Abstract
Primary DNA damage sensing in mammalian global genome nucleotide excision repair (GG-NER) is performed by the xeroderma pigmentosum group C (XPC)/HR23B protein complex. HR23B and HR23A are human homologs of the yeast ubiquitin-domain repair factor RAD23, the function of which is unknown. Knockout mice revealed that mHR23A and mHR23B have a fully redundant role in NER, and a partially redundant function in embryonic development. Inactivation of both genes causes embryonic lethality, but appeared still compatible with cellular viability. Analysis of mHR23A/B double-mutant cells showed that HR23 proteins function in NER by governing XPC stability via partial protection against proteasomal degradation. Interestingly, NER-type DNA damage further stabilizes XPC and thereby enhances repair. These findings resolve the primary function of RAD23 in repair and reveal a novel DNA-damage-dependent regulation mechanism of DNA repair in eukaryotes, which may be part of a more global damage-response circuitry.
Keywords: mHR23A/B, XPC regulation, ubiquitin/proteasome pathway, DNA damage response, GG-NER
Multiple, complementary DNA repair systems have evolved to protect the genome against the detrimental effects of DNA lesions. Nucleotide excision repair (NER) primarily focuses on helix-distorting injuries, including UV-induced cyclobutane pyrimidine dimers (CPD) and pyrimidine (6–4) pyrimidone photoproducts (6–4PP), as well as numerous chemical DNA adducts (Friedberg et al. 1995). Inherited defects in NER are the cause of the cancer-prone syndrome xeroderma pigmentosum (XP). Patients are characterized by extreme sun sensitivity, sun-induced pigmentation anomalies, and a >2000-fold predisposition to UV-induced skin cancer. XP is a multigenetic disease, involving seven genes (XPA to XPG; Bootsma et al. 2001).
NER entails a multistep reaction and requires the coordinated action of ∼30 proteins implicated in damage detection, helix opening, lesion verification, dual incision of the damaged strand bracketing the injury, removal of the 25–30-base damage-containing oligonucleotide, gap-filling DNA synthesis, and ligation (Hoeijmakers 2001; Wood et al. 2001). Two NER subpathways exist: global genome NER (GG-NER), operating genome wide, and transcription-coupled repair (TCR), focusing on transcription-blocking lesions in the transcribed strand of active genes (Hanawalt 2000; Wood et al. 2000). Most XP genes are implicated in both NER subpathways, but XPC-deficient cells are unique in being selectively deficient in GG-NER (Venema et al. 1990).
Previously, we have cloned two human homologs of the Saccharomyces cerevisiae NER gene RAD23, designated hHR23A and hHR23B (Masutani et al. 1994) and found part of hHR23B to be tightly complexed with XPC and a recently identified protein, centrin 2 (CEN2; Araki et al. 2001). This heterotrimeric complex performs the crucial, damage-sensing step within GG-NER, triggering subsequent association of TFIIH, XPG, XPA, RPA, and ERCC1/XPF (Sugasawa et al. 1998; Yokoi et al. 2000; Volker et al. 2001). hHR23B stimulates the NER activity of XPC in vitro (Sugasawa et al. 1996) for which the 60-amino-acid XPC-binding domain alone is sufficient (Masutani et al. 1997). In whole-cell extracts, virtually all XPC is complexed with hHR23B (van der Spek et al. 1996a), whereas a trace amount copurifies with hHR23A (Araki et al. 2001). Only a minority of hHR23B and hHR23A is bound to XPC, suggesting that both proteins have additional functions (Sugasawa et al. 1996).
HR23 proteins contain a ubiquitin-like (UbL) N terminus and two ubiquitin-associated (UBA) domains (van der Spek et al. 1996b), suggesting a link to the ubiquitin/proteasome-mediated protein degradation pathway. The UbL domain of yeast RAD23 is important for UV survival and for interaction with the 26S proteasome (Watkins et al. 1993; Schauber et al. 1998), whereas the UBA domains enable binding to ubiquitin (Bertolaet et al. 2001; Chen et al. 2001; Wilkinson et al. 2001; Rao and Sastry 2002). However, the functional relationship between RAD23, NER, and the ubiquitin system is unclear. To investigate the role of HR23 in mammalian NER and other processes and the enigmatic NER–HR23–ubiquitin connection, we have generated mouse mutants for both HR23A and HR23B by gene targeting. Unlike other NER mouse mutants, mHR23B-deficient mice show a severe phenotype including impaired embryonic development and high rates of intrauterine death. Surviving animals display retarded growth, male sterility, and facial dysmorphology, but are NER-proficient (Ng et al. 2002). These data show that mHR23B is essential for normal development of the mouse and suggest an additional role of mHR23B outside NER. Apparently, this role is not or only partly compensated for by mHR23A, whereas HR23A is able to substitute for HR23B in NER. Here we report the generation and analysis of mHR23A–/– mice, the generation of mouse embryonic fibroblasts (MEFs) with a complete mHR23 deficiency, and the use of these double-mutant cells to elucidate the function of mammalian RAD23 homologs in NER.
Results
Generation of mHR23A-deficient mice and cells
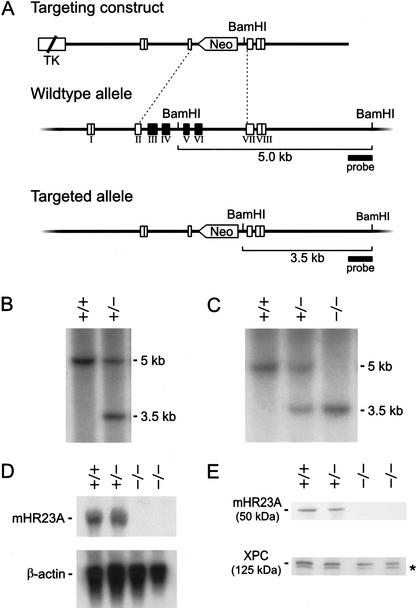
To generate a mouse model for mHR23A, we created a targeting construct in which the mHR23A exons III to VI, and part of exons II and VII (encoding residues 55–288 of the mHR23A protein), were replaced by the neomycin-resistance marker. Gene targeting creates an mHR23A allele encoding a severely truncated protein in which >85% of the coding sequence is deleted (even truncating the UbL domain) and thus can be considered a null allele (Fig. 1A). Two correctly targeted clones (obtained at a frequency of 16%; Fig. 1B) were used for blastocyst injections. Heterozygous offspring from matings between germ-line chimeric males and C57BL/6 female mice were intercrossed to generate homozygous mutant mHR23A animals (Fig. 1C), as well as embryonic day 13.5 (E13.5) embryos for isolation of MEFs. Neither the mHR23A mRNA nor the 50-kD mHR23A protein could be detected in mHR23A–/– MEFs (Fig. 1D,E). We conclude that we have generated mHR23A-null mice. The two independent mouse lines were biochemically and phenotypically indistinguishable for all parameters tested, indicating that the findings presented in this study are the result of genuine mHR23A inactivation.
Figure 1.
Targeted disruption of the mHR23A gene by homologous recombination. (A) Genomic organization and disruption strategy for mHR23A depicting the gene, the targeting construct, and the targeted mHR23A allele. Exons III to VI (and part of exons II and VII) were replaced by the dominant selectable neomycin-resistance marker transcribed in an antisense orientation. (B) Southern blot analysis of BamHI-digested DNA from ES cells showing the 5.0-kb and 3.5-kb fragments representing the wild-type and targeted allele of mHR23A, respectively. (C) Southern blot analysis of BamHI-digested tail DNA from mHR23A+/+, mHR23A+/–, and mHR23A–/– mice. (D, top) RNA blot analysis of mHR23A mRNA from mHR23A+/+, mHR23A+/–, and mHR23A–/– MEFs using full-length mHR23A cDNA as a probe. (Bottom) As a loading control for the amount of RNA, the blot was reprobed with β-actin cDNA. (E) Immunoblot analysis of mHR23A protein in cellular extracts from mHR23A+/+, mHR23A+/–, and mHR23A–/– MEFs loaded in equal amounts. Polyclonal antibodies against the human HR23A protein (top panel) and the human XPC protein (bottom panel) were used. The asterisk indicates an aspecific cross-reacting band.
mHR23A–/– animals and MEFs are NER proficient
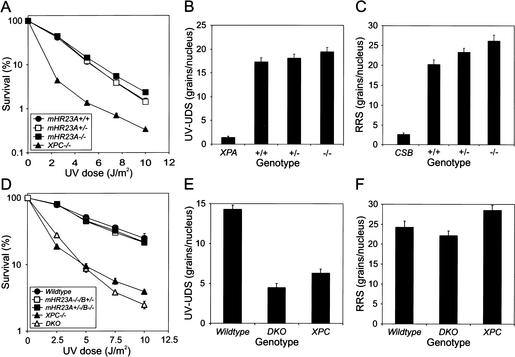
Because of the expected role of both mHR23 variants (A and B) in NER as suggested by in vitro experiments (Masutani et al. 1994) we assessed key repair parameters in mHR23A–/– MEFs. As shown in Figure 2A–C, UV survival, UV-induced unscheduled DNA synthesis (UDS), and RNA synthesis recovery after UV exposure were all in the wild-type range, indicating that global as well as transcription-coupled NER are unaffected, mimicking the situation in the mHR23B mutant (Ng et al. 2002). These data suggest that mHR23A and mHR23B are functionally redundant for NER in vivo, extending our in vitro observations (Sugasawa et al. 1997).
Figure 2.
