Abstract
B-lymphocyte development involves sequential DNA rearrangements of immunoglobulin (Ig) heavy (μ) and light (κ, λ) chain loci and is dependent on transient expression of μ containing pre-antigen receptor complexes (pre-BCR). To date, genetic analysis has not identified transcription factors that coordinate the pre-B-to-B transition. We demonstrate that the related interferon regulatory factors IRF-4 (Pip) and IRF-8 (ICSBP) are required for Ig light but not heavy-chain gene rearrangement. In the absence of these transcription factors, B-cell development is arrested at the cycling pre-B-cell stage and the mutant cells fail to down-regulate the pre-BCR. On the basis of molecular analysis, we propose that IRF-4,8 function as a genetic switch to down-regulate surrogate light-chain gene expression and induce conventional light-chain gene transcription and rearrangement.
Keywords: IRF-4, IRF-8, Ets—IRF complexes, lymphocyte development
IRF-4,8 are two specialized members of the interferon regulatory factor (IRF) family that have been implicated in regulating immunoglobulin (Ig) light-chain gene transcription during B-lymphocyte development (Pongubala et al. 1992; Eisenbeis et al. 1995; Shaffer et al. 1997; Brass et al. 1999; Muljo and Schlissel 2002). They are more highly related to one another than to other IRFs and are selectively expressed in the lymphoid and myeloid lineages of the immune system (for review, see Taniguchi et al. 2001). IRF-4,8 bind very weakly to DNA-containing IRF sites, but are recruited to their binding sites via interactions with other transcription factors. In particular, the related Ets family transcription factors PU.1 and Spi-B have been shown to recruit IRF-4 or IRF-8 to Ets—IRF composite elements (EICE) in Ig κ 3′ and λ enhancers (Pongubala et al. 1992; Eisenbeis et al. 1995; Brass et al. 1999; Escalante et al. 2002). These heterodimeric protein complexes have been implicated in the control of Ig light-chain gene transcription. PU.1 is required for early B-cell development and Spi-B is important for B-cell activation (Scott et al. 1994; Su et al. 1997). It remains to be determined whether PU.1/Spi-B regulate Ig light-chain gene expression during the pre-B-to-B transition. IRF-4-/- mutant mice exhibit profound defects in B- and T-cell activation that are manifested in the failure to mount antibody, cytotoxic, or antitumor responses (Mittrucker et al. 1997; Rengarajan et al. 2002). IRF-8-/- mutant mice are impaired in macrophage development, highly susceptible to viral infections, and manifest a chronic myelogenous leukemia-like syndrome (Holtschke et al. 1996). Loss of either IRF-4 or IRF-8 does not block the generation of B lymphocytes. Given their structural relatedness and similar molecular properties, we sought to examine whether they function redundantly to regulate Ig light-chain gene expression and the pre-B-to-B transition.
Results and Discussion
B-cell development is arrested at the pre-B stage in IRF-4,8-/- mice
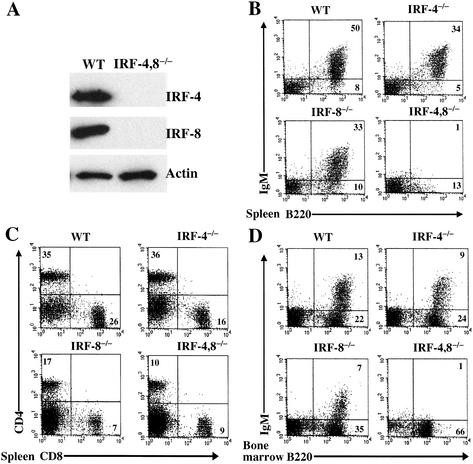
IRF-4,8-/- compound mutants were derived from the single knockouts at the expected Mendelian ratio. Loss of IRF-4,8 expression was confirmed by Western blot analysis of mutant splenocytes (Fig. 1A). Strikingly, the double mutant mice lacked sIgM+ peripheral B lymphocytes (Fig. 1B). In contrast, splenic T lymphocytes, (CD4+ or CD8+) were present in the double mutant mice, albeit with reduced numbers (Fig. 1C). We note that loss of IRF-4 and IRF-8 does not impair T-cell development in the thymus (data not shown). We next examined B-cell generation in the bone marrow. Immature IgM+ B cells were absent in the IRF-4,8-/- bone marrow (Fig. 1D). These results establish that the combined loss of IRF-4,8 causes a lineage-specific block to B-cell development. It should be noted that as is the case for IRF-8-/- mice, there was an increase in the numbers of myeloid cells (Mac-1+, Gr-1+) in the bone marrow and spleen of IRF-4,8-/- mice (data not shown).
