoprecipitation (ChIP) assays
Embryos were collected 12–22 h AEL and decholionated. Chromatin immunoprecipitation assays were carried out as described (Orlando et al. 1998). Primers used were 5′-GCCATA ACGGCAGAACCAAAGTGCC-3′ and 5′-CTGCAGAGGCAG CGACTGCGCC-3′ for the Ubx bxd region, 5′-CGAATGAAT GAACGAGAGGCGCCC-3′ and 5′-ACCAAATCGCAGTTGC CAGTGTTTT-3′ for the Ubx promoter region, 5′-ACCGCC AGCAGCGCAATGAACTC-3′ and 5′-TGCCCAGCGAGAGA GGGAATCCTC-3′ for the Ubx exon 1, 5′-GTACAAATGGTC TGCGAAGACGCG-3′ and 5′-TGATCTGCCGCTCCGTCAG GCATA-3′ for the Ubx exon 4, 5′-CAAAGAGAGTTGGAAA GAGTATTGG-3′and 5′-CGCTCTCTTGCTTACCAATACAT T-3′ for the Abd-B iab-6 region, and 5′-GCAGCAAGTGGT AACCGACTACG-3′ and 5′-GCAGCATCATTTCACTTTTTA CCGG-3′ for the mbf1 coding region. Amplification of 0.02%, 0.1%, and 0.5% of input DNA established that the PCR reactions were quantitative, but we show only the results of the 0.1% input in Figure 6.
Genetic analyses
To analyze genetic interactions among Ubx, Trl, and spt16, Ubx1/TM3 or Ubx130/TM3 females were crossed with Trl13C/TM3y+, Δspt16/TM3y+, Δspt16 Trl13C/TM3y+, or P[w+,hsp83–spt16+]; Δspt16 Trl13C/TM3y+ males. Halteres of the progenies that did not carry the balancer chromosomes were observed under a microscope. Sex combs and bristles on A6 were observed using +/TM3y+, Trl13C/TM3y+, Δspt16/TM3y+, Δspt16 Trl13C/TM3y+, or P[w+,hsp83–spt16+]; Δspt16 Trl13C/TM3 y+ males.
Accession number
The DDBJ/EMBL/GenBank accession number for dSPT16 cDNA is AB083008.
Acknowledgments
We thank D. Reinberg, G. Orphanides, and C. Wu for helpful suggestions, and J.M. Mason, S. Hayashi, P. Schedl, and C. Wu for kindly supplying fly lines, DNAs, and antibodies. We also thank K. Ura for her suggestion on the preparation of mononucleosomes and K. Maenaka for his hospitality to T.S. This work was supported by Grants in aid for Scientific Research from the Ministry of Education, Science, Sports, Culture and Technology of Japan and a grant from the Naito Foundation. T.S. and M.O. were supported by a Center of Excellence Program of Japan and the Japan Society for the Promotion of Science, respectively.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1086803.
Corresponding author.
References
- Agianian B., Leonard, K., Bonte, E., Zandt, H.V., Becker, P.B., and Tucker, P.A. 1999. The glutamine-rich domain of the Drosophila GAGA factor is necessary for amyloid fibre formation in vitro, but not for chromatin remodeling. J. Mol. Biol. 285: 527–544. [DOI] [PubMed] [Google Scholar]
- Akhtar A. and Becker, P.B. 2000. Activation of transcription through histone H4acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5: 367–375. [DOI] [PubMed] [Google Scholar]
- Bantignies F., Goodman, R.H., and Smolik, S.M. 2000. Functional interaction between the coactivator Drosophila CREB-binding protein and ASH1, a member of the trithorax group of chromatin modifiers. Mol. Cell. Biol. 2: 9317–9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P.B. and Wu, C. 1992. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol. Cell. Biol. 12: 2241–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Mueller, L., Xu, N., Pappano, M., Gao, J., Mosammaparast, M., Conklin, D., Granok, H., Craig, C., and Elgin, S. 1997. Multiple isoforms of GAGA factor, a critical component of chromatin structure. Nucl. Acids Res. 25: 3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K.M., Farkas, G., Karch, F., Gyurkovics, H., Gausz, J., and Schedl, P. 1996. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development 122: 1113–1124. [DOI] [PubMed] [Google Scholar]
- Biggin M.D. and Tjian, R. 1988. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell 53: 699–711. [DOI] [PubMed] [Google Scholar]
- Brewster N.K., Johnston, G.C., and Singer, R.A. 1998. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem. 273: 21972–21979. [DOI] [PubMed] [Google Scholar]
- Cavalli G. and Paro, R. 1999. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 286: 955–958. [DOI] [PubMed] [Google Scholar]
