Abstract
Streptomycetes exhibit a complex morphological differentiation. After a submerged mycelium has been formed, filaments grow into the air to septate into spores. A class of eight hydrophobic secreted proteins, ChpA–H, was shown to be instrumental in the development of Streptomyces coelicolor. Mature forms of ChpD–H are up to 63 amino acids in length, and those of ChpA–C are larger (±225 amino acids). ChpA–C contain two domains similar to ChpD–H, as well as a cell-wall sorting signal. The chp genes were expressed in submerged mycelium (chpE and chpH) as well as in aerial hyphae (chpA–H). Formation of aerial hyphae was strongly affected in a strain in which six chp genes were deleted (ΔchpABCDEH). A mixture of ChpD–H purified from cell walls of aerial hyphae complemented the ΔchpABCDEH strain extracellularly, and it accelerated development in the wild-type strain. The protein mixture was highly surface active, and it self-assembled into amyloid-like fibrils at the water–air interface. The fibrils resembled those of a surface layer of aerial hyphae. We thus conclude that the amyloid-like fibrils of ChpD–H lower the water surface tension to allow aerial growth and cover aerial structures, rendering them hydrophobic. ChpA–C possibly bind ChpD–H to the cell wall.
Keywords: Streptomycetes, aerial hyphae, surface protein, chaplins, amyloid, surface activity
Streptomyces coelicolor is a representative of the group of soil-dwelling, filamentous bacteria responsible for producing most natural antibiotics used today in medicine. It has become a model organism to study bacterial differentiation with the great advantage of the availability of the complete genome sequence (Bentley et al. 2002). Its life cycle resembles that of filamentous fungi. After a feeding submerged mycelium is established, aerial hyphae are formed that eventually septate into chains of spores. These spores are dispersed by wind or insects to give rise to a new vegetative mycelium. Surface characteristics change dramatically when submerged hyphae grow into the air. Surfaces of submerged hyphae are hydrophilic, whereas those of aerial hyphae and spores are hydrophobic. This change in surface hydrophobicity may prevent backgrowth of hyphae into the moist substrate, facilitate dispersal of spores, and mediate attachment to hydrophobic surfaces such as that of a host.
Aerial growth and differentiation in S. coelicolor has been defined genetically by the isolation of bld mutants that lack aerial hyphae and, therefore, have a “bald” appearance (for reviews, see Chater 1998, 2001; Kelemen and Buttner 1998; Wösten and Willey 2000). This phenotype correlates with the failure to produce and secrete SapB (Willey et al. 1991, 1993). SapB is a small peptide of 18 amino acids with a high surface activity. By lowering the water surface tension of the aqueous environment from 72 to 32 mJ/m2, SapB enables hyphae to overcome the physical barrier posed by the medium–air interface (Tillotson et al. 1998). SapB is produced on complex agar media but not on minimal media supplemented with mannitol or arabinose (Willey et al. 1991). On the latter media, other molecules are expected to fulfill a role similar to that of SapB.
Application of purified SapB to growing colonies of S. coelicolor bld mutants restores the capacity to form aerial hyphae that, however, do not sporulate (Tillotson et al. 1998). The vegetative nature of these aerial hyphae indicates that SapB enables hyphae to grow into the air but has no role as a signaling molecule in differentiation. A function of SapB, once hyphae have escaped the aqueous environment, is also not envisaged because SapB could only be detected in the medium but not at surfaces of aerial hyphae and spores (Wösten and Willey 2000). Thus, the change in surface characteristics accompanying aerial growth should be attributed to other molecules.
Aerial hyphae and spores of streptomycetes possess a surface layer, called the rodlet layer, with a typical ultrastructure of a mosaic of 8–10-nm-wide parallel rods (Wildermuth et al. 1971; Smucker and Pfister 1978; Claessen et al. 2002). Two homologous proteins, the rodlins RdlA and RdlB, are necessary for formation of this layer (Claessen et al. 2002). RdlA and RdlB are secreted into the cell wall of growing aerial hyphae, where they form a highly stable insoluble surface layer. Deletion of both rdl genes resulted in loss of the rodlet layer (Claessen et al. 2002). Interestingly, the water-repellent nature of aerial hyphae and spores was not affected, implying the existence of other hydrophobic compounds. In this study we describe the identification and characterization of these molecules that comprise a novel class of proteins, called chaplins. Chaplins lower the water surface tension of the aqueous environment, thus enabling hyphae to grow into the air, and coat aerial structures, making them hydrophobic. They function by self-assembly into amyloid-like fibrils.