Figure 1.
B-cell development is blocked in IRF-4,8-/- mice. (A) Western blot analysis of IRF-4 and IRF-8 expression in splenocytes from wild-type and IRF-4,8-/- mice. (B–D) FACS analysis of B- and T-lymphocyte populations in spleen and bone marrow of wild-type and IRF-4,8 mutant mice. Cells were examined using a lymphocyte gate and antibodies directed against B220, IgM, CD4, and CD8. Numbers in quadrants indicate percentage of cells in the gated population.
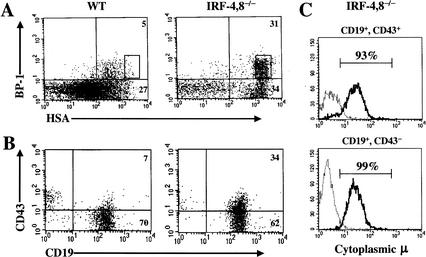
B-lineage intermediates are characterized on the basis of sequential DNA rearrangements of Ig loci as well as the expression of specific cell-surface markers (for review, see Hardy and Hayakawa 2001). The pro-B compartment can be analyzed using a combination of anti-B220, CD43, HSA, and BP-1 antibodies (Hardy et al. 1991). The IRF-4,8-/- B-lineage cells progressed up to the stage delineated by the coexpression of B220, CD43, BP-1, and HSA (Fig. 2A). The levels of HSA expression on the mutant B220+, CD43+, BP-1+, B-lineage cells were particularly high, suggesting that they represent transitional pre-B cells, which are a subset of fraction C cells in the nomenclature of Hardy et al. (1991). The disproportionate increase in the percentage fraction C cells in the IRF-4,8-/- bone marrow (Fig. 2A) was also reflected in a substantial increase in their absolute numbers. Wild-type bone marrow was found to contain 0.07 ± 0.01 × 106 fraction C cells. In contrast, the IRF-4,8-/- bone marrow contained 0.32 ± 0.14 × 106 fraction C cells, representing a four- to fivefold increase. The pre-B compartment was analyzed on the basis of CD19 and CD43 expression. The mutant cells were uniformly CD19+, and a majority had down-regulated CD43 expression (Fig. 2B). This cell-surface phenotype is consistent with a block at the pre-B stage. The hallmark of a pre-B cell is the productive rearrangement of the IgH locus resulting in the expression of μ protein, most of which is retained in the endoplasmic reticulum (for review, see Hardy and Hayakawa 2001). To verify the nature of the block at the pre-B stage, we determined whether the mutant cells were expressing μ protein in their cytoplasm. The vast majority of CD19+, CD43+, and CD19+, CD43- mutant B-lineage cells expressed μ protein in the cytoplasm. The former subset likely represents the transitional pre-B cells described above. These results suggest that IRF-4,8-/- mutant pre-B cells are strongly selected and undergo expansion in the bone marrow.
Figure 2.
B-cell development is arrested at the pre-B stage in IRF-4,8-/- mice. (A) Bone marrow cells were analyzed for expression of B220, CD43, BP-1, and HSA using four-color FACS. The BP-1 and HSA profiles were obtained after gating on B220+, CD43+ cells. (B) Bone marrow cells were analyzed for CD19 and CD43 expression after gating on B220+ lymphocytes. (C) Intracellular μ expression in CD19+ CD43+, or CD19+ CD43- IRF-4,8-/- mutant B-lineage cells. Myeloid-lineage-depleted cells (Mac-1-, Gr-1-) from IRF-4,8-/- bone marrow were stained with anti-CD19 and anti-CD43 antibodies. After fixation and permeabilization, the cells were reacted with an anti-IgM antibody (solid line) or an isotype control (broken line), and then analyzed by FACS.