Repair characteristics of mHR23A–/– E13.5 and DKO E8.5 MEFs. (A) UV survival curves of primary mHR23A+/+, mHR23A+/–, and mHR23A–/– E13.5 MEFs. XPC–/– fibroblasts were included as a negative control. Cells were exposed to different doses of UV (254 nm). After 4–5 d, the number of proliferating cells was estimated from the amount of radioactivity incorporated during a 3-h pulse with [3H]thymidine. For each genotype, identical results were obtained with three other cell lines (data not shown). (B) Global genome repair (UDS) in primary mHR23A+/+, mHR23A+/–, and mHR23A–/– E13.5 MEFs. Cells were irradiated with 16 J/m2 of UV (254 nm) and labeled with [3H]thymidine. Incorporation of radioactivity was measured by autoradiography and grain counting (average of 50 nuclei per cell line; the standard error of the mean is indicated). XPA–/– fibroblasts were measured as a negative control. For each genotype, consistent results were obtained with three other independent cell lines (data not shown). (C) RNA synthesis recovery (RRS) after UV exposure of primary mHR23A+/+, mHR23A+/–, and mHR23A–/– E13.5 MEFs. Cells were UV-irradiated (10 J/m2, 254 nm) and allowed to recover for 16 h. After a 1-h pulse labeling with [3H]uridine, cells were processed for autoradiography. The relative rate of RNA synthesis was expressed as the quotient of the number of autoradiographic grains over the UV-exposed nuclei and the number of grains over the nuclei of unirradiated cells (average of 50 nuclei per cell line; the standard error of the mean is indicated). CSB–/– cells were used as a negative control. For each genotype, three other independent lines were assayed with similar outcomes (data not shown). (D) UV survival of E8.5 MEF lines of wild-type, XPC–/–, mHR23A–/–/B+/–, mHR23A+/–/B–/–, and mHR23A–/–/B–/– (DKO). (E) UV-induced UDS in wild-type, XPC–/–, and DKO E8.5 MEFs. (F) RNA synthesis recovery after UV irradiation of wild-type, XPC–/–, and DKO E8.5 MEFs. For details for panels D–F, see legends to panels A–C, respectively, and Materials and Methods. Two independent experiments using two other DKO cell lines (before the cultures extinguished; data not shown) showed a similar effect on UDS and RNA synthesis recovery.
In striking contrast to mHR23B–/– animals, mHR23A–/– mice were born with Mendelian frequency and appeared indistinguishable from wild-type and heterozygous littermates for all parameters tested (including morphology, main pathology, and growth rate up to at least 18 mo). mHR23A–/– male and female mice were fertile, and their mating activity and litter size were normal. Apparently, mHR23A is not essential for mouse development, and mHR23B can compensate for any additional functions of mHR23A.
Total mHR23 deficiency is incompatible with animallife
To investigate the effect of a total mHR23 deficiency, we tried to generate mHR23A–/–/B–/– animals (hereafter referred to as: DKO for “double knockout”) and to obtain corresponding MEFs, by double heterozygous matings. Remarkably, out of 427 newborns analyzed, no DKOs were found (Table 1). This indicates that inactivation of mHR23A aggravates the severe developmental defects caused by a mHR23B deficiency (Ng et al. 2002) to a level incompatible with life. Whereas we obtained phenotypically normal mHR23A–/–/B+/– mutant mice at Mendelian ratios (71/427 found and 83/427 expected), surprisingly, mHR23A+/–/B–/– animals were not born (0/427). However, we were able to isolate E13.5 mHR23A+/–/B–/– mutant MEFs, although they showed poor growth. Apparently, loss of even one allele of mHR23A in a completely mHR23B-null background causes lethality in embryogenesis.
Table 1.
Genotype analysis of DKO (mHR23A-/-/B-/-) embryos and offspring
| Stage | Analyzed | Expecteda (if Mendelian) | Found |
|---|---|---|---|
| E8.5 | 43 | 7 | 3b |
| E10.5 | 14 | 1.8 | 0 |
| E13.5 | 77 | 9.1 | 0 |
| Newborn | 427 | 41.4 | 0 |
Derived from different mHR23A+/-/B+/- and mHR23A-/-/B+/- intercrosses.
One cell line established.
To investigate embryonic lethality caused by a complete mHR23 deficiency, we isolated embryos at various stages of development. No DKO embryos were present at E13.5 and E10.5, but growth-retarded mHR23-deficient embryos were observed at E8.5. Importantly, three DKO MEF lines were isolated from E8.5 embryos (3/43; see Table 1). Compared with wild-type and double-heterozygous mutant MEFs, these cells displayed reduced rates of proliferation, which resulted in the loss of two lines. Nevertheless, we succeeded in establishing one DKO cell line after 30 wk of culturing, which permitted functional characterization of a total mHR23 deficiency.
Totally mHR23-deficient cells show an XPC-like repair phenotype
Cell survival experiments revealed that DKO MEFs are remarkably similar to the unique NER phenotype of XPC–/– cells in terms of UV survival (Fig. 2D), deficiency of UV-induced UDS, and proficiency of RNA synthesis recovery after UV exposure (Fig. 2E,F). In contrast, MEFs retaining only one mHR23A or mHR23B allele were NER competent (Fig. 2D). Apparently, one out of four mHR23 copies is sufficient for normal NER activity.
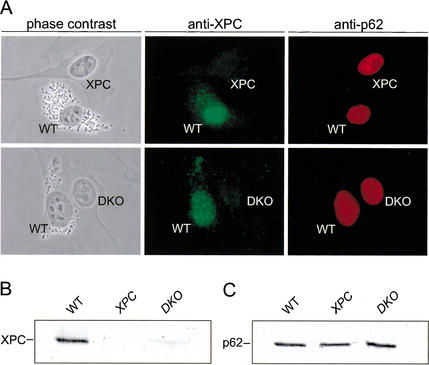
The resemblance to the XPC–/– repair phenotype prompted us to examine the status of the XPC protein in the DKO MEFs. Interestingly, steady-state levels of XPC appeared strongly reduced in DKO MEFs compared with wild-type and mHR23A–/– (Fig. 1E) and mHR23B–/– single KO cells (data not shown), as shown by comparative immunofluorescence (Fig. 3A) and immunoblot analysis of whole-cell extracts (Fig. 3B). Thus, in the absence of both mouse RAD23 proteins, XPC is either hardly expressed at RNA or protein level, or unstable. In view of the direct physical interaction between HR23 proteins and XPC, the latter option seems most plausible.
Figure 3.
XPC expression in DKO E8.5 MEFs. (A) Phase contrast (left panels) and epifluorescence (middle and right panels) images of fixed wild-type (WT, labeled with latex beads), XPC–/– (XPC), and mHR23A–/–/B–/– (DKO) MEFs. Cells were fixed by paraformaldehyde, permeabilized by 0.1% Triton X-100, and subsequently immunolabeled with affinity-purified polyclonal antibodies against the human XPC protein (middle panels; stained green with goat anti-rabbit Alexa 488-labeled secondary antibody). Monoclonal antibodies recognizing the p62 subunit of TFIIH (right panels; stained red with goat antimouse Cy3-labeled secondary antibody) were used as an internal control. All images were taken at the same magnification. (B,C) Immunoblot analysis of XPC protein in cellular extracts from wild-type, XPC–/–, and DKO E8.5 MEFs using polyclonal anti-human XPC antibodies (B). (C) Monoclonal anti-p62 antibodies were used as an internal reference for the amount of protein in each lane.
hHR23B and hXPC-GFP rescue the UV sensitivity of DKO cells
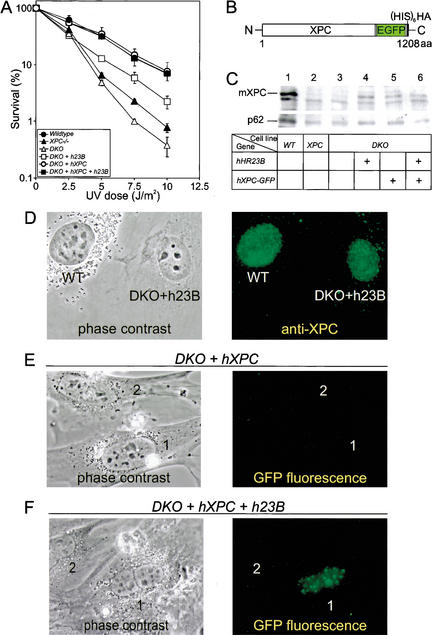
To provide direct evidence that the XPC-like phenotype of DKO cells is specifically caused by the mHR23 defect, we stably transfected human hHR23B cDNA into DKO MEFs. The UV sensitivity of DKO cells expressing hHR23B was only partly rescued, perhaps because of human–mouse differences (Fig. 4A). Importantly, expression of hHR23B induced an increase in the total amount of endogenous mouse mXPC, as shown by both immunoblot (Fig. 4C, lane 4) and immunofluorescence analysis (Fig. 4D).
Figure 4.
Characterization of DKO cells expressing hHR23B and XPC-GFP. (A) UV survival of wild-type, XPC–/–, DKO, and DKO MEFs cotransfected with hHR23B (h23B), human XPC-GFP (hXPC), and h23B and hXPC-GFP cDNAs. Cells were exposed to different doses of UV (254 nm). After 4–5 d, the number of proliferating cells was estimated from the amount of radioactivity incorporated during a 3-h pulse with [3H]thymidine. For details, see Materials and Methods. For each cDNA construct, similar results were obtained with at least two other independent stably transfected cell lines (data not shown). (B) Schematic representation of XPC-EGFP–His6HA-N3 fusion protein (1208 amino acids). Indicated are the human XPC protein (940 amino acids), the enhanced green fluorescent protein tag (EGFP; 238 amino acids), and the hexameric histidine-hemagglutinin double-epitope tag (His6HA; 17 amino acids). (C, top) Immunoblot analysis of XPC expression in cellular extracts of wild-type (WT; lane 1), XPC (lane 2), DKO (lane 3), and DKO MEFs cotransfected with h23B (lane 4), hXPC-GFP (lane 5), and h23B and hXPC-GFP (lane 6) cDNAs, using a polyclonal antibody against the C terminus of human XPC. (Bottom) Monoclonal anti-p62 antibodies were used as a loading control. (D) Phase contrast (left) and epifluorescence (right) images of fixed WT (labeled with latex beads) and DKO cells cotransfected with hHR23B cDNA. Cells were fixed by paraformaldehyde, followed by 0.1% Triton X-100 permeabilization, and subsequently immunolabeled with affinity-purified polyclonal anti-human XPC (right; stained green with goat anti-rabbit Alexa 488-labeled secondary antibody). Monoclonal anti-p62 antiserum was used as an internal control (stained red with goat anti-mouse Cy3-labeled secondary antibody; data not shown). Images were taken at the same magnification. Similar results were obtained with DKO cells cotransfected with hXPC-GFP, and hHR23B and hXPC-GFP cDNAs (data not shown). (E,F) Phase contrast (left) and epifluorescence (right) images of living DKO cells cotransfected with hXPC-GFP (E) or hHR23B and hXPC-GFP (F) cDNAs. All images were taken at the same magnification.
