Abstract
Long-range interactions between the 5′ and 3′ ends of mRNA molecules have been suggested to play a role in the initiation of translation and the regulation of gene expression. To identify such interactions and to study their molecular evolution, we used phylogenetic analysis to generate a model of mRNA higher-order structure in the Adh transcript of Drosophila melanogaster. This model predicts long-range, tertiary contacts between a region of the protein-encoding sequence just downstream of the start codon and a conserved sequence in the 3′ untranslated region (UTR). To further examine the proposed structure, site-directed mutations were generated in vitro in a cloned D. melanogaster Adh gene, and the mutant constructs were introduced into the Drosophila germ line through P-element mediated transformation. Transformants were spectrophotometrically assayed for alcohol dehydrogenase activity. Our results indicate that transformants containing a silent mutation near the start of the protein-encoding sequence show an ≈15% reduction in alcohol dehydrogenase activity relative to wild-type transformants. This activity can be restored to wild-type levels by a second, compensatory mutation in the 3′ UTR. These observations are consistent with a higher-order structure model that includes long-range interactions between the 5′ and 3′ ends of the Adh mRNA. However, our results do not fit the classical compensatory substitution model because the second mutation by itself (in the 3′ UTR) did not show a measurable reduction in gene expression.
There is a growing body of evidence for functional, long-range interactions between the 5′ and 3′ ends of eukaryotic mRNA molecules. Tarun and Sachs (1) have shown that a protein which binds to the 3′ end of yeast mRNA is involved in the initiation of translation which occurs at the 5′ end. Similarly, in Drosophila, proteins that bind to an mRNA’s 3′ untranslated region (UTR) have been shown to affect levels of translation (2, 3). In addition to these studies of protein–protein interactions, several sources have suggested direct RNA–RNA pairings between the two ends of mRNA molecules. Konings et al. (4) used a free-energy minimization algorithm to predict folding patterns for 38 eukaryotic mRNAs. Their results indicate a common pattern of mRNA folding, where the 3′ UTR forms contacts with the coding region just downstream of the start codon. Stephan and Kirby (5) used a phylogenetic comparison method to predict mRNA secondary structures in Drosophila and found evidence for long-range pairings between the 3′ UTR and the protein-encoding region. These findings raise a number of important questions. For instance, which nucleotides are involved in RNA–RNA interactions and how are they arranged into secondary and tertiary pairing regions? Do the currently available models describe the evolution of compensatory mutations adequately? To begin to address these questions, we have focused on identifying elements of the Adh mRNA higher-order structure in Drosophila melanogaster.
Adh produces two developmentally regulated transcripts, which differ only in their 5′-untranslated leader sequences (Fig. 1; ref. 6). The two transcripts are initiated from separate promoters, each having its own enhancer sequence (7). Transcripts from a proximal promoter are found predominantly in larvae, while transcripts from a distal promoter are found predominantly in adult flies (6). P-element-mediated transformation experiments have shown that all of the cis-acting sequence elements required for proper Adh expression are contained within a 8.6-kb SacI–ClaI genomic Adh fragment (7, 8) and that a single replacement substitution can alter the catalytic efficiency of the alcohol dehydrogenase (ADH) enzyme (9). In addition, it has been demonstrated that nonreplacement sites must also play a role in determining the level of Adh expression (9, 10). For example, a complex substitution polymorphism within the first (adult) intron has been shown to affect the level of ADH protein in adult flies (10).
Figure 1.
Restriction map of the 8.6-kb Adh fragment used for transformation experiments. The mRNA-encoding region is shown as a box, with the solid portions representing the protein-encoding regions. An enlargement of the transcriptional unit is shown above, with exons represented as boxes and introns as solid lines. The locations of the adult and larval 5′ leader sequences are indicated. The 3′ UTR and the portion of the 5′ UTR shared by both adult and larval transcripts are shown as shaded boxes. The Adh probe used to identify single-insert lines is shown underneath as a hatched box.