- Celniker S.E., Sharma, S., Keelan, D.J., and Lewis, E.B. 1990. The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 9: 4277–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona D.F.V., Clapier, C.R., Becker, P.B., and Tamkun, J.W. 2002. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 31: 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R., Fanti, L., Armstrong, J.A., Sarte, M., Papoulas, O., Prestel, M., Daubresse, G., Verardo, M., Moseley, S.L., Berloco, M., et al. 2000. The ISWI chromatin-demodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5: 355–365. [DOI] [PubMed] [Google Scholar]
- Espinás M.L., Jiménez-Garcia, E., Vaquero, A., Canudas, S., Bernués, J., and Azorin, F. 1999. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J. Biol. Chem. 274: 16461–16469. [DOI] [PubMed] [Google Scholar]
- Evans D.R.H., Brewster, N.K., Xu, Q., Rowley, A., Altheim, B.A., Johnston, G.C., and Singer, R.A. 1998. The yeast protein complex containing Cdc68 and Pob3 mediates core-promoter repression through the Cdc68 N-terminal domain. Genetics 150: 1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas G., Gausz, J., Galloni, M., Reuter, G., Gyurkovics, H., and Karch, F. 1994. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature 371: 806–808. [DOI] [PubMed] [Google Scholar]
- Formosa T., Eriksson, P., Wittmeyer, J., Ginn, J., Yu, Y., and Stillman, D.J. 2001. Spt16–Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20: 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galant R. and Carroll, S.B. 2002. Evolution of a transcriptional repression domain in an insect Hox protein. Nature 415: 910–913. [DOI] [PubMed] [Google Scholar]
- Horabin J.I. and Schedl, P. 1993. Sex-lethal autoregulation requires multiple cis-acting elements upstream and down-stream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol. Cell. Biol. 13: 7734–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T., King, D.L., LaBonne, C., and Kafatos, F.C. 1993. A Drosophila single-strand DNA/RNA-binding factor contains a high-mobility-group box and is enriched in the nucleolus. Proc. Natl. Acad. Sci. 90: 6488–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S., Howe, L., Tafrov, S.T., Grant, P.A. Sternglanz, R., and Workman, J.L. 2001. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAFII30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)–FACT complex. Genes & Dev. 14: 1196–1208. [PMC free article] [PubMed] [Google Scholar]
- Kal A.J., Mahmoudi, T., Zak, N.B., and Verrijzer, C.P. 2000. The Drosophila Brahma complex is an essential coactivator for the trithorax group protein Zeste. Genes & Dev. 14: 1058–1071. [PMC free article] [PubMed] [Google Scholar]
- Katsani K.R., Hajibagheri, M.A.N., and Verrijzer, C.P. 1999. Cooperative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 18: 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.E., Stokes, D.G., and Perry, R.P. 1999. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma 108: 10–25. [DOI] [PubMed] [Google Scholar]
- Kennison J.A. 1995. The Polycomb and trithorax group proteins of Drosophila: Trans-regulators of homeotic gene function. Annu. Rev. Genetics 29: 289–303. [DOI] [PubMed] [Google Scholar]
- Kimura H. and Cook, P.R. 2001. Kinetics of core histones in living human cells: Little exchange of H3 and H4and some rapid exchange of H2B. J. Cell Biol. 153: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs J.E., Fry, C.J., Samuels, M.L., and Peterson, C.L. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102: 587–598. [DOI] [PubMed] [Google Scholar]
- Li F.-Q., Ueda, H., and Hirose, S. 1994. Mediators of activation of fushi tarazu gene transcription by BmFTZ-F1. Mol. Cell. Biol. 14: 3013–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D.L. and Zimm, G.G. 1992. The genome of Drosophila melanogaster, pp. 99, 759–760, 785. Academic Press, Inc., San Diego, CA.