Because the absence of mHR23 causes a strong reduction in mXPC, we reasoned that (over)expression of “exogenous” XPC might bypass the repair defect of DKO cells. Therefore, we generated double-mutant MEFs that stably express human hXPC tagged with GFP (and additional His6 and HA tags; Fig. 4B), to allow direct observation in living cells (Houtsmuller et al. 1999). Functionality of the hXPC-GFP was demonstrated after microinjection and transfection of the cDNA construct in XPC-deficient cells (data not shown). Although hXPC-GFP was undetectable by fluorescence microscopy (Fig. 4E), stable transformants (verified for the presence of hXPC-GFP cDNA by DNA blotting) had largely regained wild-type UV resistance (Fig. 4A), indicating that the repair defect was rescued. Unfortunately, our XPC antibodies (raised against the C terminus of the protein) failed to recognize the hXPC-GFP fusion protein because of interference by the C-terminal tag. However, introduction of hXPC-GFP appeared to restore endogenous mXPC levels as shown by immunoblot (Fig. 4C, lane 5) and immunofluorescence analysis (data not shown). Apparently, hXPC-GFP has a trans-effect on mXPC stability.
To investigate the stabilizing effect of mHR23B on XPC, we cotransfected hHR23B with hXPC-GFP cDNA into DKO cells. Stably transfected clones exhibited wild-type UV resistance (Fig. 4A) and normalized levels of endogenous mXPC (Fig. 4C, lane 6; data not shown). In contrast to MEFs expressing only hXPC-GFP, a significant fraction (>40%) of the double cotransfected cells displayed green fluorescent nuclei (Fig. 4F), which, however, decreased to <10% upon culturing. This is probably caused by a level of hXPC-GFP expression still below the detection limit in most cells, because immunofluorescence using anti-HA monoclonals revealed that most cells expressed the tagged transgene (data not shown). Cells with green nuclei also showed bright foci, corresponding with sites containing a local high DNA concentration visible after DAPI staining (data not shown). These data indicate that the cotransfected hHR23B acts as a stabilizing factor for both hXPC-GFP and endogenous mXPC.
DNA damage causes accumulation of hXPC-GFP
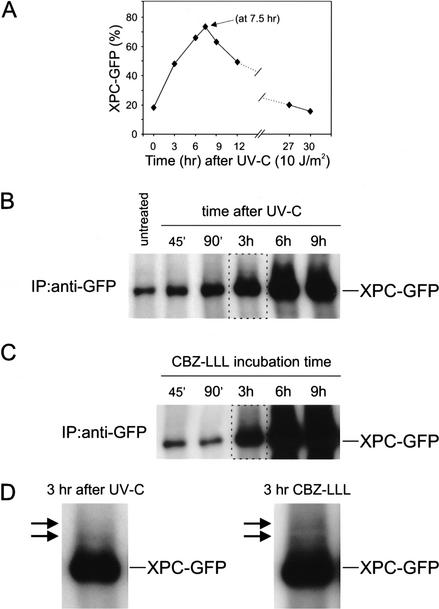
The hXPC-GFP/hHR23B DKO cell line provided a convenient tool to monitor the effect of DNA damage on XPC levels and mobility in living cells. Interestingly, UV irradiation (5, 10, and 15 J/m2) strongly increased the percentage of green cells and the intensity of the GFP signal. Kinetic analysis of UV exposure revealed a time-dependent reversible accumulation of XPC-GFP in the majority of the cells (Fig. 5A). In addition, immunoprecipitation studies using an anti-GFP antiserum showed a significant increase in time after UV irradiation of the amount of precipitated fusion protein (Fig. 5B). Monitoring individual cells in time after UV irradiation corroborated these findings (Fig. 6A). Because this phenomenon was specific for DKO cells transfected with hXPC-GFP/hHR23B (data not shown), these results indicate that XPC levels are responsive to UV in an HR23-dependent fashion.
Figure 5.
Analyses of XPC-GFP expression after UV irradiation and treatment with proteasome inhibitor CBZ-LLL. (A) Kinetic analysis of living DKO cells expressing XPC-GFP/hHR23B after exposure to 10 J/m2 of UV-C in time over a period of 30 h. Percentage XPC-GFP, the percentage of GFP-expressing fluorescent cells of the total number of cells. (B) Immunoprecipitation study of UV-irradiated DKO cells expressing XPC-GFP/hHR23B in time. XPC-GFP was immunoprecipitated (IP) from WCEs with monoclonal anti-GFP antibodies. The precipitates were analyzed by immunoblotting using polyclonal anti-XPC antiserum. Untreated WCE, and extracts from cells isolated at 45 min, 90 min, 3 h, 6 h, and 9 h after exposure to 10 J/m2 of UV-C. The amount of expressed XPC-GFP was also visualized by immunoblot analysis of the total cell lysate with anti-XPC or anti-GFP antibodies, and with a monoclonal antibody against the p62 subunit of TFIIH as a loading control (data not shown). XPC-GFP expression was also dose-dependent (4, 8, 12, and 16 J/m2 of UV-C; data not shown). (C) Immunoprecipitation analysis of DKO cells expressing XPC-GFP/hHR23B after CBZ-LLL treatment. IPs of WCEs from cells treated with 10 μM CBZ-LLL for 45 min, 90 min, 3 h, 6 h, and 9 h, were performed as in B. XPC-GFP expression was confirmed by immunoblot analysis of the total cell lysate using anti-XPC or anti-GFP antibodies, and anti-p62 as a loading control (data not shown). (D) Large magnification of the higher migrating XPC species (arrows) detected in the 3-h samples after UV (left, see panel B) and CBZ-LLL treatment (right, see panel C).
Figure 6.
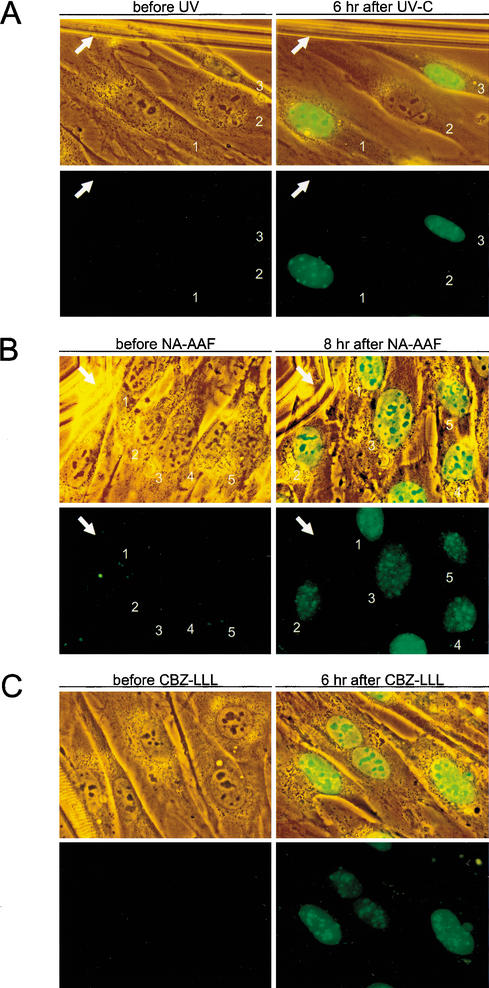
Effect of UV, NA-AAF, and CBZ-LLL on hHR23B-dependent XPC-GFP levels in living DKO cells. (A) Combined phase contrast (transmission light) and fluorescence (green) images (top panels), and epifluorescence images (bottom panels) of the same living DKO cells expressing XPC-GFP/hHR23B before UV (left panels) and 6 h after 10 J/m2 of UV-C (right panels). White arrows indicate the scratch mark on glass coverslips for proper orientation. Numbers represent the same living cells before and after UV exposure. Identical results were obtained with two other independent DKO cell lines expressing XPC-GFP/hHR23B (data not shown). All images were taken at the same magnification. (B) Combined phase contrast (transmission light) and fluorescence (green) images (top panels), and epifluorescence images (bottom panels) of living DKO cells expressing XPC-GFP/hHR23B before NA-AAF (left panels) and 8 h after 50 μM NA-AAF (right panels). White arrows indicate the scratch on glass coverslips for proper comparison. The numbers represent the corresponding living cells on coverslips before and after NA-AAF treatment. Identical results were obtained with two other independent DKO cell lines expressing XPC-GFP/hHR23B (data not shown). All images were taken at the same magnification. (C) Combined phase contrast (transmission light) and fluorescence (green) images (top panels), and only epifluorescence images (bottom panels) of living DKO cells expressing XPC-GFP/hHR23B before treatment with proteasome inhibitor CBZ-LLL (left panels) and 6 h after 10 μM CBZ-LLL (right panels). All images were taken at the same magnification.
To investigate whether XPC accumulation is specific for NER-type DNA damage or just stress-related, cells were exposed to different kinds of genotoxic agents. N-acetoxy-2-acetylaminofluorene (NA-AAF, 50 and 100 μM), which induces bulky adducts processed by NER, elicited a very potent UV-like response in all cells within 6–8 h (Fig. 6B). In contrast, γ-rays (6 and 10 Gy) and mitomycin C (MMC, 1.2 and 2.4 μg/mL), inducing mainly strand breaks and interstrand cross-links, respectively (which are dealt with by other repair pathways), failed to provoke detectable XPC accumulation (data not shown). Also, heat shock (41°C, analyzed for up to 12 h) failed to boost fluorescence. The possibility that UV and NA-AAF evoke a general accumulation of protein was ruled out because cells expressing GFP alone do not exhibit a significant increase in fluorescence after genotoxic insults. This indicates that lesions specifically recognized by the NER pathway enhance the level of hXPC-GFP in an HR23-dependent manner.
One of the direct consequences of UV- and NA-AAF-induced DNA damage is a temporary block of transcription. To investigate whether hXPC-GFP accumulation requires transcription or is induced by a DNA-damage-independent blockage of transcription, mRNA synthesis in DKO cells expressing hXPC-GFP/hHR23B was reversibly arrested by incubation with 5,6-dichloro-1β-D-ribofuranosyl-benzimidazole (DRB, 100 μM) and α-amanitin (10 μg/mL). No induction of XPC-GFP fluorescence was observed: instead, preincubation with DRB (2–3 h) prior to UV treatment prevented UV-induced XPC-GFP accumulation (data not shown). Consistent with this result, no enhanced XPC fluorescence was found in cells treated with the translational inhibitor cycloheximide (30 and 50 μg/mL), demonstrating the requirement for de novo RNA and protein synthesis. However, hHR23B-mediated XPC-GFP accumulation is strictly dependent on the presence of DNA damage, because in unchallenged conditions the steady-state level of XPC remains low.