Phylogenetic comparisons have suggested that epistatic selection is acting on nucleotide sites within the Adh transcriptional unit to maintain possible pairing stems involved in RNA secondary structures. Kirby et al. (11) used phylogenetic analysis to identify hairpin structures in the Adh pre-mRNA of Drosophila and suggest that selective maintenance of these structures is responsible for the clusters of linkage disequilibria observed in natural Drosophila pseudoobscura populations (12). While these structures involve short RNA stretches of less than 50 nt, Stephan and Kirby (5) presented preliminary phylogenetic evidence that RNA–RNA interactions may extend over the total length of the Adh primary transcript. To further investigate these long-range interactions, we have extended the phylogenetic analysis of Stephan and Kirby (5). Furthermore, because long-range, compensatory evolution is expected to be very slow (13) and the predicted pairing regions are short, we have followed up the phylogenetic approach by mutation experiments. Here we describe the results of our phylogenetic analysis and report the effects on Adh gene expression of site-directed mutations at both a silent codon position just downstream of the start codon and in the 3′ UTR.
MATERIALS AND METHODS
Adh Sequence Alignment and Covariation Search.
For phylogenetic comparison, Adh sequences were aligned for 10 species from the subfamily Drosophilinae, covering three subgenera. The alignment of these 10 sequences was essentially the same as that described previously (5). The Drosophila species used for the alignment (followed by their GenBank/EMBL accession numbers) are as follows: D. melanogaster (M14802), Drosophila teissieri (X54118), Drosophila erecta (X54116), D. pseudoobscura (M60982), and Drosophila ambigua (X54813) from the subgenus Sophophora; Drosophila hydei (X58694), Drosophila mulleri (X03048), Drosophila affinidisjuncta (X13812), and Drosophila silvestris (M63291) from the subgenus Drosophila; and Drosophila lebanonensis (M97637) from the subgenus Scaptodrosophila. In addition, two Adh sequences (Z30194, Z30195) from the Mediterranean fruit fly Ceratitis capitata were newly included in the comparison. Due to the high level of sequence divergence in noncoding regions, 3′ UTR sequences were aligned manually based on pairwise and multiple alignments within the subgenera (11). Although the complete 3′ UTR alignment is somewhat ambiguous, a conserved sequence of 8 nt of the 3′ UTR could be unambiguously aligned for all 10 Drosophila species (coordinates 1762–1769; see Fig. 2). This 8-bp motif is also conserved in the medfly sequences, although not perfectly. Once the alignment of Fig. 2 was established, the DNA sequences were searched for compensatory substitutions (“covariations”) by preparing a covariation matrix using the program of Han and Kim (14) on complementary, pairwise aligned sequences. The predicted pairings were then examined using a likelihood ratio test (11, 15).
Figure 2.
Sequence alignment of the protein-encoding region of exon 2 and the conserved portion of the 3′ UTR of Adh alleles from 10 different Drosophila species. mel, D. melanogaster; tei, D. teissieri; ere, D. erecta; psu, D. pseudoobscura; amb, D. ambigua; hyd, D. hydei; mul, D. mulleri; aff, D. affinidisjuncta; sil, D. silvestris; leb, D. lebanonensis. The two medfly sequences are shown at the bottom (med1 and med2). The D. melanogaster sequence is that of the Wa-F allele used in our experiments. Numbering is from Kreitman (17). Gaps in the alignment are indicated by dashes. Regions that showed too much divergence for alignment are indicated by dots. The brackets at the bottom indicate the phylogenetically predicted pairing regions. Covarying sites are underlined. Note that some of the pairings are not conserved in the medfly (see text).
Plasmid Construction and Mutagenesis.
Basic molecular techniques were carried out using the procedures of Sambrook et al. (16). All Adh constructs were derived from an 8.6-kb SacI–ClaI fragment of the Wa-F allele [described by Kreitman (17)]. A 3.2-kb SalI–ClaI fragment containing the entire Adh coding region was subcloned into a pUC18 plasmid and used for mutagenesis. Site-directed mutations were made using the Transformer system (CLONTECH). Single nucleotide substitutions were made at positions 819 (C to T) and 1756 (G to A) to create two mutant constructs designated as mutC819T and mutG1756A, respectively [all coordinates are from Kreitman (17)]. Following mutagenesis, a restriction fragment (HpaI–BamHI for mutC819T, BamHI–ClaI for mutG1756A) was used to replace the corresponding fragment in the original 8.6-kb SacI–ClaI clone. At this point, the entire region subjected to mutagenesis was sequenced using a cycle sequencing method (Life Technologies, Gaithersburg, MD) to ensure that the desired mutation (and no other changes) was present. A construct containing both mutations (mutC819T–G1756A) was generated in the same fashion, using the mutC819T HpaI–BamHI fragment to replace the corresponding fragment in a mutG1756A SacI–ClaI clone.