IRF-4,8-/; pre-B cells are cycling and express high levels of the pre-B-cell receptor (pre-BCR)
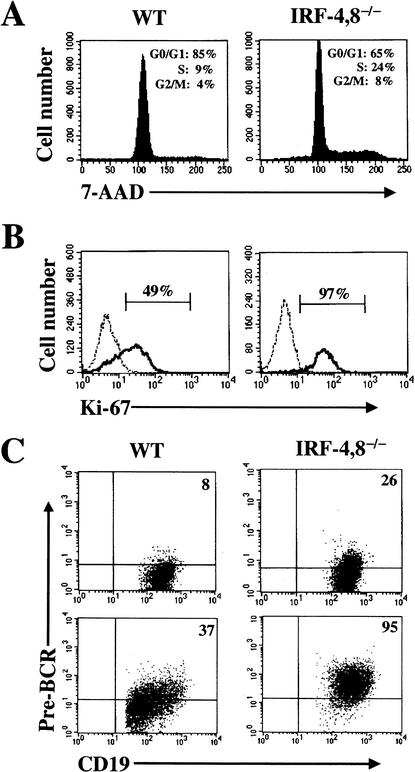
The μ chain in pre-B cells associates with surrogate light chains λ5 and VpreB along with the signaling subunits Igα and β to form the pre-BCR, which signals survival and cell expansion. Down-regulation of the pre-BCR has been linked to exit from the cell cycle and the induction of Ig light-chain (IgL) gene rearrangement (for review, see Melchers et al. 2000; Burrows et al. 2002). We examined the cell cycle status of the IRF-4,8-/- cells by analyzing genomic DNA content with 7-AAD and also by measuring intracellular levels of the proliferation associated antigen Ki-67. In contrast to their wild-type counterparts, a much larger fraction of the mutant pre-B cells (32%) were in the S and G2/M phases of the cell cycle (Fig. 3A). Consistent with this result, the vast majority of mutant pre-B cells expressed Ki-67 (Fig. 3B). Thus, loss of IRF-4,8 appears to result in a failure of pre-B cells to exit the cell cycle. Because cycling of pre-B cells is dependent on signaling from the pre-BCR, we examined its expression in the mutant cells using an antibody that recognizes the assembled receptor. Intriguingly, pre-BCR complexes were seen to be expressed at higher levels on the surface of both freshly isolated (Fig. 3C, top) as well as cultured pre-B cells (Fig. 3C, bottom). This observation strongly suggests that the mutant pre-B cells fail to down-regulate pre-BCR in the absence of IRF-4,8, thereby remaining in a proliferative state. Cycling pre-B cells can also be enumerated by a colony-forming assay using semisolid medium containing IL-7. In wild-type bone marrow, pre-B CFU were detected at a frequency of 30± 18/105 cells. In contrast, IRF-4,8-/- bone marrow was found to have an eightfold increase in pre-B CFU (240± 118/105 cells). This observation is consistent with the conclusion that loss of IRF-4,8 results in accumulation of cycling pre-B cells in the bone marrow. Interestingly, some of these mutant pre-B cells may exit the bone marrow as they were also detected using the pre-B CFU assay in peripheral lymphoid organs such as the spleen and lymph nodes (data not shown).
Figure 3.
IRF-4,8-/- pre-B cells have an increased proliferation index and express higher levels of the pre-BCR. (A,B) Cell cycle status of IRF-4,8-/- pre-B cells. Bone marrow cells after enriching for sIgM—B-lineage cells were stained with anti-CD19 and anti-CD43 antibodies. After fixation and permeabilization, the cells were incubated with either 7-AAD or anti-Ki-67 antibody. FACS analysis was performed by gating on CD19+ CD43- cells. Ki-67 staining was assessed using an isotype-matched antibody as control (broken line). (C) Expression of pre-BCR on IRF-4,8-/- pre-B cells. (Top) Freshly isolated bone marrow cells were analyzed by four-color FACS using anti-CD19, CD43, IgM, and pre-BCR antibodies. The FACS profiles were obtained by gating on CD19+, CD43- and IgM-, and cells. (Bottom) The same FACS analysis was performed using pre-B cells that were expanded in culture using IL-7. For each panel, the pre-BCR quadrant was set using an isotype control.