Results
Identification and characterization of the chaplins of S. coelicolor
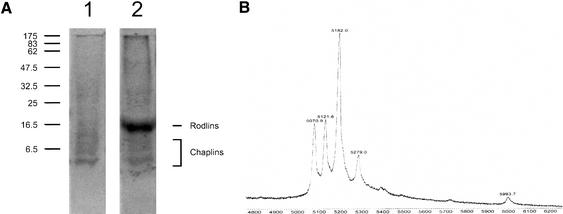
Recently, RdlA and RdlB of S. coelicolor were identified as proteins extractable with trifluoroacetic acid (TFA) that form an SDS-insoluble layer at the outer surface of aerial hyphae. Deleting the rdlA and rdlB genes resulted in the absence of the outer rodlet layer but did not affect surface hydrophobicity (Claessen et al. 2002). To isolate the molecules that are involved in the surface hydrophobicity of aerial structures, we performed a TFA extraction on SDS-extracted cell walls from a 5-day-old sporulating culture of an S. coelicolor strain in which both rdlA and rdlB were deleted (M145ΔrdlAB). SDS-PAGE of the TFA extract, followed by staining with Coomassie Brilliant Blue (CBB), revealed a faint smear between 5 and 10 kD (Fig. 1A). The intensity of staining slightly increased by staining with silver (data not shown). A similar smear was observed in wild-type extracts that contained the rodlins RdlA and RdlB as the major proteins (Fig. 1A). Masses of the proteins in the TFA extracts of the SDS-extracted cell walls of S. coelicolor wild-type and ΔrdlAB strains were determined by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectroscopy. Five discrete peaks could be identified in extracts of the ΔrdlAB strain with molecular masses ranging from 5071 to 5994 (Fig. 1B). Peptides with identical masses were identified in the wild-type extract that also contained proteins with masses corresponding to those of RdlA and RdlB (data not shown). The 5-kD peptides were not observed in cell walls of 1-day-old cultures growing submerged only. They appeared at day 2, coinciding with formation of aerial hyphae and were shown to be still present in 7-day-old cultures that completed sporulation. Amounts of these proteins in cell walls of differentiating cultures were considerable, comprising ∼1% of the dry weight.
Figure 1.
(A) SDS-PAGE analysis of TFA extracts of SDS-treated cell walls of sporulating cultures of S. coelicolor M145ΔrdlAB (lane 1) and wild-type strain M145 (lane 2). (B) A faint smear is observed between 5 and 10 kD, consisting of a mixture of five small proteins, called ChpD–H, with masses ranging from 5071 to 5994 Da as determined by MALDI-TOF mass spectrometry.