P-Element Mediated Germ-Line Transformation.
For each transformation construct, the appropriate SacI–ClaI fragment was inserted into a ClaI site introduced into the polycloning region of the YES vector, which is a P-element vector containing the D. melanogaster yellow (y) gene as a selectable marker (18). As a control, the wild-type SacI–ClaI fragment was placed into a YES vector and used for transformation. Germ-line transformation was achieved by embryo microinjection (19, 20). Injected embryos were from a y w; Adhfn6;Δ2-3,Sb/TM6 Drosophila line. Adhfn6 is a null allele that produces no detectable ADH protein due to a splicing defect (21). The Δ2-3 P insertion on the third chromosome was used as a source of transposase (22). Following microinjection, adult flies were crossed to a y w; Adhfn6 stock and transformed offspring were identified by body color.
Some insertions on the X and third chromosomes were mobilized to new locations by crossing the transformed line to the y w; Adhfn6;Δ2-3,Sb/TM6 stock and then crossing the offspring containing both a YES insertion and the Δ2-3 source of transposase to the y w; Adhfn6 stock. Transformants with inserts at new locations were identified as y+ offspring where the y+ marker was not segregating with the same chromosome as the original insert.
Identification of Single Insert Lines.
Genomic DNA was prepared from each transformed line, digested with three six-cutter restriction enzymes (BglII, SalI, and StuI) and separated on a 1 % agarose gel. The DNA fragments were then blotted onto a nitrocellulose membrane and hybridized with an Adh-specific probe. The probe was made from a SalI fragment that spans ≈1.5 kb of the Adh 5′ flanking sequence (Fig. 1). The probe hybridizes to a fragment of constant size for the genomic Adh gene and a fragment of unique size for each Adh insertion. Only lines containing a single Adh insert were used for further analysis. In addition, insert DNA from two independent lines of each transformant class was amplified by PCR and sequenced to ensure the correct haplotype with respect to positions 819 and 1756.
ADH Assays.
For analysis of ADH enzymatic activity, transformed males (or females in the case of X chromosome inserts) were crossed to a y w; Adhfn6 stock to produce offspring heterozygous for the Adh insertion. For each cross, five males and five females were placed in each of two vials, and the pooled progeny of each cross were collected at age 6–8 days and used for ADH assays. Two separate assays were performed on each transformed line following the procedure of Maroni (23), using five male flies for each assay and isopropanol as the substrate. Units of ADH activity were measured as μmol of NAD reduced per minute per milligram of total protein. Total protein was estimated using the method of Lowery et al. (24). The entire assay procedure was repeated at two different time blocks, which resulted in a total of four measurements for each transformed line. Differences in ADH activity between mutant and wild-type transformants were tested by ANOVA using a model that accounts for chromosomal and position effects (25).
RESULTS
Phylogenetic Analysis Reveals a Conserved Region in the 3′ UTR That May Pair with a Segment Downstream of the Start Codon. Adh sequences were aligned for 10 Drosophila species, spanning a variety of genetic distances.
The alignment included sequences from three different subgenera (Sophophora, Drosophila, and Scaptodrosophila). In addition, the two Adh sequences of the Mediterranean fruit fly C. capitata were aligned with the Drosophila sequences. While the protein-encoding region was sufficiently conserved over all species to allow for an unambiguous alignment (Fig. 2), the correct alignment was not clear for much of the noncoding regions. The 3′ UTR Drosophila sequences could be aligned within each subgenus, but the complete alignment for all 10 species was not obvious. A portion of the 3′ UTR could be easily aligned, however, due to a highly conserved region found in all 10 species (Fig. 2). The conserved region consists of 8 nt that are perfectly conserved over all Drosophila species. In the medfly sequences, however, this 8-bp sequence is less conserved: 5 of 8 nt were conserved in med1 and 7 of 8 in med2. In addition, the 6 nt preceding the 8-bp motif (starting at position 1756) are conserved in all species from the subgenus Sophophora.