IRF-4,8-/- pre-B cells are impaired for light-chain gene transcription and rearrangement
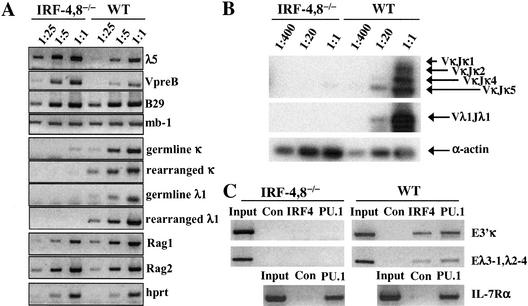
Recent evidence suggests that down-regulation of the pre-BCR may be initiated by the inhibition of surrogate light-chain gene expression (Wang et al. 2002). We therefore examined the expression levels of the λ5 and VpreB genes in CD19+,CD43-IRF-4,8-/- pre-B cells. These two genes were expressed at approximately fivefold higher levels in the mutant pre-B cells than in their wild-type counterparts (Fig. 4A). Importantly, expression of the mb-1 and B29 genes that encode the Ig α and β subunits of the pre-BCR was unaffected by the loss of IRF-4,8. These results suggest that the increased levels of the pre-BCR in the IRF-4,8-/- cells is due to a specific failure to inhibit surrogate light-chain gene expression. It remains to be determined whether IRF-4,8 directly repress the transcription of the λ5 and VpreB genes. In striking contrast, germ line as well as rearranged transcripts from the conventional Ig light-chain loci (κ and λ) were barely detectable in the mutant cells, reduced at least 25-fold when compared with their levels in wild-type pre-B cells. In addition to defects in IgL-chain gene transcription, the IRF-4,8-/- pre-B cells evidenced a profound block in DNA rearrangements at these loci (Fig. 4B).
Figure 4.
IRF-4,8-/- pre-B cells express high levels of surrogate light-chain genes (λ5, VpreB) and are impaired for conventional light-chain (κ, λ) transcription and rearrangement. (A) Reverse transcriptase PCR (RT—PCR) analysis of B-lineage genes expressed in IRF-4,8-/- pre-B cells. CD19+, CD43-, and IgM- pre-B cells were isolated by sorting after enrichment of B-lineage cells from bone marrow. Serial dilutions of total cDNA were used for RT—PCR with primers for the indicated genes. (B) PCR analysis of κ and λ DNA rearrangements. Serial dilutions of genomic DNA from the cells described in A were analyzed using primers that detect the indicated κ and λ rearrangements. (C) Chromatin cross-linking and immunoprecipitation analysis of IRF-4 and PU.1 binding to the κ 3′ and λ enhancers. IL-7 expanded pre-B cells, as described in Figure 3C, were cross-linked with formaldehyde, and then chromatin fragments were immunoprecipitated using anti-IRF-4 or anti-PU.1 antibodies. Rabbit IgG antibodies were used as a control. Indicated DNA fragments were amplified by PCR. Input represents an equivalent amount of cross-linked chromatin not subjected to immunoprecipitation.
Ig light-chain gene transcription and recombination are dependent on the activity of several enhancer elements (Hagman et al. 1990; Gorman et al. 1996; Xu et al. 1996; Inlay et al. 2002). The κ locus has two distinct enhancers (Eiκ and E3′κ), whereas the λ locus has two duplicated enhancers (Eλ2-4, Eλ3-1). Experiments performed with B-lineage cell lines have shown that IRF-4 or IRF-8 coassemble with the ets family transcription factors PU.1 or Spi-B to regulate the activity of Ig κ 3′ and λ enhancers (Pongubala et al. 1992; Eisenbeis et al. 1995; Brass et al. 1999). We used chromatin cross-linking and immunoprecipitation assays with bone marrow derived pre-B cells to determine whether PU.1 and IRF-4 are bound to the 3′ κ and λ enhancers in vivo (Fig. 4C). Both transcription factors were bound to these enhancers in wild-type pre-B cells. Intriguingly, loss of IRF-4,8 resulted in diminished PU.1 cross-linking, suggesting that IRF-4,8 may be required to stabilize the binding of PU.1 to Ig enhancer DNA in vivo. Alternatively, the lack of PU.1 binding to these enhancers may reflect the failure of IRF-4,8-/- pre-B cells to differentiate to a stage in which the chromatin of Ig light-chain loci becomes accessible for PU.1 binding. Importantly, PU.1 cross-linking to a binding site in the IL-7Rα gene was not affected by loss of IRF-4,8 (Fig. 4C, bottom). This site is not a composite element (DeKoter et al. 2002) and, therefore, PU.1 binding to it in vivo is not perturbed by the absence of IRF-4,8. We suggest that loss of IRF-4,8 results in a severe impairment in the functioning of the κ 3′ and λ enhancers in vivo. This is consistent with defects in Ig κ germ line and λ transcription in IRF-4,8-/- pre-B cells, as they have been shown to depend on E3′κ and λ enhancers, respectively (Brass et al. 1999; Inlay et al. 2002; Muljo and Schlissel 2003).