Regulation of hXPC-GFP levels involves proteasome-dependent proteolysis
To further examine the HR23-dependent XPC stabilization, DKO cells expressing hXPC-GFP/hHR23B were incubated with the proteasomal proteolysis inhibitor N-CBZ-LEU-LEU-LEU-AL (CBZ-LLL, 5 and 10 μM; Wiertz et al. 1996). Similar to UV irradiation and NA-AAF, all cells displayed a striking XPC-GFP accumulation in time (Fig. 6C), which was reversible upon drug removal (data not shown). Longer exposure to CBZ-LLL further increased XPC-GFP levels, as demonstrated by immunoblot analysis (Fig. 5C). These findings indicate that degradation of XPC-GFP occurs via ubiquitin/proteasome-dependent proteolysis and suggest that the NER-damage-specific accumulation of XPC-GFP is due to inhibition of degradation in combination with de novo mRNA and protein synthesis.
Ubiquitination is an important step in targeting proteins to the 26S proteasome for proteolysis (Pickart 2002), and Saccharomyces cerevisiae RAD23 was found to be involved in translocating ubiquitinated RAD4 (the yeast ortholog of XPC) to the proteasome (Lommel et al. 2002). Therefore, we examined whether XPC is also subject to (poly)ubiquitination. Immunoprecipitation of XPC-GFP (using anti-GFP) and subsequent immunoblot analysis with XPC antibodies from cells exposed to UV-C light or CBZ-LLL revealed a clear time-dependent accumulation of XPC-GFP (Fig. 5B,C). Interestingly, in addition to the expected XPC-GFP protein band, a number of slower-migrating XPC species were identified (Fig. 5D), consistent with the idea that they are (poly)ubiquitination products of XPC-GFP. Unfortunately, none of the different anti-ubiquitin anti-sera used was sensitive enough to detect those species above background, precluding unequivocal assessment of the identity of these XPC modifications.
Application of local UV damage to hXPC-GFP-expressing cells
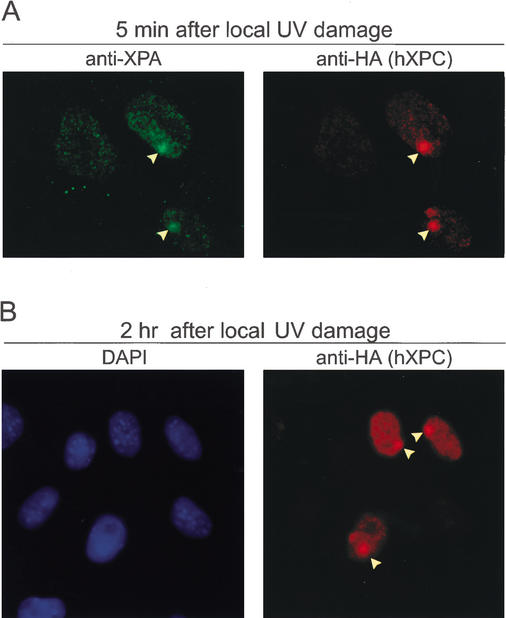
Two mechanisms may explain the transient stabilization of hXPC-GFP. The binding of XPC to DNA damage per se might protect it from proteolysis. Alternatively or in addition, DNA damage may trigger a specific response such as XPC modification that inhibits degradation. To explore the mechanism by which hXPC-GFP is stabilized, we used a recently developed method for induction of DNA damage in a restricted part of the nucleus. For this purpose, a monolayer of DKO cells expressing hXPC-GFP/hHR23B was covered with a UV-light-shielding isopore polycarbonate filter (pore diameter ∼5 μm). Upon UV irradiation, only at the position of pores is UV damage induced, as detected with antibodies that specifically recognize CPD and 6–4PP lesions. These locations attract all NER proteins tested thus far, including XPC (Volker et al. 2001; W. Vermeulen, unpubl.). Cells were fixed at different time points after UV to allow simultaneous immunostaining with antibodies against various proteins and GFP fluorescence microscopy (Fig. 7). Nonirradiated nuclei and undamaged regions within partly irradiated nuclei serve as internal controls. Very rapidly (<2 min) after UV exposure, GFP fluorescence (data not shown) and anti-HA immunostaining revealed high local accrual of hXPC-GFP(His6HA) in part of the nuclei, which colocalized with XPA (Fig. 7A) and the p62 subunit of TFIIH (data not shown). These findings demonstrate that in living cells, the GFP-tagged XPC protein translocates very rapidly to sites containing UV lesions. If XPC stabilization only occurs when bound to the damage, we expect an increase in fluorescent signal predominantly at the damaged sites. On the other hand, with an (additional) overall stabilization of hXPC, it is expected that in time a concomitant increase of fluorescence over the entire nucleus (in addition to the damaged area) would be observed in comparison to undamaged nuclei. The increase of hXPC-GFP (Fig. 7A) initially occurs only at the locally damaged sites, but after 2 h also in the remainder of locally damaged nuclei, a clearly higher signal is noted when compared to unexposed nuclei in the vicinity (Fig. 7B). These findings suggest an overall intranuclear stabilization of hXPC-GFP triggered by binding to lesions.
Figure 7.
Local UV damage induces overall XPC stabilization in nuclei of DKO cells expressing XPC-GFP/hHR23B. (A,B) DKO cells expressing XPC-GFP/hHR23B were exposed to 64 J/m2 of UV-C through 5.0-μm pore filters and fixed 5 min (A) and 2 h (B) later with paraformaldehyde. Double immunofluorescent labeling using antibodies against XPA (A, left panel; stained green with goat anti-rabbit Alexa 488-labeled secondary antibody) and HA epitope (A, right panel; stained red with goat anti-rat Alexa 594-labeled secondary antibody); DAPI stained (B, left panel) and HA antibody labeling (B, right panel). Arrowheads indicate the site of UV-induced local damage in the nuclei of DKO cells expressing hXPC-GFP/hHR23B. Note: Compare the increased fluorescence signal over the entire nucleus of damaged cells to the signal of undamaged nuclei for overall stabilization of XPC (B).
High levels of XPC mediate a transient enhancement of DNA repair
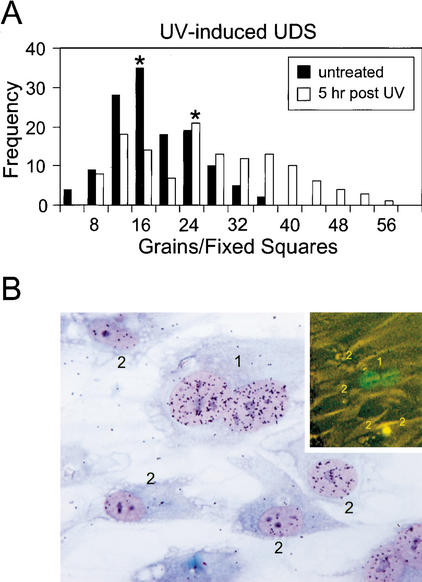
To investigate the biological significance of DNA-damage-induced stabilization of XPC, we tested the DNA repair capacity (UV-induced UDS) in DKO cells expressing XPC-GFP/hHR23B prechallenged with UV light. The mean UDS level (as determined by 1 h of 3H-thymidine pulse-labeling immediately after a dose of 16 J/m2) 5 h after UV irradiation (10 J/m2) was increased 1.5-fold compared with cells assayed in parallel that were not preirradiated (Fig. 8A). UV-induced XPC-GFP accumulation was confirmed microscopically (data not shown) just prior to the UDS assay. The increase in UDS is not derived from the additional effect of NER still dealing with lesions remaining from the first UV dose, because in a separate UDS experiment without the second UV irradiation, no significant UDS was observed (data not shown). The 1.5-fold increase in UDS is an underestimation because not all cells respond to the UV challenge (see also Figs. 5A, 6A). When corrected for the nonresponding cell fraction, UDS levels were more than two-fold enhanced. These data suggest that UV-induced accumulation of XPC-GFP caused a concomitant increase in GG-NER. Enhanced repair by increased levels of XPC was confirmed by microinjection of XPC-GFP cDNA into homopolykaryons of wild-type human fibroblasts. Microinjected cells expressing XPC-GFP (Fig. 8B, top right panel) exhibit a higher UDS compared with neighboring, noninjected monokaryons (Fig. 8B). Interestingly, when a cocktail of XPC-GFP and hHR23B cDNA was injected, UDS in the majority of the cells was significantly lower and injection of this cocktail appeared highly toxic (data not shown). These data indicate that large amounts of stabilized XPC (as a result of overexpressed hHR23B) may reduce cell viability and suggest that the XPC level is under a delicate control.
Figure 8.
Enhanced DNA repair correlates with high levels of XPC in UV-induced UDS in DKO cells expressing XPC-GFP/hHR23B. (A) Histogram of UV-induced UDS in DKO cells expressing XPC-GFP/hHR23B. Five hours after exposure to 10 J/m2 of UV-C, cells were subsequently irradiated with 16 J/m2 of UV-C and labeled with [3H]thymidine for 1 h (white columns, mean of UDS level is 25 ± S.E.M. 1). In parallel, nonprechallenged cells only exposed to 16 J/m2 of UV-C were used as controls (black columns, mean of UDS level is 16 ± S.E.M. 0.6). Asterisks indicate the mean values of the UDS levels. Incorporation of radioactivity was measured by autoradiography and grain counting (130 fixed squares counted per cell line; each square represented ∼50% of the nucleus surface). UV-induced UDS of wild-type (mean 17 ± S.E.M. 0.8) fibroblasts were measured as a control (data not shown). (B) Effect of microinjection of XPC-GFP cDNA on UV-induced UDS in human wild-type (C5RO) fibroblasts. Shown is a micrograph of a wild-type homodikaryon (numbered 1) microinjected with XPC-GFP in one of the nuclei and subjected to UV-induced UDS. Prior to UDS, fluorescence images were captured (inset in B). The injected cell has a considerably larger number of grains above its nuclei than the noninjected, surrounding mononuclear cells (numbered 2).