We searched for covariations in exon 2 and the aligned sequences of the 3′ UTR simultaneously using the method of Han and Kim (14). In exon 2, where the start codon is located, we found four pairings that are conserved among all Drosophila sequences and showed at least one covariation (Table 1; Figs. 2, 3A). The results of a likelihood ratio test (15) suggest that the evolution of the sequences in these four regions is subject to secondary structure constraints. Indeed, the values of the pairing parameter λ (Table 1) for the Drosophila sequences are relatively high, except for the putative 853–855/859–861 helix. However, if the two medfly sequences are also included in the comparison, the λ value of the predicted 793–797/833–807 pairing drops considerably, suggesting that this pairing is not conserved in the medfly.
Table 1.
Results of phylogenetic analysis
| Pairing | Covariations | λ | LRT |
|---|---|---|---|
| Exon 2 | |||
| 793–797/833–837 | 1 | 5.00 (2.13) | 18.01 (6.82) |
| 803–805/813–815 | 1 | 3.65 (3.60) | 8.27 (11.22) |
| 838–841/845–848 | 2 | 5.77 (3.20) | 15.77 (13.06) |
| 853–855/859–861 | 2 | 2.46 (2.44) | 4.71 (6.54) |
| Exon 2/3′ UTR | |||
| 810–812/1762–1764 | 1 | 3.54 (3.82) | 7.88 (12.83) |
| 817–819/1756–1758 | 1 | 4.34 (2.10) | 8.34 (4.20) |
| Entire structure | 8 | 3.94 (2.69) | 61.39 (51.53) |
The first column shows the coordinates of the pairing regions [according to Kreitman (17)]. The second column contains the number of covariations found for the 12 sequences aligned in Fig. 2. The third and fourth columns give the values of the pairing parameter (λ) and of the likelihood-ratio-test (LRT) statistic, respectively (15). Values are given for an alignment of the 10 Drosophila sequences and for the 10 Drosophila plus the two medfly sequences (in parantheses). The values for the entire structure (with six pairings) are presented in the last line. Note that when sites evolve independently, the pairing parameter is λ = 1; λ > 1 indicates Watson–Crick base pairing.
Figure 3.
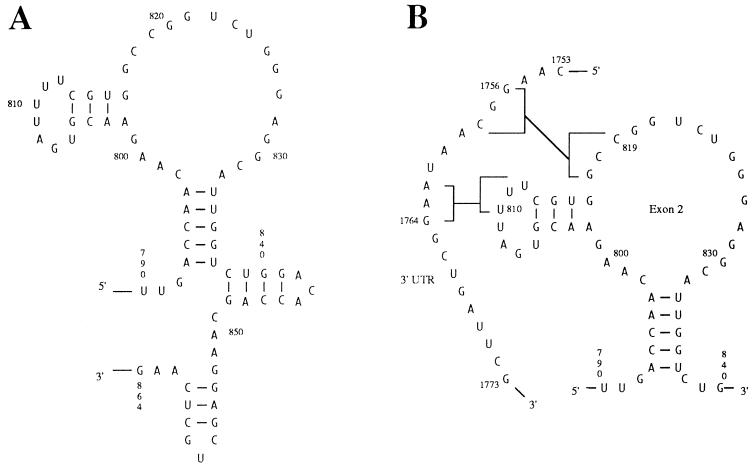
Model of the RNA structure for portions of the Adh transcript. (A) Secondary structure proposed for exon 2. Pairing regions are shown connected by straight lines. All pairing stems were determined by phylogenetic analysis (see Table 1). Note that this figure shows only the portion of exon 2 where the four conserved pairings were found. (B) Potential tertiary contacts between the conserved portion of the 3′ UTR and a region of exon 2. Positions 819 and 1756, which showed a covariation in our alignment and were the targets of site-directed mutagenesis, are indicated.