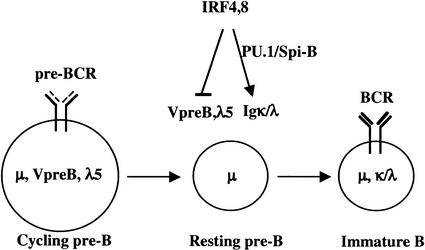
A model for transcriptional control of the pre-B-to-B transition
The strong block to Ig κ locus rearrangement observed in IRF-4,8-/- pre-B cells cannot be simply accounted for by inactivity of the κ 3′ enhancer. It has been established that endogenous κ rearrangement can be mediated by either the κ intron or 3′ enhancer (Gorman et al. 1996; Xu et al. 1996; Inlay et al. 2002). We note that the intron enhancer does not contain an Ets—IRF composite element that can be recognized by PU.1/Spi-B and IRF-4/8 complexes. Ig DNA rearrangement is also dependent on the proliferative state of pre-B, as Rag-1,2 gene expression and Rag-2 protein stability are cell-cycle regulated (for review, see Desiderio et al. 1996). The mutant pre-B cells express fivefold lower levels of Rag-1 transcripts (Fig. 4A). Therefore, we suggest that the profound block to Ig light-chain gene rearrangements in the mutant pre-B cells is due to defects in enhancer activity as well as the inability to exit the cell cycle, and therefore induce Rag gene and protein expression. These results lead us to propose a regulatory model for the pre-B-to-B transition in which IRF-4,8 appear to function as a genetic switch (Fig. 5). In this model, IRF-4,8 coordinate the pre-B-to-B transition in part by down-regulating surrogate light-chain gene expression, thereby facilitating cell-cycle exit and up-regulation of Rag gene and protein expression. Concomitantly, IRF-4,8 directly induce enhancer-dependent light-chain transcription and recombination, thereby enabling pre-B-cell differentiation.
Figure 5.
A model for transcriptional control of the pre-B-to-B transition. IRF-4,8 are depicted as components of a genetic switch whose function is to down-regulate expression of the surrogate light-chain genes, thereby inhibiting pre-BCR-dependent proliferation of pre-B cells. Concomitantly, IRF-4,8 promote pre-B-cell differentiation by inducing conventional light-chain gene transcription and rearrangement. These latter processes are proposed to depend on binding of IRF-4/8 to κ and λ enhancers with the transcription factors PU.1/Spi-B.
Signaling by the pre-BCR appears to regulate survival, proliferation, and possibly differentiation of pre-B cells (for review, see Melchers et al. 2000; Muljo and Schlissel 2000; Burrows et al. 2002). Knock-out mutations in components of the pre-BCR such as μ heavy chain, λ5 and Ig β result in inhibition of the pro-B to pre-B transition (Kitamura et al. 1991, 1992; Gong and Nussenzweig 1996). A similar developmental block is observed with mutations in genes encoding the signaling molecules Syk and PI3K, which are required for pre-BCR-dependent survival and proliferation (Cheng et al. 1995; Turner et al. 1995; Fruman et al. 1999). Recently, the adapter molecule BLNK (SLP65) has been implicated in regulating the pre B-to-B transition (Jumaa et al. 1999; Flemming et al. 2003). BLNK-/- pre-B cells show enhanced proliferation capacity in culture and express high levels of pre-BCR. We note that IRF-4,8-/- pre-B cells express wild-type levels of BLNK (data not shown). These results raise an intriguing possibility that IRF-4,8 could function as nuclear effectors of a pre-BCR-initiated signaling pathway, modulated by BLNK, that feeds back to limit pre-B-cell expansion and induces differentiation.
Materials and methods
Generation of IRF-4,8-/- mice
The IRF4,8-/- mice were generated by initially crossing IRF-4-/- and IRF-8-/- mice of mixed B6x129 background. The resulting compound heterozygotes (IRF-4+/-, IRF-8+/-) mice were intercrossed to generate IRF-4,8-/- mice. All mice were used at 4–6 wk of age and were maintained in a pathogen-free facility.