- Liu Q.-X., Jindra, M., Ueda, H., Hiromi, Y., and Hirose, S. 2003. Drosophila MBF1 is a coactivator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development 130: 719–728. [DOI] [PubMed] [Google Scholar]
- McGinnis W. and Krumlauf, R. 1992. Homeobox genes and axial patterning. Cell 68: 283–302. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J., Fan, H.-Y., and Kingston, R.E. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487. [DOI] [PubMed] [Google Scholar]
- Okada M. and Hirose, S. 1998. Chromatin remodeling mediated by Drosophila GAGA factor and ISWI activates fushi tarazu gene transcription in vitro. Mol. Cell. Biol. 18: 2455–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuhara K., Ohta, K., Seo, H., Shioda, M., Yamada, T., Tanaka, Y., Dohmae, N., Seyama, Y., Shibata, T., and Murofushi, H. 1999. A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr. Biol. 9: 341–350. [DOI] [PubMed] [Google Scholar]
- Orlando V., Jane, E.P., Chinwalla, V., Harte, P.J., and Paro, R. 1998. Binding of Trithorax and Polycomb proteins to the bithorax complex: Dynamic changes during early Drosophila embryogenesis. EMBO J. 17: 5141–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., LeRoy, G., Chang, C.H., Luse, D.S., and Reinberg, D. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Wu, W.H., Lane, W.S., Hampsey, M., and Reinberg, D. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400: 284–288. [DOI] [PubMed] [Google Scholar]
- Papoulas O., Beek, S.J., Moseley, S.L., McCallum, C.M., Sarte, M., Shearn, A., and Tamkun, J.W. 1998. The Drosophila trithorax group proteins BRM, ASH1, and ASH2 are subunits of distinct protein complexes. Development 125: 3955–3966. [DOI] [PubMed] [Google Scholar]
- Petruk S., Sedkov, Y., Smith, S., Tillib, S., Kraevski, V., Nakamura, T., Canaani, E., Croce, C.M., and Mazo, A. 2001. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294: 1331–1335. [DOI] [PubMed] [Google Scholar]
- Poux S., Horard, B., Sigrist, C.J.A., and Pirrotta, V. 2002. The Drosophila Trithorax protein is a coactivator required to prevent reestablishment of Polycomb silencing. Development 129: 2483–2493. [DOI] [PubMed] [Google Scholar]
- Simic R., Lindstrom, D.L., Tran, H.G., Roinick, K.L., Costa, P.J., Johnson, A., Hartzog, G.A., and Arndt, K. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22: 1846–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R.H. and Felsenfeld, G. 1979. A new procedure for purifying histone pairs H2A + H2B and H3 + H4from chromatin using hydroxylapatite. Nucl. Acids. Res. 6: 689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeller W.C., Oh, C.E., and Kornberg, T.B. 1993. Isolation of cDNAs encoding the Drosophila GAGA transcription factor. Mol. Cell. Biol. 13: 7961–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A. 1989. Reconstitution of chromatin from purified components. Methods Enzymol. 170: 585–603. [DOI] [PubMed] [Google Scholar]
- Tie F., Furuyama, T., Prasad-Sinha, J., Jane, E., and Harte, P.J. 2001. The Drosophila Polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128: 275–286. [DOI] [PubMed] [Google Scholar]
- Topol J., Dearolf, C.R., Prakash, K., and Parker, C.S. 1991. Synthetic oligonucleotides recreate Drosophila fushi tarazu zebra-stripe expression. Genes & Dev. 5: 855–867. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T. and Wu, C. 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83: 1011–1020. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Becker, P.B., and Wu, C. 1994. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367: 525–533. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Daniel, C., Tamkun, J., and Wu, C. 1995. ISWI, a member of the SW12/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83: 1021–1026. [DOI] [PubMed] [Google Scholar]
- Ueda H., Sonoda, S., Brown, L., Scott, M.P., and Wu, C. 1990. A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes & Dev. 4: 624–635. [DOI] [PubMed] [Google Scholar]
- Vignali M., Hassan, A.H., Neely, K.E., and Workman, J.L. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20: 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Champion, L.E., Biessmann, H., and Mason, J.M. 1994. Mapping a mutator, mu2, which increases the frequency of terminal deletions in Drosophila melanogaster. Mol. Gen. Genet. 245: 598–607. [DOI] [PubMed] [Google Scholar]
- Wilkins R.C. and Lis, J.T. 1997. Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucl. Acids Res. 25: 3963–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P. and Brown, D.D. 1986. DNA replication in vitro erases a Xenopus 5S RNA gene transcription complex. Cell 47: 217–227. [DOI] [PubMed] [Google Scholar]
- Xiao H., Sandaltzopoulos, R., Wang, H.M., Hamiche, A., Ranallo, R., Lee, K.M., Fu, D., and Wu, C. 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8: 531–543. [DOI] [PubMed] [Google Scholar]