The evidence for pairing between sequences of exon 2 and the 3′ UTR is less striking. Based on the alignment shown in Fig. 2, we identified a covariation in the putative pairing region 810–812/1762–1764 at positions 810/1762, and a covariation in 817–819/1756–1758 at positions 819/1756. These two pieces of evidence may support long-range interactions involving the 5′ and 3′ regions of the Adh transcript. The two pairings between exon 2 and 3′ UTR sequences are summarized in the model shown in Fig. 3B. They involve sequences in the loops of two hairpins identified in exon 2 and are therefore tertiary contacts. In both cases, however, the alignment of the 3′ UTR sequences is somewhat ambiguous, and the values of the likelihood-ratio-test statistic are rather low (because of the short lengths of the tertiary contacts). Furthermore, in the second case the pairing is not conserved in three of the 10 Drosophila and the two medfly sequences (Fig. 2), although there are some alternate pairing opportunities in the immediate neighborhood. To overcome these difficulties, which are inherent in phylogenetic comparisons, mutation analysis was used to further investigate the hypothesized long-range interactions. In this paper we concentrate on the pairing 817–819/1756–1758.
A Silent Mutation Reduces ADH Activity.
To test the possibility that there may be a long-range RNA–RNA interaction between a region of exon 2 and the conserved region of the 3′ UTR, we made a site-directed mutation in the putative pairing sequence of exon 2. This mutation would disrupt the potential for a Watson–Crick base pairing in the proposed model (Fig. 3B). Because the sequence in question was within the ADH protein-encoding region, the mutation was made in a degenerate third codon position (a C to T change at position 819) so that the ADH amino acid sequence would not be affected. This particular mutation was suggested by phylogenetic analysis, which indicates that site 819 is covarying with site 1756 such that a C-G pair in Sophophora is replaced by U-A in the two Hawaiian species, D. affinidisjuncta and D. silvestris (see above and Fig. 2). Transformed lines were generated by P-element transformation and the ADH activity of the transformants was measured spectrophotometrically. As a control, transformants containing the wild-type Adh sequence were also generated. ADH activity units were measured as μmol of NAD reduced per minute per milligram total protein (multiplied by 100). The average ADH activity of 12 wild-type transformed lines was 108.5 units, while the average activity for 14 mutC819T lines was 92.9 units (Fig. 4). This difference in activity level was significant (P = 0.01) when tested by ANOVA. Because the ADH protein of the wild-type and mutant transformants was identical at the amino acid level, the difference in measured activity must be due to a difference in the amount of enzyme produced (i.e., Adh gene expression).
Figure 4.
ADH activity of all lines (Upper) and autosomal insert lines (Lower) for the different transformant classes. Activity is given in units of μmol of NAD reduced per minute per milligram of total protein (multiplied by 100). Error bars represent the least significant difference at the 5% level. Least significant differences were calculated for individual comparisons, with mutC819T being compared with wild type, and all other lines being compared with mutC819T.
A Compensatory Mutation in the 3′ UTR Restores ADH Activity to Wild-Type Levels.
If the reduction in ADH activity in mutC819T is a result of the disruption of a Watson–Crick base pair involved in an RNA higher-order structure, then it is expected that the level of activity could be increased by making a compensatory mutation at a second site that restores the potential for pairing. To test this we made a second site-directed mutation, mutG1756A, within the 3′ UTR. This mutation was introduced into the mutC819T background and transformants were generated by P-element transformation. Transformants containing both mutations (mutC819T–G1756A) showed wild-type levels of ADH activity (Fig. 4). The average activity of the 10 double-mutant lines was 104.4 units, which was not significantly different from wild-type. The activity of mutC819T–G1756A was, however, significantly higher than that of mutC819T (P = 0.05). Thus, the mutation at position 1756 appears to compensate for the reduction in ADH activity caused by the mutation at position 819, resulting in an almost 15% increase in ADH activity.
The 3′ UTR Mutation Has No Effect by Itself.
Transformants containing just the single mutation in the 3′ UTR at position 1756 (mutG1756A) were also generated through P-element transformation. The average ADH activity of 11 mutG1756A lines was 109.0 units and was not significantly different from that of wild-type transformants (Fig. 4). Thus mutG1756A does not have a measurable effect on its own when placed in a wild-type background, but does have an effect when placed in the mutC819T background.