Antibodies and flow cytometry
Flow cytometry analysis was performed on single-cell suspensions washed in Dulbecco's PBS containing 5 mM EDTA and 0.5% BSA (Roche). Cells were stained with fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)-, allophycocyanin (APC)-conjugated or biotin-conjugated antibodies. In the latter case, streptavidin (SA)-PE-Cy7 was used as a secondary reagent. Anti-mouse monoclonal antibodies were purchased from Pharmingen unless otherwise indicated and used according to supplier's recommendation. The antibodies used included RA3-6B2(B220); S7(CD43); 1D3(CD19); M1/69(HSA); 6C3(BP-1); RB6-8C5-biotin (Gr-1); M1/70-biotin (Mac-1); Ter119-biotin; II/41-Biotin (IgM); 145-2C11-biotin (CD3ε); SL-156-biotin (pre-BCR); anti-Ki-67; anti-IgM-RPE (Southern Biotechnology Associates, Inc). FACS analysis was performed using the FACS Calibur flow cytometer.
Intracellular staining and cell-cycle analysis with 7-AAD
Intracellular staining was performed as described by Medina et al. (2001). Bone marrow pro-B/pre-B cells were enriched by negative selection using biotinylated anti-Gr-1, Mac-1, Ter119, CD3ε, and IgM antibodies, strepavidin-conjugated magnetic beads and MACS columns (Militenyi Biotec). The enriched cells were incubated with anti-CD19(APC), CD43(PE) antibodies and fixed in 1% paraformaldehyde at 4°C overnight. After permeabilization with 70% ethanol for 40 min at -20°C, the cells were incubated either with anti-Ki-67(FITC) or anti-IgM(FITC) for 30 min on ice, and then analyzed by FACS. For cell-cycle analysis, the enriched pro-B/pre-B cells were incubated with anti-CD19(FITC) and CD43(PE) antibodies prior to fixation. Cells were stained with 7-AAD (Pharmingen) and analyzed by FACS.
Pre-B-cell cultures
B220+ cells were isolated from bone marrow using a MACS separation column and were cultured in IL-7 (R&D Systems) at 5 ng/mL for 5–10 d in OptiMEM (GIBCO-BRL) medium supplemented with 5% FCS, 50 μM β-mercaptoethanol, 2 mM L-glutamine, and 100 U/mL penicillin-streptomycin. Cultures contained >95% μ+ pre-B cells.
Pre-B-cell colony formation assay
Freshly isolated bone marrow, lymph node, and spleen cells were mixed with Methocult M3630 medium (StemCell Technologies) in a 1:10(v/v) ratio. Cells (5 × 104) were dispensed in triplicate into a 6-well plate. Colonies were scored after 7 d of growth at 37°C.
Analysis of transcription and Ig light-chain gene recombination in pre-B cells
After lineage depletion (see above), bone marrow-derived pre-B cells (CD19+,CD43-,IgM-) were isolated using a MoFlo cell sorter. Total RNA and DNA were isolated using Trizol reagent (GIBCO-BRL). RNA was reverse transcribed using a first-strand cDNA synthesis kit (Amersham Biosciences). PCR primers used to analyze B-lineage gene expression and Ig light-chain gene rearrangement have been described previously (Sun and Storb 2001; DeKoter et al. 2002; Inlay et al. 2002; Wang et al. 2002). Serial dilutions of cDNA or genomic DNA were used for the PCR reactions. DNA rearrangement PCR reactions were analyzed by Southern blotting after agarose gel electorphoresis.
Chromatin cross-linking and immunoprecipitation assay
IL-7 cultured pre-B cells (see above) were used for ChIP assays as described previously (DeKoter et al. 2002). The following antibodies: anti-IRF-4 (Santa Cruz), anti-PU.1 (Santa Cruz), and rabbit IgG (Santa Cruz) were used for immunoprecipitation of formaldehyde cross-linked chromatin. The sequences of primer pairs used for the amplification are as follows: κ 3′enhancer, sense (GTTATTGATTTTCATGGTGACTG GC), antisense (AGCAGGTGTATGAGGCTTTGGAAAC); λ enhancers, sense (CACCCTTGAGAGTCCACAAGC), antisense (GTCCCTAAC ATTTAGATGC); IL-7Rα primers are described in DeKoter et al. (2002).