Discussion
Involvement and function of HR23A and HR23B proteins in NER
The aim of this work was to shed light on the enigmatic role of the two mammalian RAD23 orthologs (HR23A and HR23B) in NER and to reveal their biological impact by the generation of mHR23A and mHR23B mouse mutants. Because single mutants failed to exhibit any detectable NER defect, the most logical interpretation is that for NER, these proteins are functionally redundant despite their >40% amino acid sequence divergence and size difference (Masutani et al. 1994). Indeed, double-mutant cells, which we were able to establish, notwithstanding early embryonic lethality and poor growth properties during adaptation to in vitro culturing, exhibited an NER-deficient phenotype. The ability of each of the HR23 genes to rescue this defect unequivocally established their involvement and fully overlapping function in NER in vivo. DKO cells carried a selective impairment of the GG-NER subpathway and apparently normal TC-NER, as deduced from the fact that recovery of RNA synthesis after UV irradiation is unaffected. This is strikingly similar to the repair phenotype of XPC, which hitherto was unique among the NER mutants, but deviates from the S. cerevisiae RAD23 prototype mutant and its Schizosaccharomyces pombe equivalent, which carry a combined GG-NER and TC-NER deficiency (Verhage et al. 1996; Lombaerts et al. 2000). This unresolved mammalian–yeast difference is also registered for XPC (Venema et al. 1990) and its yeast counterpart RAD4, underlining the parallels between HR23 and XPC.
HR23 proteins controlproteasome-mediated degradation of XPC
The remarkable correspondence with XPC mutants prompted us to examine the XPC status in DKO cells. The virtual absence of XPC in immunoblots and immunofluorescence (Fig. 3) directly explained the HR23 phenocopy of XPC: in the absence of HR23, the XPC protein is apparently unstable. This function of HR23 reveals a novel mechanism of NER regulation. In view of the involvement of the proteasome in XPC degradation indicated by the CBZ-LLL treatment, these observations put a functional perspective to the growing number of links emerging between RAD23/HR23 and the ubiquitin system (see above). Although our anti-ubiquitin antibodies were not sensitive enough to directly demonstrate ubiquitination of XPC, we were able to detect a minor XPC fraction migrating as discrete higher-molecular-weight species consistent with a set of (poly)ubiquitinated XPC modifications. This suggests that the steady-state levels of this type of intermediate are very low and that XPC targeted for degradation is rapidly processed.
The results derived from the CBZ-LLL experiments are also relevant in relation to controversies on the functional implications of the RAD23/NER-19S/26S proteasome connection. In the absence of proteasome function, yeast conditional 19S proteasome mutants were reported to display enhanced repair, suggesting that proteins involved in (regulation of) NER are degraded by the 26S proteasome (Lommel et al. 2000). Consistent with this idea, evidence was reported for RAD4 ubiquitination and degradation via the 26S proteasome, involving RAD23 (Lommel et al. 2002), although the specific function in DNA repair remained unresolved. In addition, in vitro studies using GST-ΔUbL rad23 (RAD23 lacking the UbL domain) indicate that the UbL domain targets (multiubiquitinated) protein substrates for proteolysis (Ortolan et al. 2000). On the other hand, overexpression of tagged wild-type RAD23 caused stabilization of artificial substrates in vivo by preventing multiubiquitination (Chen and Madura 2002). Physiological levels of RAD23 were speculated to facilitate substrate degradation, but might also transiently stabilize (Chen and Madura 2002). In contrast, other investigators (Russell et al. 1999; Gillette et al. 2001) showed that the 19S regulatory complex represses NER independently of RAD23 and apparently independently of proteolysis. Finally, interactions between HR23B and the 26S proteasome were also observed in human cells (Hiyama et al. 1999). Our studies of the HR23A/B double mutant clearly demonstrate that XPC is intrinsically unstable and is degraded by the 26S proteasome and that HR23 proteins partly stabilize XPC in vivo. Consistent with this finding, we were able to alleviate the NER defect caused by HR23 deficiency, by forced overexpression of exogenous hXPC compensating for the short half-life of endogenous XPC in the absence of HR23. The severity of the NER defect in DKO cells correlating with the low cellular XPC content suggests that XPC stabilization is the main in vivo NER function of HR23 proteins.
Effect of DNA damage on XPC/HR23
Interestingly, both UV and NA-AAF induced a dramatic accumulation of XPC-GFP (Fig. 6A,B), causing strong nuclear fluorescence within a few hours. This phenomenon clearly depends on (helix-distorting) lesions that are substrates for GG-NER and is not due to a general stress-related response or to any type of DNA damage. XPC-GFP induction occurred with grossly similar kinetics when proteasome-mediated proteolysis is inhibited, providing further evidence for the idea that protein stabilization is responsible for the XPC-GFP increase. Interestingly, very recently a twofold increase in the interaction of RAD23 with ubiquitinated proteins was observed upon treatment with the genotoxin 4-NQO (Chen and Madura 2002).
Infliction of local DNA damage (Fig. 7) revealed a very rapid (within minutes) accumulation of XPC-GFP in the damaged regions, which is found for all NER factors and is caused by actual engagement in DNA repair events (Volker et al. 2001; Hoogstraten et al. 2002; W. Vermeulen, unpubl.). However, the additional enhancement of XPC-GFP over the entire nucleus (2 h postirradiation) supports the argument against the possibility that only lesion-bound molecules are protected from proteolysis. It is possible that participation of XPC/HR23 in NER events triggers its stabilization and that the subsequent dissociation of XPC/HR23 from the damage (when repair is accomplished) provides a pool of modified/stabilized protein that distributes over the entire nucleus. In view of the fact that XPC is involved in the initial lesionsensing step, the DKO cells stably expressing functional GFP-tagged XPC allow direct visualization of the effect of DNA damage in living cells.
A novel regulation mechanism of GG-NER
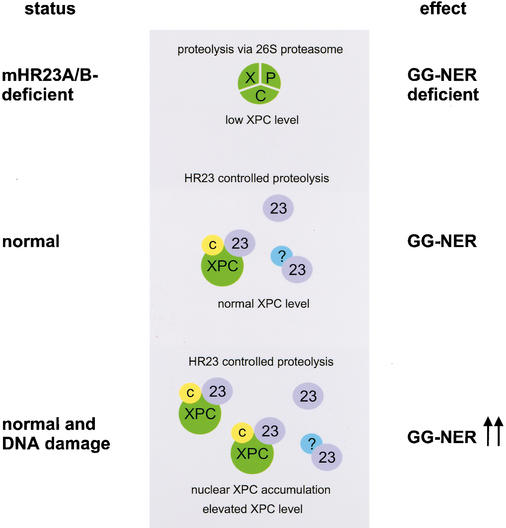
The level of XPC appears to modulate GG-NER. In the absence of HR23, XPC concentrations are strongly reduced, and as a consequence GG-NER is severely affected. Conversely, elevated amounts of XPC, both by UV irradiation of DKO cells expressing XPC-GFP/hHR23B and by microinjection of XPC-GFP in control cells, augmented the repair capacity by 1.5- to 2-fold (Fig. 8). These findings are in line with the observations made in yeast, where overexpression of RAD4 confers accelerated rates of NER (Lommel et al. 2000). The maximal twofold increase is consistent with our previous in vivo findings that under uninduced conditions, maximal NER capacity is associated with damage-dependent immobilization of 40%–45% of the ERCC1/XPF (Houtsmuller et al. 1999) and TFIIH (Hoogstraten et al. 2002) NER complexes. Thus, these factors become limiting when NER stimulation exceeds approximately twofold. A unifying model for all our findings on HR23, XPC, and proteolysis is depicted in Figure 9. As the main initiator of GG-NER (Sugasawa et al. 1998), XPC constitutes an ideal focal point for the regulation of the entire pathway, which involves HR23. Absence of HR23 proteins reveals that XPC on its own is highly unstable because of proteolysis via the 26S proteasome. Under normal conditions, HR23 complex formation with XPC results in a significant reduction of XPC proteolysis and consequently in increased steady-state levels of the protein complex. This correlates with proficient GG-NER. As mentioned above, a protecting role of Rad23/HR23 via inhibition of polyubiquitination was already postulated from various other studies (Schauber et al. 1998; Lommel et al. 2000, 2002; Ortolan et al. 2000). Under conditions of a high level of DNA damage, involvement in NER stimulates the protective role of HR23. Particularly after prolonged higher damage load, this would lead in normal cells to gradual up-regulation of XPC and consequently the entire GG-NER pathway. This rheostat model for adapting XPC levels to the amount of damage provides a novel type of regulation of DNA repair capacity in eukaryotes. Intriguingly, overexpression of exogenous hXPC costabilized endogenous mXPC at the same time (Fig. 4). One interpretation of this trans-effect is that the pathway responsible for degradation of mXPC becomes saturated by high production of exogenous hXPC driven by the strong SV40 promoter. This points to a quite specific proteolysis mechanism that keeps XPC concentrations low in the absence of DNA damage.
Figure 9.
Model for the DNA damage and HR23-dependent regulation of XPC and GG-NER. In the total absence of the HR23 proteins (mHR23A/B-deficient), XPC is intrinsically unstable and targeted for ubiquitin-dependent proteolysis via the 26S proteasome. As a consequence, the steady-state level of XPC is decreased, resulting in reduced GG-NER capacity (top panel). Under normal conditions, HR23 proteins (indicated as 23) control XPC degradation, leading to partial stabilization of XPC [in a complex with HR23 and CEN2 (C)]. Higher steady-state levels of XPC result in proficient GG-NER (middle panel). NER-type DNA damage (e.g., UV irradiation) induces a transient further increase in XPC/HR23/CEN2 protein levels through nuclear accumulation of XPC bound to lesions, and accordingly enhances GG-NER capacity (bottom panel). A comparable HR23-mediated stabilization mechanism may hold for other factors (here indicated by “?”) and cellular pathways in which HR23 proteins are implicated (see Discussion for further explanation).
Down-regulation of XPC
Why are XPC levels not constitutively high? At first sight, it would seem only advantageous to allow GG-NER always to operate at maximal capacity instead of time-consuming up-regulation as revealed here for XPC. However, several observations indicate that RAD4 and its mammalian equivalent XPC are highly toxic when expressed at too high levels in homologous and heterologous systems. Even low-copy RAD4 expression in Escherichia coli interfered with growth, and only defective derivatives of the gene could be propagated (Siede and Eckardt-Schupp 1986; Wei and Friedberg 1998). These findings and the results of XPC microinjection (see Results) suggest that the protein interferes with a vital process, likely DNA metabolism, by its ability to bind to a wide range of (aberrant) DNA structures. XPC detects helix distortions on the basis of disrupted base pairing but also binds to regular mismatches, the normal substrate for mismatch repair (Sugasawa et al. 2001). This highlights a fundamental dilemma of damage sensors: some normally occurring DNA conformations resemble DNA lesions, which may lead to disruption of important cellular processes. Thus, titrating XPC to low levels may reduce untargeted repair interfering with other DNA transactions.