Comparison of X Chromosome and Autosomal Insertions.
Because some of the transposable element insertions occurred on the X chromosome and male flies were used for ADH assays, there is a possibility of dosage compensation mechanisms affecting levels of ADH activity. In transformation experiments using a different P-element vector, Laurie-Ahlberg and Stam (25) found that X chromosome Adh inserts showed partial dosage compensation. To check for possible effects of dosage compensation in our transformed lines, we compared the activities of X chromosome insert lines and autosomal insert lines within each transformant class by ANOVA. While there was a significant level of position effect variation within each transformant class (P < 0.0001), there was no significant difference between X and autosomal insert lines for wild-type, mutG1756A, and mutC819T–G1756A transformed lines, where the number of X chromosome inserts were five, four, and three, respectively. For mutC819T, X chromosome insert lines had a significantly higher level of ADH activity than autosomal insert lines (P < 0.0001). There were, however, only two X chromosome inserts out of 14 mutC819T lines. When the complete data set was analyzed using only autosomal insert lines, the results were essentially the same as when using all transformed lines (Fig. 4). mutC819T lines had a level of ADH activity significantly lower than both wild-type (P = 0.005) and mutC819T-mutG1756A lines (P = 0.02).
DISCUSSION
We have used site-directed mutagenesis followed by P-element mediated germ-line transformation to provide the first experimental evidence for long-range RNA–RNA interactions in a protein-encoding gene of Drosophila. Our results show that a silent mutation within the protein-encoding region of Adh (mutC819T) results in an approximate 15% reduction in Adh expression. A second, compensatory mutation in the 3′ UTR can restore expression to wild-type levels. These observations are consistent with the model of Adh higher-order structure shown in Fig. 3B. This model consists of relatively simple secondary structural elements formed in exon 2 and two long-range tertiary pairings. The model predicts a tertiary contact between a region of exon 2 at coordinates 817–819 and a highly conserved region of the 3′ UTR at 1756–1758, for which a covariation was found at positions 819/1756. In addition, the model indicates a tertiary pairing 810–812/1762–1764, which is supported phylogenetically by a covariation at positions 810/1762. Our data, however, do not fit Kimura’s classical compensatory substitution model (26) because the second mutation by itself (in the 3′ UTR) did not show a measurable reduction in gene expression. According to the classical model, the single mutation (G to A) in the 3′ UTR creates a C/A intermediate and should therefore be less stable than the C to T change at position 819 (which leads to a U/G wobble pair). As discussed below, Kimura’s model appears to be too simple in that it considers compensatory interactions between only two nucleotide sites without taking the rest of the molecule into account.
How realistic is the proposed higher-order structure model given the data? We first discuss mutC819T, a mutation at a silent codon position that led to a significant reduction in Adh gene expression. One possible alternative explanation for the effect on gene expression caused by a silent substitution is that the two synonymous codons may not be equivalent in their effects on translation. This idea could explain the high level of nonrandom codon usage observed in some D. melanogaster genes, including Adh (27). Adh is a highly expressed gene in D. melanogaster, accounting for ≈1–2% of the translational activity of mRNA in adult flies (28). Highly expressed genes have been shown to be positively correlated with levels of codon bias in both bacteria and yeast (29–31). In multicellular eukaryotes such as Drosophila it is more difficult to quantify expression levels, but it appears that highly expressed Drosophila genes also show high levels of codon bias (27). These observations indicate that codon preference may be under the influence of natural selection. In our case, mutC819T changes an alanine codon from GCC to GCT, which is a change from a preferred codon to an unpreferred codon (27). This mutation also occurs near the start codon (the 14th codon from the start), which may be important if unpreferred codons near the ribosomal assembly site can reduce the efficiency of translation initiation.