Acknowledgments
We thank Tak Mak (OCI) and Keiko Ozato (NIH) for the IRF-4 and IRF-8 knockout mice, respectively. We acknowledge assistance for flow cytometry services and animal care by UCCRC and ARC core facilities. We thank Eric Bertolino for helpful discussions. R.L. is a fellow of the Leukemia and Lymphoma Society, K.M. is a fellow of the Irvington Institute, and H.S. is an Investigator with the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1104803.
References
- Brass A.L., Zhu, A.Q., and Singh, H. 1999. Assembly requirements of PU.1–Pip (IRF-4) activator complexes: Inhibiting function in vivo using fused dimers. EMBO J. 18: 977–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows P.D., Stephan, R.P., Wang, Y.H., Lassoued, K., Zhang, Z., and Cooper, M.D. 2002. The transient expression of pre-B cell receptors governs B cell development. Semin. Immunol. 14: 343–349. [DOI] [PubMed] [Google Scholar]
- Cheng A.M., Rowley, B., Pao, W., Hayday, A., Bolen, J.B., and Pawson, T. 1995. Syk tyrosine kinase required for mouse viability and B-cell development. Nature 378: 303–306. [DOI] [PubMed] [Google Scholar]
- DeKoter R.P., Lee, H.J., and Singh, H. 2002. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity 16: 297–309. [DOI] [PubMed] [Google Scholar]
- Desiderio S., Lin, W.C., and Li, Z. 1996. The cell cycle and V(D)J recombination. Curr. Top. Microbiol. Immunol. 217: 45–59. [DOI] [PubMed] [Google Scholar]
- Eisenbeis C.F., Singh, H., and Storb, U. 1995. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes & Dev. 9: 1377–1387. [DOI] [PubMed] [Google Scholar]
- Escalante C.R., Brass, A.L., Pongubala, J.M., Shatova, E., Shen, L., Singh, H., and Aggarwal, A.K. 2002. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol. Cell 10: 1097–1105. [DOI] [PubMed] [Google Scholar]
- Flemming A., Brummer, T., Reth, M., and Jumaa, H. 2003. The adaptor protein SLP-65 acts as a tumor suppressor that limits pre-B cell expansion. Nat. Immunol. 4: 38–43. [DOI] [PubMed] [Google Scholar]
- Fruman D.A., Snapper, S.B., Yballe, C.M., Davidson, L., Yu, J.Y., Alt, F.W., and Cantley, L.C. 1999. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85α. Science 283: 393–397. [DOI] [PubMed] [Google Scholar]
- Gong S. and Nussenzweig, M.C. 1996. Regulation of an early developmental checkpoint in the B cell pathway by Ig β. Science 272: 411–414. [DOI] [PubMed] [Google Scholar]
- Gorman J.R., van der Stoep, N., Monroe, R., Cogne, M., Davidson, L., and Alt, F.W. 1996. The Ig(κ) enhancer influences the ratio of Ig(κ) versus Ig(λ) B lymphocytes. Immunity 5: 241–252. [DOI] [PubMed] [Google Scholar]
- Hagman J., Rudin, C.M., Haasch, D., Chaplin, D., and Storb, U. 1990. A novel enhancer in the immunoglobulin λ locus is duplicated and functionally independent of NF κ B. Genes & Dev. 4: 978–992. [DOI] [PubMed] [Google Scholar]
- Hardy R.R. and Hayakawa, K. 2001. B cell development pathways. Annu. Rev. Immunol. 19: 595–621. [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Carmack, C.E., Shinton, S.A., Kemp, J.D., and Hayakawa, K. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtschke T., Lohler, J., Kanno, Y., Fehr, T., Giese, N., Rosenbauer, F., Lou, J., Knobeloch, K.P., Gabriele, L., Waring, J.F., et al. 1996. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87: 307–317. [DOI] [PubMed] [Google Scholar]
- Inlay M., Alt, F.W., Baltimore, D., and Xu, Y. 2002. Essential roles of the κ light chain intronic enhancer and 3′ enhancer in κ rearrangement and demethylation. Nat. Immunol. 3: 463–468. [DOI] [PubMed] [Google Scholar]
- Jumaa H., Wollscheid, B., Mitterer, M., Wienands, J., Reth, M., and Nielsen, P.J. 1999. Abnormal development and function of B lymphocytes in mice deficient for the signaling adaptor protein SLP-65. Immunity 11: 547–554. [DOI] [PubMed] [Google Scholar]
