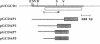Abstract
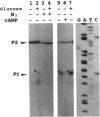
DNA sequence and expressional analyses of the gcd gene of Escherichia coli K-12 W3110 revealed that two promoters that were detected were regulated negatively by cyclic AMP and positively by oxygen. Sequence conservation of the gcd gene between E. coli K-12 W3110 and PPA42 suggests that glucose dehydrogenase is required for the E. coli cells, even though it ordinarily exists as an apoprotein.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988 Apr 20;200(4):709–723. doi: 10.1016/0022-2836(88)90482-2. [DOI] [PubMed] [Google Scholar]
- Chepuri V., Gennis R. B. The use of gene fusions to determine the topology of all of the subunits of the cytochrome o terminal oxidase complex of Escherichia coli. J Biol Chem. 1990 Aug 5;265(22):12978–12986. [PubMed] [Google Scholar]
- Cleton-Jansen A. M., Goosen N., Fayet O., van de Putte P. Cloning, mapping, and sequencing of the gene encoding Escherichia coli quinoprotein glucose dehydrogenase. J Bacteriol. 1990 Nov;172(11):6308–6315. doi: 10.1128/jb.172.11.6308-6315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Ebright Y. W., Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1989 Dec 25;17(24):10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erni B. Glucose transport in Escherichia coli. FEMS Microbiol Rev. 1989 Jun;5(1-2):13–23. doi: 10.1016/0168-6445(89)90004-1. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nakazawa T., Inouye S., Nakazawa A. Physical and functional mapping of RP4-TOL plasmid recombinants: analysis of insertion and deletion mutants. J Bacteriol. 1980 Oct;144(1):222–231. doi: 10.1128/jb.144.1.222-231.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma P. W., Broekhuizen C. P., Geerse R. H. The role of the PEP: carbohydrate phosphotransferase system in the regulation of bacterial metabolism. FEMS Microbiol Rev. 1989 Jun;5(1-2):69–80. doi: 10.1016/0168-6445(89)90010-7. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Hartzell G. W., 3rd Identifying protein-binding sites from unaligned DNA fragments. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Yamada M., Nakazawa A. Factors necessary for the export process of colicin E1 across cytoplasmic membrane of Escherichia coli. Eur J Biochem. 1984 Apr 16;140(2):249–255. doi: 10.1111/j.1432-1033.1984.tb08095.x. [DOI] [PubMed] [Google Scholar]
- Yamada M., Saier M. H., Jr Glucitol-specific enzymes of the phosphotransferase system in Escherichia coli. Nucleotide sequence of the gut operon. J Biol Chem. 1987 Apr 25;262(12):5455–5463. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Schie B. J., Hellingwerf K. J., van Dijken J. P., Elferink M. G., van Dijl J. M., Kuenen J. G., Konings W. N. Energy transduction by electron transfer via a pyrrolo-quinoline quinone-dependent glucose dehydrogenase in Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter calcoaceticus (var. lwoffi). J Bacteriol. 1985 Aug;163(2):493–499. doi: 10.1128/jb.163.2.493-499.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]