Several observations, however, argue against the effects of mutC819T being caused by a switch from a preferred to an unpreferred codon. First, codon bias does not explain how a mutation in the 3′ UTR (mutG1756A) could compensate for the reduction in ADH activity seen in mutC819T lines, restoring the activity to wild-type levels. Second, Choudhary and Laurie (9) performed similar experiments where they altered a threonine codon from ACC to ACG (a change from preferred to unpreferred) and reported no difference in ADH activity. Although their mutation involved a codon for a different amino acid and at a location much further from the start of translation, their results show that a change from a preferred to an unpreferred codon in Adh does not result in a measurable change in gene expression. Finally, the power of selection on individual synonymous sites is expected to be quite weak (Nes < 1, where Ne is the effective population size and s is the coefficient of selection) in D. melanogaster (27). For these reasons, it seems unlikely that a change from a preferred to an unpreferred codon would result in an expression difference as large as the 15% difference measured in our experiments.
If the 15% reduction in ADH activity cannot be explained by codon usage bias, could it then be accounted for by an alteration in RNA higher-order structure? A disruption of an RNA secondary structure by a nucleotide substitution may lead to the formation of an alternate structure that could affect levels of gene expression. In mutational experiments using yeast, Libri et al. (32) found that nucleotide substitutions may result in complex rearrangements of RNA secondary structures and that these structures may influence expression levels. We have identified a potential alternative pairing within exon 2 (792–796/819–823) which would be facilitated by mutC819T. A C to T change at position 819 may create a stable 5-bp-long stem in exon 2 that could alter the phylogenetically predicted structure shown in Fig. 3A and thus make a tertiary contact between exon 2 and 3′ UTR sequences impossible. Although such a change in secondary structure cannot be determined phylogenetically, we have used the free-energy minimization method of Jaeger et al. (33) to predict the secondary structure of the D. melanogaster exon 2 sequence. Using this method, we indeed see a change in conformation such that the predicted wild-type pairing (793–797/833–837) is replaced by the alternate paring (792–796/819–823) when position 819 is changed from C to T. The two predicted structures are equivalent in their predicted free-energy changes (ΔG = −14.4 kcal/mol).
The strongest support for a long-range base pairing interaction in Adh comes from the observation that mutC819T, which disrupts a Watson–Crick base pair in the proposed model (Fig. 3B), reduces ADH activity, while a compensatory mutation in the 3′ UTR which restores the potential for Watson–Crick pairing (mutG1756A) restores the ADH activity to wild-type levels. Using the method of Freier et al. (34), the estimated free-energy change (ΔG) for the short pairing stem 817–819/1756–1758 in the wild-type sequence is −6.7 kcal/mol. The ΔG values for mutC819T and mutC819T–G1756A are −3.8 and −4.7 kcal/mol, respectively. These estimates are qualitatively consistent with our experimental results. The RNA structure model, however, also predicts that mutG1756A (ΔG = −2.9 kcal/mol) should cause a reduction in ADH activity when placed into a wild-type background. Our results show that transformants containing only mutG1756A have wild-type levels of ADH activity. An explanation for this observation that is in line with the above discussion on alternate pairings in exon 2 may be as follows. Our phylogenetic analysis shows that position 1756 is the least conserved site within the proposed pairing region 817–819/1756–1758. Hence, mutating this nucleotide site under wild-type conditions may affect levels of gene expression only slightly. On the other hand, in a mutC819T background, this site may be more important for proper function of the molecule, possibly because an alternate structure that may have formed in exon 2 following transcription (discussed above) has to be displaced by a binding between the appropriate sequences in exon 2 and those of the 3′ UTR. To increase the binding affinity, a perfect match of 3 nt forming the pairing region may be necessary. While such long-range interactions have not previously been reported within mRNA molecules, there is experimental evidence for local secondary structures affecting trans-interactions between short RNA–RNA contacts involved in translation initiation and RNA processing (35, 36).
Our model of long-range contacts within the Adh transcript (Fig. 3B) is supported both phylogenetically and experimentally. This model makes testable predictions and thus provides a framework for future mutation experiments to explore the importance of mRNA structure in gene expression and molecular evolution.