Other targets for HR23
Next to XPC, several other proteins have been found to associate with HR23 and may be subject to a similar mechanism of transient stabilization. An interesting HR23 binding partner is 3-methyladenine (3Me-A) DNA glycosylase (MAG or MPG), an enzyme that initiates base excision repair (BER) of a number of DNA alkylated base damages (Memisoglu and Samson 2000). Binding of HR23 in vitro elevates the rate of excision of specific substrates (Miao et al. 2000), which opens the possibility that this branch of BER is regulated in an analogous fashion to GG-NER. Consistent with this model, the human small ubiquitin-related modifiers SUMO-1 and SUMO-2/3 physically interact with the human BER initiator thymine-DNA glycosylase and actively modulate its enzymatic turnover in base release assays (Hardeland et al. 2002). All SUMOs are known to modify target proteins covalently by an enzymatic pathway analogous to ubiquitin conjugation (Jesenberger and Jentsch 2002).
Moreover, multiple engagements between RAD23/HR23 and cell cycle regulation are apparent: (1) RAD23 has a partially redundant role with and binds to RPN10 in the G2/M transition (Lambertson et al. 1999); (2) RAD23 is involved in spindle assembly and S-phase checkpoints together with the RAD23-like protein DDI1 in PDS1-dependent mitotic control (Clarke et al. 2001); and (3) RAD23, together with DSK2, has a role in spindle pole duplication (Biggins et al. 1996). The link with spindle pole duplication was recently strengthened by the discovery of the centrosome factor CEN2 as the third component of the XPC/HR23 complex (Araki et al. 2001). Furthermore, the damage-signaling tumor-suppressor protein p53 is partly regulated by hHR23A via inhibition of the CREB (cyclic AMP-responsive element binding) protein, which acts a coactivator of p53 transcription (Zhu et al. 2001). Finally, HR23 proteins themselves appear regulated in a cell-cycle-dependent manner with specific degradation during S phase (Kumar et al. 1999).
The principal mechanism for regulation of XPC and GG-NER by HR23 may also be applicable to many other HR23 partners and corresponding processes (such as cell cycle control and BER). These mechanisms represent important pathways in the total network of genomic maintenance, which might be functionally connected via HR23. A common regulatory mechanism, such as binding of HR23 to primary damage sensors (XPC and MAG), would enable coordinated control of major cellular DNA-damage-response pathways, including DNA repair, cell cycle progression, and checkpoints. This response is in its effect reminiscent (but with important mechanistic differences) to the well-known SOS system in E. coli. Whereas the E. coli SOS system mainly works via RecA/LexA regulon at the transcriptional level, the HR23-dependent regulation acts even within the posttranslational stage (for review, see Sutton et al. 2000). However, XPC transcription may also be under p53 control (Adimoolam and Ford 2002; Amundson et al. 2002), extending the parallel with the E. coli paradigm. In addition, the multiple engagements of HR23 proteins provide an explanation for the deviant, compared with other NER mutants, phenotype of mHR23B-deficient mice and embryonic lethality of DKOs.
In conclusion, the generation of mammalian cells deficient in mHR23 (starting from mHR23A and mHR23B single-mutant mice) allowed the elucidation of the main NER function of RAD23, revealed an ingenious DNA damage-controlled mechanism of GG-NER regulation, and clarified an important link between NER and the ubiquitin system. The multiple engagements of mHR23 proteins may connect stress-response mechanisms including cell cycle control to known damage sensors like XPC. Finally, the cell line expressing GFP-tagged XPC will be useful for rapid screening and detection of genotoxic or carcinogenic agents.
Materials and methods
Construction of mHR23A targeting vector
An Ola129 mHR23A targeting construct was generated by converting the BglII site in exon II of clone pG7M23Ag1 (containing a 4-kb genomic EcoRI fragment subcloned in pGEM7) into a ClaI site, which (because of a ClaI site in the polylinker) allowed deletion of sequences downstream of the BglII site in exon II (clone pG7M23Ag7). Next, the remaining EcoRI site was removed by filling in the overhangs with Klenow, resulting in clone pG7M23Ag9. After changing the BstXI site into an SalI site, the 3-kb XhoI–SalI fragment was cloned into SalI-digested pGEM5, resulting in clone pG5M23Ag17. Next, the 3′ arm of the construct, consisting of a Klenow-blunted 1.5-kb SmaI–XbaI fragment starting at the SmaI site in exon VII, was inserted into the blunted NdeI site of pG5M23Ag17 (giving pG5M23Ag20), followed by insertion of a Neo marker cassette in antisense orientation in the ClaI site (giving pG5M23Ag24). Finally, the NotI–NsiI insert of pG5M23Ag24 was recloned into a pGEM-9Zf(–)-based vector containing a 2.8-kb thymidine kinase (TK) marker cassette (giving pG5M23Ag30).
ES cell culture and transfection
The Ola129-derived ES cell line E14 was electroporated with the mHR23A targeting construct and cultured on dishes treated with gelatin as described previously (Ng et al. 2002). G418 (Geneticin, GIBCO; final concentration, 200 μg/mL) was added 24 h after electroporation, and cells were maintained under selection for 6–8 d. Genomic DNA from G418-resistent clones was digested with BamHI and subjected to Southern blot analysis using a 0.6-kb XbaI–RsaI fragment (3′ external to the construct) as a probe. Targeted clones were subsequently screened with a Neo cDNA probe (ClaI fragment) to confirm proper homologous recombination in the 5′ arm.
Generation of the mHR23A–/– and mHR23A–/–/B–/– (DKO) mice and fibroblasts
Cells from two independent targeted clones with 40 chromosomes were injected into 3.5-day-old blastocysts isolated from pregnant C57BL/6 females (Ng et al. 2002). Male chimeric mice were mated with C57BL/6 females to obtain heterozygous animals. Germ-line transmission was observed in the coat color of F1 offspring. Males and females heterozygous for mHR23A were interbred to generate mHR23A+/+, mHR23A+/–, and mHR23A–/– mice. For the generation of double-mutant mHR23A/B mice, male and female animals heterozygous for both mHR23A and mHR23B (Ng et al. 2002) were interbred. Genotyping was performed by Southern blot or PCR analysis of genomic DNA from tail biopsies of 10–14-day-old pups.
Primary mHR23A MEFs (three independent lines per genotype) were isolated from E13.5 embryos obtained from matings between mHR23A+/– mice. Double-mutant mHR23A/B MEFs were isolated from E8.5 embryos derived from different crossings between mHR23A+/–/B+/– and mHR23A–/–/B+/– mice. Part of each embryo was used for genotyping, and the remaining tissue was minced and immersed in a thin layer of F10/DMEM culture medium (GIBCO-BRL) supplemented with 15% fetal calf serum, 2 mM glutamate, and 50 μg/mL penicillin and streptomycin. Spontaneously immortalized (established) cell lines were obtained by continuous subculturing of primary MEFs.
For the genotyping of E8.5 embryos, the yolk sac was used as described (Gurtner et al. 1995). In short, the yolk sac was collected in 20 μL of water and immediately frozen in dry ice. Samples were heated at 95°C for 5 min and incubated with 1 μL of proteinase K (10 mg/mL) at 55°C for 1 h. Proteinase K was heat-inactivated at 95°C for 5 min. PCR analysis was performed using the three-primer sets described below for 30 cycles (93°C, 1 min; 55°C, 1 min; 72°C, 90 sec) using mHR23A and mHR23B primers. Primer set 1: mHR23Ap1 (5′-ATGGGACT TGGGCATAGGTGA-3′), mHR23Ap2 (5′-TCTTCAGCCAG GCCTCTTAC-3′), and anti-sense neo (5′-ATCTGCGTGTTC GAATTCGCCAATG-3′), giving 243- and 350-bp PCR fragments from the wild-type and targeted allele, respectively. Primer set 2: mHR23Bp1 (5′-GTAAAGGCATTGAAAGAG AAG-3′), mHR23Bp2 (5′-CTACAGTCTTGTTTCTGACAG-3′), and anti-sense pgk3 (5′-TAGGGGAGGAGTAGAAGGTG-3′), giving 202- and 600-bp PCR fragments from the wild-type and targeted allele, respectively.
DNA repair assays and microneedle injection
UV sensitivity was determined as described (Ng et al. 2002). MEF cultures were exposed to different doses of UV-C light (254 nm, Philips TUV lamp) and allowed to grow for another 3–5 d, before reaching confluence. The number of proliferating cells was estimated by scintillation counting of the radioactivity incorporated during a 3-h pulse with [3H]thymidine (5 μCi/mL, specific activity, s.a.: 50 Ci/mmole; Amersham). Cell survival was expressed as the ratio of 3H incorporation in irradiated and unirradiated cells.
UV-induced global genome repair was assayed using the UDS method as described (Vermeulen et al. 1994). Cells were exposed to 16 J/m2 of 254-nm UV light and labeled with [methyl-3H]thymidine (10 μCi/mL, s.a.: 50 Ci/mmole). Repair capacity was quantified by grain counting after autoradiography.
RNA synthesis recovery after UV-irradiation was measured according to Ng et al. (2002). Cells were exposed to 10 J/m2 of 254-nm UV light, allowed to recover for 16 h, labeled with [5,6-3H]uridine (10 μCi/mL, s.a.: 50 Ci/mmole), and processed for autoradiography. The relative rate of RNA synthesis was expressed as the number of autoradiographic grains over the UV-exposed nuclei divided by the number of grains over the nuclei of unirradiated cells on parallel slides.
Microneedle injection of control cells (C5RO) was performed as described previously (Vermeulen et al. 1994). After injection of at least 50 homopolykaryons, cells were cultured for the desired time in normal culture medium before they were assayed for their repair capacity by means of UV-induced UDS.
RNA and protein analysis
Total RNA was isolated from mHR23A MEFs using an RNeasy Mini Kit (QIAGEN). Twenty micrograms of total RNA was separated on a 0.9% agarose gel and transferred to Hybond-N+ membrane (Amersham Pharmacia Biotech). RNA blots were hybridized using full-length mHR23A and β-actin 32P-labeled cDNA probes.