- Kitamura D., Roes, J., Kuhn, R., and Rajewsky, K. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 350: 423–426. [DOI] [PubMed] [Google Scholar]
- Kitamura D., Kudo, A., Schaal, S., Muller, W., Melchers, F., and Rajewsky, K. 1992. A critical role of λ 5 protein in B cell development. Cell 69: 823–831. [DOI] [PubMed] [Google Scholar]
- Medina K.L., Garrett, K.P., Thompson, L.F., Rossi, M.I., Payne, K.J., and Kincade, P.W. 2001. Identification of very early lymphoid precursors in bone marrow and their regulation by estrogen. Nat. Immunol. 2: 718–724. [DOI] [PubMed] [Google Scholar]
- Melchers F., ten Boekel, E., Seidl, T., Kong, X.C., Yamagami, T., Onishi, K., Shimizu, T., Rolink, A.G., and Andersson, J. 2000. Repertoire selection by pre-B-cell receptors and B-cell receptors, and genetic control of B-cell development from immature to mature B cells. Immunol. Rev. 175: 33–46. [PubMed] [Google Scholar]
- Mittrucker H.W., Matsuyama, T., Grossman, A., Kundig, T.M., Potter, J., Shahinian, A., Wakeham, A., Patterson, B., Ohashi, P.S., and Mak, T.W. 1997. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science 275: 540–543. [DOI] [PubMed] [Google Scholar]
- Muljo S.A. and Schlissel, M.S. 2000. Pre-B and pre-T-cell receptors: Conservation of strategies in regulating early lymphocyte development. Immunol. Rev. 175: 80–93. [PubMed] [Google Scholar]
- ____. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat. Immunol. 4: 31–37. [DOI] [PubMed] [Google Scholar]
- Pongubala J.M., Nagulapalli, S., Klemsz, M.J., McKercher, S.R., Maki, R.A., and Atchison, M.L. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin κ 3′ enhancer activity. Mol. Cell. Biol. 12: 368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J., Mowen, K.A., McBride, K.D., Smith, E.D., Singh, H., and Glimcher, L.H. 2002. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195: 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E.W., Simon, M.C., Anastasi, J., and Singh, H. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265: 1573–1577. [DOI] [PubMed] [Google Scholar]
- Shaffer A.L., Peng, A., and Schlissel, M.S. 1997. In vivo occupancy of the κ light chain enhancers in primary pro- and pre-B cells: A model for κ locus activation. Immunity 6: 131–143. [DOI] [PubMed] [Google Scholar]
- Su G.H., Chen, H.M., Muthusamy, N., Garrett-Sinha, L.A., Baunoch, D., Tenen, D.G., and Simon, M.C. 1997. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 16: 7118–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T. and Storb, U. 2001. Insertion of phosphoglycerine kinase (PGK)neo 5′ of Jλ1 dramatically enhances VJλ1 rearrangement. J. Exp. Med. 193: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Ogasawara, K., Takaoka, A., and Tanaka, N. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19: 623–655. [DOI] [PubMed] [Google Scholar]
- Turner M., Mee, P.J., Costello, P.S., Williams, O., Price, A.A., Duddy, L.P., Furlong, M.T., Geahlen, R.L., and Tybulewicz, V.L. 1995. Perinatal lethality and blocked B-cell development in mice lacking the tyrosine kinase Syk. Nature 378: 298–302. [DOI] [PubMed] [Google Scholar]
- Wang Y.H., Stephan, R.P., Scheffold, A., Kunkel, D., Karasuyama, H., Radbruch, A., and Cooper, M.D. 2002. Differential surrogate light chain expression governs B-cell differentiation. Blood 99: 2459–2467. [DOI] [PubMed] [Google Scholar]
- Xu Y., Davidson, L., Alt, F.W., and Baltimore, D. 1996. Deletion of the Ig κ light chain intronic enhancer/matrix attachment region impairs but does not abolish V κ J κ rearrangement. Immunity 4: 377–385. [DOI] [PubMed] [Google Scholar]