Acknowledgments
We thank C. Laurie for providing Adh clones and an Adhfn6 Drosophila stock, P. Geyer for providing the YES vector, and K. Chang, an undergraduate in the Howard Hughes Medical Institute Research Program, for technical assistance in the laboratory. Furthermore, we are grateful to D. Kirby, S. Mount, and two reviewers for very helpful comments on the manuscript; to S. Mount and S. Woodson for help with the free-energy calculations; to S. Muse for assistance with the likelihood analysis; and to M. Green for handling the review process very quickly. This research was supported in part by National Science Foundation Grant DEB-9407226 to W.S. and by a GRB Summer Research Award to S.T.
Footnotes
Abbreviations: UTR, untranslated region; ADH, alcohol dehydrogenase.
References
- 1.Tarun S Z, Sachs A B. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 2.Dubnau J, Struhl G. Nature (London) 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 3.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring W J, Jäckle H. Nature (London) 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 4.Konings D A M, Van Duijn L P, Voorma H O, Hogeweg P. J Theor Biol. 1987;127:63–78. doi: 10.1016/s0022-5193(87)80161-3. [DOI] [PubMed] [Google Scholar]
- 5.Stephan W, Kirby D A. Genetics. 1993;135:97–103. doi: 10.1093/genetics/135.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benyajati C, Spoerel N, Haymerle H, Ashburner M. Cell. 1983;22:125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- 7.Corbin V, Maniatis T. Genetics. 1990;124:637–646. doi: 10.1093/genetics/124.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg D A, Posakony J W, Maniatis T. Cell. 1983;34:59–73. doi: 10.1016/0092-8674(83)90136-8. [DOI] [PubMed] [Google Scholar]
- 9.Choudhary M, Laurie C C. Genetics. 1991;129:481–488. doi: 10.1093/genetics/129.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurie C C, Stam L F. Genetics. 1994;138:379–385. doi: 10.1093/genetics/138.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirby D A, Muse S V, Stephan W. Proc Natl Acad Sci USA. 1995;92:9047–9051. doi: 10.1073/pnas.92.20.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaeffer S W, Miller E L. Genetics. 1993;135:541–552. doi: 10.1093/genetics/135.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephan W. Genetics. 1996;144:419–426. doi: 10.1093/genetics/144.1.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han K, Kim H-J. Nucleic Acids Res. 1993;21:1251–1257. doi: 10.1093/nar/21.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muse S V. Genetics. 1995;139:1429–1439. doi: 10.1093/genetics/139.3.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Kreitman M. Nature (London) 1983;304:412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- 18.Patton J S, Gomes X V, Geyer P K. Nucleic Acids Res. 1992;20:5859–5860. doi: 10.1093/nar/20.21.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spradling A C, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 20.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 21.Benyajati C, Place A R, Wang N, Pentz E, Sofer W. Nucleic Acids Res. 1982;10:7261–7272. doi: 10.1093/nar/10.22.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson H M, Preston C R, Phillis R W, Johnson-Schlitz D M, Benz W K, Engels W R. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maroni G. Biochem Genet. 1978;16:509–523. doi: 10.1007/BF00484215. [DOI] [PubMed] [Google Scholar]
- 24.Lowery O H, Rosebrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 25.Laurie-Ahlberg C C, Stam L F. Genetics. 1987;115:129–140. doi: 10.1093/genetics/115.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura M. J Genet. 1985;64:7–19. [Google Scholar]
- 27.Akashi H. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benyajati C, Wang N, Reddy A, Weinberg E, Sofer W. Nucleic Acids Res. 1980;23:5649–5667. doi: 10.1093/nar/8.23.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gouy M, Gautier C. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shields D C, Sharp P M. Nucleic Acids Res. 1987;15:8023–8040. doi: 10.1093/nar/15.19.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennetzen J L, Hall B D. J Biol Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- 32.Libri D, McCarthy T, Rosbash M. RNA. 1995;1:425–436. [PMC free article] [PubMed] [Google Scholar]
- 33.Jaeger J A, Turner D H, Zuker M. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 34.Freier S M, Kierzek R, Jaeger J A, Sugimoto N, Caruthers T N, Turner D H. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Smit M H, van Duin J. J Mol Biol. 1994;244:144–150. doi: 10.1006/jmbi.1994.1714. [DOI] [PubMed] [Google Scholar]
- 36.Madhani H D, Guthrie C. Genes Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]