Immunoblot analysis was performed on fibroblast extracts obtained by sonification (5 × 106 cells in 300 μL of phosphate-buffered saline, PBS) or extraction. In the latter case, NP lysis buffer [25 mM Tris-HCl at pH 8.0, 1 mM EDTA, 10% glycerol, 0.01% Nonidet P-40 (NP-40), 1 mM dithiothreitol (DTT), 0.25 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor mix (chymostatin, leupeptin, antipain, and pepstatin A)] was added to a monolayer of MEFs. After 30 min on ice, the lysate was collected with a cell scraper and clarified by 2× centrifugation at 4°C. NP lysis buffer containing 0.3 M NaCl was added to the cell pellet and homogenized by sonification.
SDS–polyacrylamide gel electrophoresis was performed by loading 25–50 μg of total cellular protein per lane on 6%–8% gels. Proteins were blotted to nitrocellulose membranes (Schleicher & Schuell) and probed with polyclonal antibodies recognizing human HR23A or XPC, or with monoclonal antibodies recognizing the HA epitope (HA.11, BAbCO) or p62 subunit of TFIIH (3C9; kindly provided by J.M. Egly, Institut de Genetique et de Biologie Moleculaire et Cellulaire, Illkirch Cedex, France). Proteins were visualized using alkaline phosphatase-labeled goat anti-rabbit or peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies.
Immunoprecipitation
Immunoprecipitations (IP) were performed on whole-cell extracts (WCE). Cells were harvested in lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mM DTT, 0.25 mM PMSF, and complete protease inhibitors; Roche) containing 10 mM iodacetamine (Sigma). WCEs were kept on ice for 30 min and centrifuged at 4°C prior to each precipitation. Supernatants of WCE were transferred to a fresh microtube, mouse monoclonal anti-GFP antibodies (7.1 + 13.1; Roche) were added, and the mixture was incubated at 4°C for 3–4 h with mild rotation. Subsequently, 50 μL of protein-G sepharose beads (4 Fast Flow; Amersham), equilibrated with lysis buffer, were added and incubated at 4°C overnight with mild rotation. Beads were collected (5000 rpm, 1 min) and washed three times with lysis buffer at 4°C for 10 min and once with wash buffer (50 mM Tris-HCl at pH 7.5, 0.25 mM NaCl, 0.1% NP-40, 0.05% sodium deoxycholate). SDS sample buffer was added to the beads, and samples were boiled for 10 min. Immunoprecipitated samples were separated on 6% SDS-polyacrylamide gel and transferred to nitrocellulose membranes. Immunodetection was performed with rabbit polyclonal anti-human XPC or mouse monoclonal anti-GFP (7.1 + 13.1; Roche).
Immunofluorescence labeling
Cells were grown on glass coverslips at 60%–80% confluency. After washing twice with PBS, cells were fixed with 2% paraformaldehyde in PBS at room temperature (RT) for 10 min and permeabilized with 0.1% Triton X-100 in PBS at RT twice for 10 min. After extensive washing (three times of 5 min each) with PBS+ (PBS supplemented with 0.15% glycine and 0.5% BSA), cells were incubated with affinity-purified primary antibodies in PBS+ in a moist chamber for at RT for 90 min. After washing five times in PBS+, cells were incubated with the secondary antibodies in PBS+ in a moist chamber at RT for 90 min. Following five washes with PBS+ and one with PBS, coverslips were preserved with Vectashield Mounting Medium (Vector Laboratories) containing 4′-6-diamidino-2-phenylindole (DAPI, 1.5 μg/μL) to visualize the nuclei.
The primary antibodies used were affinity-purified, rabbit polyclonal anti-human XPC; rabbit polyclonal anti-human ERCC1; rabbit polyclonal anti-XPA (a kind gift from K. Tanaka, Division of Cellular Genetics, Institute for Molecular and Cellular Biology, Osaka University and Core Research for Evolutional Science and Technology, Japan); mouse monoclonal anti-p62 of TFIIH subunit (3C9; J.M. Egly, Institut de Genetique et de Biologie Moleculaire et Cellulaire, Illkirch Cedex, France); and high-affinity, rat monoclonal anti-HA (3F10; Boehringer). The secondary antibodies were goat anti-rat and goat antirabbit Alexa 594-conjugated, and goat anti-rat and goat antirabbit Alexa 488-conjugated antibodies (Molecular probes); and goat anti-mouse Cy3-conjugated antibodies (Jackson ImmunoResearch Laboratories).
Generation of XPC-GFP fusion cDNA construct and cotransfection studies
Full-length human XPC cDNA (ScaI–Asp718I fragment) was cloned in an EcoRI–Asp718I-digested eukaryotic expression vector pEGFP-N3 (Clontech) containing a 3′-histidine-hemaglutinine tag (generated by insertion of a double-stranded oligonucleotide in SspBI–NotI-digested pEGFP-N3; kindly provided by D. Hoogstraten, Erasmus Medical Center, Department of Cell Biology and Genetics, Medical Genetics Center, Rotterdam, The Netherlands). For simplicity, the resulting tagged cDNA construct hXPC-EGFP–His6HA-N3 is referred to as hXPC-GFP.
Full-length cDNAs of the hHR23B (in a pSLM vector; Pharmacia Biotech) and hXPC-GFP were cotransfected into DKO MEFs using puromycin as a selectable marker. The transfection was performed using SuperFect Transfection Reagent (QIAGEN); puromycin was added 24 h after transfection to a final concentration of 1 μg/mL, and the cells were maintained under selection for 20–40 d. Stable puromycin-resistant clones were isolated, and integration of the cDNA construct was confirmed by DNA blotting (data not shown).
Exposure of cells to DNA-damaging agents
Cells stably expressing hXPC-GFP/hHR23B were rinsed with PBS, exposed to UV-C light (254 nm; Philips TUV lamp, dose as indicated in the text), and subsequently cultured at 37°C for various time periods (as indicated in the text). XPC was detected either by immunoblot analysis or by visualization in living cells using fluorescence microscopy. A similar approach was used to study the effect of N-acetoxy-2-acetylaminofluorene (NA-AAF, final concentration 50 or 100 μM), mitomycin C (MMC; Sigma, final concentration 1.2 or 2.4 μg/mL), ionizing radiation (γ-rays from a 137Cs source, single dose of 6 and 10 Gy), the proteasome inhibitor N-CBZ-LEU-LEU-LEU-AL (CBZ-LLL; Sigma, final concentration 5 or 10 μM), the transcription inhibitors 5,6-dichloro-1β-D-ribofuranosyl-benzimidazole (DRB; Sigma, final concentration of 100 μM, 2–3 h) and α-amanitin (Sigma, final concentration 10 μg/mL, 1–3 h), the translation inhibitor cycloheximide (CHX; Boehringer, final concentration 30, 50, and 100 μg/mL, 1–3 h), and heat shock (39.5°C and 41°C, for 2–12 h).
Local UV irradiation was obtained by covering cells grown on glass coverslips with an isopore polycarbonate filter with pores of 5.0 μm diameter (Millipore, TMTP) during UV irradiation (4 × 16 J/m2 of UV-C). Immediately after exposure, the filter was removed, medium was added back to the cells, and culturing was continued. After various time periods (as indicated in the text), cells were processed for immunolabeling.
To identify cells in mixtures of control and mutant fibroblasts, cells were labeled with latex beads (diameter 0.79 μm; Polybead Carboxylate Microspheres, Polysciences) added to fibroblast cultures 2 d prior to mixing of the cells. Cells were thoroughly washed in PBS (three times) before trypsinization to remove the unincorporated beads and seeded in a 1:1 ratio on coverslips and cultured for 2 d.
Light microscopy and image analysis
Immunofluorescent microscopy images were obtained with either a Leitz Aristoplan microscope equipped with epifluorescene optics and a PlanApo 63×/1.40 oil immersion lens or a Leica DMRBE microscope equipped with epifluorescene optics and a PL Fluotar 100×/1.30 oil immersion lens. For the detection of GFP-tagged proteins in the living cell, we have used an Olympus IX70 microscope equipped with epifluorescence optics and an Olympus PlanApo 60×/1.40 oil immersion lens. GFP images were obtained after excitation with 455–490 and long pass emission filter (>510 nm). Cy-3 images were obtained after excitation with 515–560 and long pass emission filter (580 nm).
Acknowledgments
J.H.J.H., W.V., and G.T.J.v.d.H. are supported by the Dutch Cancer Society (EUR-1774), the Dutch Science Organization (Medical, Chemical and Life Sciences Divisions), the EC (QLRT-1999-02002), NIH (AG17242-02), a SPINOZA award, and the Louis Jeantet Foundation. This work was also supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by the Biodesign Research Program and the Bioarchitect Research Project from RIKEN.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.260003.
Corresponding author.
References
- Adimoolam S. and Ford, J.M. 2002. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc. Natl. Acad. Sci. 99: 12985–12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundson S.A., Patterson, A., Do, K.T., and Fornace Jr., A.J. 2002. A nucleotide excision repair master-switch: p53 regulated coordinate induction of global genomic repair genes. Cancer Biol. Ther. 1: 145–149. [DOI] [PubMed] [Google Scholar]
- Araki M., Masutani, C., Takemura, M., Uchida, A., Sugasawa, K., Kondoh, J., Ohkuma, Y., and Hanaoka, F. 2001. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 276: 18665–18672. [DOI] [PubMed] [Google Scholar]
- Bertolaet B.L., Clarke, D.J., Wolff, M., Watson, M.H., Henze, M., Divita, G., and Reed, S.I. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8: 417–422. [DOI] [PubMed] [Google Scholar]
- Biggins S., Ivanovska, I., and Rose, M.D. 1996. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell. Biol. 133: 1331–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootsma D., Kraemer, K.H., Cleaver, J.E., and Hoeijmakers, J.H.J. 2001. Nucleotide excision repair syndromes: Xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. In The metabolic and molecular bases of inherited disease, 8th ed. (eds. C.R. Scriver et al.), Vol. 1, pp. 677–703. McGraw-Hill, New York. [Google Scholar]
- Chen L. and Madura, K. 2002. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22: 4902–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Shinde, U., Ortolan, T.G., and Madura, K. 2001. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2: 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D.J., Mondesert, G., Segal, M., Bertolaet, B.L., Jensen, S., Wolff, M., Henze, M., and Reed, S.I. 2001. Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control. Mol. Cell. Biol. 21: 1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C., Walker, G.C., and Siede, W. 1995. DNA repair and mutagenesis. ASM Press, Washington, DC.
- Gillette T.G., Huang, W., Russell, S.J., Reed, S.H., Johnston, S.A., and Friedberg, E.C. 2001. The 19S complex of the proteasome regulates nucleotide excision repair in yeast. Genes & Dev. 15: 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner G.C., Davis, V., Li, H., McCoy, M.J., Sharpe, A., and Cybulsky, M.I. 1995. Targeted disruption of the murine VCAM1 gene: Essential role of VCAM-1 in chorioallantoic fusion and placentation. Genes & Dev. 9: 1–14. [DOI] [PubMed] [Google Scholar]
- Hanawalt P.C. 2000. DNA repair. The bases for Cockayne syndrome. Nature 405: 415–416. [DOI] [PubMed] [Google Scholar]
- Hardeland U., Steinacher, R., Jiricny, J., and Schar, P. 2002. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21: 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama H., Yokoi, M., Masutani, C., Sugasawa, K., Maekawa, T., Tanaka, K., Hoeijmakers, J.H.J., and Hanaoka, F. 1999. Interaction of hHR23 with s5a. The ubiquitin-like domain of hhr23 mediates interaction with s5a subunit of 26s proteasome. J. Biol. Chem. 274: 28019–28025. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H.J. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374. [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Nigg, A.L., Heath, H., Mullenders, L.H., van Driel, R., Hoeijmakers, J.H.J., Vermeulen, W., and Houtsmuller, A.B. 2002. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell 10: 1163–1174. [DOI] [PubMed] [Google Scholar]
- Houtsmuller A.B., Rademakers, S., Nigg, A.L., Hoogstraten, D., Hoeijmakers, J.H.J., and Vermeulen, W. 1999. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science 284: 958–961. [DOI] [PubMed] [Google Scholar]
- Jesenberger V. and Jentsch, S. 2002. Deadly encounter: Ubiquitin meets apoptosis. Nat. Rev. Mol. Cell. Biol. 3: 112–121. [DOI] [PubMed] [Google Scholar]
- Kumar S., Talis, A.L., and Howley, P.M. 1999. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J. Biol. Chem. 274: 18785–18792. [DOI] [PubMed] [Google Scholar]
- Lambertson D., Chen, L., and Madura, K. 1999. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 153: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaerts M., Goeloe, J.I., den Dulk, H., Brandsma, J.A., and Brouwer, J. 2000. Identification and characterization of the rhp23+ DNA repair gene in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 268: 210–215. [DOI] [PubMed] [Google Scholar]
- Lommel L., Chen, L., Madura, K., and Sweder, K. 2000. The 26S proteasome negatively regulates the level of overall genomic nucleotide excision repair. Nucleic Acids Res. 28: 4839–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel L., Ortolan, T., Chen, L., Madura, K., and Sweder, K.S. 2002. Proteolysis of a nucleotide excision repair protein by the 26S proteasome. Curr. Genet. 42: 9–20. [DOI] [PubMed] [Google Scholar]
- Masutani C., Sugasawa, K., Yanagisawa, J., Sonoyama, T., Ui, M., Enomoto, T., Takio, K., Tanaka, K., van der Spek, P.J., Bootsma, D., et al. 1994. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 13: 1831–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C., Araki, M., Sugasawa, K., van der Spek, P.J., Yamada, A., Uchida, A., Maekawa, T., Bootsma, D., Hoeijmakers, J.H.J., and Hanaoka, F. 1997. Identification and characterization of XPC-binding domain of hHR23B. Mol. Cell. Biol. 17: 6915–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memisoglu A. and Samson, L. 2000. Base excision repair in yeast and mammals. Mutat. Res. 451: 39–51. [DOI] [PubMed] [Google Scholar]
- Miao F., Bouziane, M., Dammann, R., Masutani, C., Hanaoka, F., Pfeifer, G.P., and O'Connor, T.R. 2000. 3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. J. Biol. Chem. 275: 28433–28438. [DOI] [PubMed] [Google Scholar]
- Ng J.M.Y., Vrieling, H., Sugasawa, K., Ooms, M.P., Grootegoed, J.A., Vreeburg, J.T., Visser, P., Beems, R.B., Gorgels, T.G., Hanaoka, F., et al. 2002. Developmental defects and male sterility in mice lacking the ubiquitin-like DNA repair gene mHR23B. Mol. Cell. Biol. 22: 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortolan T.G., Tongaonkar, P., Lambertson, D., Chen, L., Schauber, C., and Madura, K. 2000. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat. Cell. Biol. 2: 601–608. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. 2002. Ubiquitin in chains. Trends Biochem. Sci. 25: 544–548. [DOI] [PubMed] [Google Scholar]
- Rao H. and Sastry, A. 2002. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the UBA domain proteins Dsk2 and Rad23. J. Biol. Chem. 277: 11691–11695. [DOI] [PubMed] [Google Scholar]
- Russell S.J., Reed, S.H., Huang, W., Friedberg, E.C., and Johnston, S.A. 1999. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol. Cell 3: 687–695. [DOI] [PubMed] [Google Scholar]
- Schauber C., Chen, L., Tongaonkar, P., Vega, I., Lambertson, D., Potts, W., and Madura, K. 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391: 715–718. [DOI] [PubMed] [Google Scholar]
- Siede W. and Eckardt-Schupp, F. 1986. DNA repair genes of Saccharomyces cerevisiae: Complementing rad4 and rev2 mutations by plasmids which cannot be propagated in Escherichia coli. Curr. Genet. 11: 205–210. [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Masutani, C., Uchida, A., Maekawa, T., van der Spek, P.J., Bootsma, D., Hoeijmakers, J.H.J., and Hanaoka, F. 1996. HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol. Cell. Biol. 16: 4852–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Ng, J.M.Y., Masutani, C., Maekawa, T., Uchida, A., van der Spek, P.J., Eker, A.P., Rademakers, S., Visser, C., Aboussekhra, A., et al. 1997. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol. Cell. Biol. 17: 6924–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Ng, J.M.Y., Masutani, C., Iwai, S., van der Spek, P.J., Eker, A.P., Hanaoka, F., Bootsma, D., and Hoeijmakers, J.H.J. 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2: 223–232. [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Okamoto, T., Shimizu, Y., Masutani, C., Iwai, S., and Hanaoka, F. 2001. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes & Dev. 15: 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton M.D., Smith, B.T., Godoy, V.G., and Walker, G.C. 2000. The SOS response: Recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34: 479–497. [DOI] [PubMed] [Google Scholar]
- van der Spek P.J., Eker, A., Rademakers, S., Visser, C., Sugasawa, K., Masutani, C., Hanaoka, F., Bootsma, D., and Hoeijmakers, J.H.J. 1996a. XPC and human homologs of RAD23: Intracellular localization and relationship to other nucleotide excision repair complexes. Nucleic Acids Res. 24: 2551–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Spek P.J., Visser, C.E., Hanaoka, F., Smit, B., Hagemeijer, A., Bootsma, D., and Hoeijmakers, J.H.J. 1996b. Cloning, comparative mapping, and RNA expression of the mouse homologues of the Saccharomyces cerevisiae nucleotide excision repair gene RAD23. Genomics 31: 20–27. [DOI] [PubMed] [Google Scholar]
- Venema J., van Hoffen, A., Natarajan, A.T., van Zeeland, A.A., and Mullenders, L.H. 1990. The residual repair capacity of xeroderma pigmentosum complementation group C fibroblasts is highly specific for transcriptionally active DNA. Nucleic Acids Res. 18: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R.A., Zeeman, A.M., Lombaerts, M., van de Putte, P., and Brouwer, J. 1996. Analysis of gene- and strand-specific repair in the moderately UV-sensitive Saccharomyces cerevisiae rad23 mutant. Mutat. Res. 362: 155–165. [DOI] [PubMed] [Google Scholar]
- Vermeulen W., Scott, R.J., Rodgers, S., Muller, H.J., Cole, J., Arlett, C.F., Kleijer, W.J., Bootsma, D., Hoeijmakers, J.H.J., and Weeda, G. 1994. Clinical heterogeneity within xeroderma pigmentosum associated with mutations in the DNA repair and transcription gene ERCC3. Am. J. Hum. Genet. 54: 191–200. [PMC free article] [PubMed] [Google Scholar]
- Volker M., Moné, M.J., Karmakar, P., Hoffen, A., Schul, W., Vermeulen, W., Hoeijmakers, J.H.J., van Driel, R., Zeeland, A.A., and Mullenders, L.H.F. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8: 213–224. [DOI] [PubMed] [Google Scholar]
- Watkins J.F., Sung, P., Prakash, L., and Prakash, S. 1993. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol. 13: 7757–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S. and Friedberg, E.C. 1998. A fragment of the yeast DNA repair protein Rad4 confers toxicity to E. coli and is required for its interaction with Rad7 protein. Mutat. Res. 400: 127–133. [DOI] [PubMed] [Google Scholar]
- Wiertz E.J., Jones, T.R., Sun, L., Bogyo, M., Geuze, H.J., and Ploegh, H.L. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84: 769–779. [DOI] [PubMed] [Google Scholar]
- Wilkinson C.R., Seeger, M., Hartmann-Petersen, R., Stone, M., Wallace, M., Semple, C., and Gordon, C. 2001. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell. Biol. 3: 939–943. [DOI] [PubMed] [Google Scholar]
- Wood R.D., Hanawalt, P.C., Mer, G., and Cooper, P.K. 2000. Nucleotide excision repair. In Biological responses to DNA damage, pp. 173–182. Cold Spring Harb. Symp. Quant. Biol. LXV. [DOI] [PubMed]
- Wood R.D., Mitchell, M., Sgouros, J., and Lindahl, T. 2001. Human DNA repair genes. Science 291: 1284–1289. [DOI] [PubMed] [Google Scholar]
- Yokoi M., Masutani, C., Maekawa, T., Sugasawa, K., Ohkuma, Y., and Hanaoka, F. 2000. The xeroderma pigmentosum group C protein complex XPC–HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 275: 9870–9875. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Wani, G., Wani, M.A., and Wani, A.A. 2001. Human homologue of yeast Rad23 protein A interacts with p300/cyclic AMP-responsive element binding (CREB)-binding protein to down-regulate transcriptional activity of p53. Cancer Res. 61: 64–70. [PubMed] [Google Scholar]