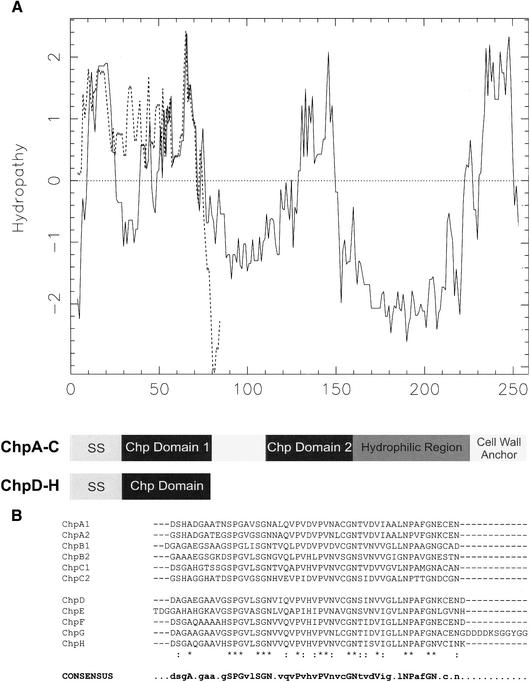
N-terminal sequencing of the proteins running at 5–10 kD in SDS-PAGE revealed one to three amino acids at each of the first six positions [(D/T)(A/D/S)(G)(A/G)(A/E/Q)(A/G/H)]. This information was used for a protein motif search in the genome sequence of S. coelicolor (http://www.sanger.ac.uk/Projects/S_coelicolor). Among the 118 hits, four proteins were identified with a molecular weight between 5 and 10 kD. These protein sequences were blasted against the complete genome sequence of S. coelicolor, resulting in the identification of eight homologous genes, which we called chaplins, for coelicolor hydrophobic aerial proteins, with gene designations chpA–H. The genes, except for chpB, are located in the core region of the linear chromosome of S. coelicolor. Genes chpC and chpH, as well as chpA and chpD, are located next to each other, the latter genes being adjacent to the rodlin genes rdlA and rdlB. The chp genes encode proteins (Table 1, Fig. 2) with putative signal sequences for secretion, as determined with SignalP (http://www.cbs.dtu.dk/services/SignalP-2.0). Experimental masses and N-terminal sequences of the proteins in the TFA extract of the ΔrdlAB strain agreed with those predicted for ChpD–H, assuming cleavage of their putative signal sequences (Table 1). Thus, ChpA–C were not identified in the cell wall extract. These chaplins are predicted to be larger than those that were extracted. In fact, they consist of two sequences very similar to those of the extractable chaplins. These sequences are separated by a stretch of 27–30 amino acids and are followed by a hydrophilic region (Fig. 2A). The C terminus of ChpA–C contains an LAXTG sequence, followed by a membrane-spanning domain and a charged C-terminal tail. This is a typical motif for anchoring surface proteins to the peptido-glycan in the cell wall (Mazmanian et al. 1999; Pallen et al. 2001). The similarity between the mature forms of the small chaplins and the corresponding sequences in the large chaplins is high: 33% of the amino acids are identical with an additional similarity of 18% when all homologous sequences are compared (Fig. 2B). Hydropathy patterns show that these sequences are quite hydrophobic (Fig. 2A). Genomic DNA of Streptomyces tendae, Streptomyces griseus, and the potato pathogen Streptomyces scabies hybridized with the coding sequence of chpD, indicating the presence of chp genes in other streptomycetes as well (data not shown).
Table 1.
Chaplins encoded in the genome of S. coelicolor
| Name | Number of amino acids (with/without signal sequence) | Predicted N-terminal sequence of the mature protein | Predicted molecular mass of the mature protein (Dalton) | Determined molecular mass of the mature protein (Dalton) |
|---|---|---|---|---|
| ChpA (SCO2716) | 252/232 | DSHADG | 19,876 a | ND |
| ChpB (SCO7257) | 237/208 | SDGAGA | 16,595 a | ND |
| ChpC (SCO1674) | 259/231 | DSGAHG | 18,863 a | ND |
| ChpD (SCO2717) | 75/52 | DAGAEG | 5071 | 5071 |
| ChpE (SCO1800) | 82/55 | TDGGAH | 5274 | 5279 |
| ChpF (SCO2705) | 79/52 | DSGAQA | 5182 | 5182 |
| ChpG (SCO2699) | 90/63 | DAGAAG | 5994 | 5994 |
| ChpH (SCO1675) | 77/52 | DSGAQG | 5121 | 5122 |
ND, not detected.
Molecular masses were calculated based on the assumption that the peptides are cleaved between the threonine and glycine contained in the LAXTG motif (Mazmanian et al. 1999; Pallen et al. 2001).
Figure 2.
(A) The hydropathy patterns of ChpC (solid line) and ChpG (broken line) illustrate the hydrophobic nature of the large (ChpA–C) and small (ChpD–H) chaplins. All chaplins have a signal sequence for secretion (SS). In ChpA–C, the two regions that conform to the sequences of the small chaplins (dark boxes) are followed by a hydrophilic region and a putative cell wall anchor containing an LAXTG motif and a stretch of hydrophobic amino acids. (B) Alignment of the homologous sequences present in the eight chaplins. The consensus sequence shows residues that are present in >50% of the sequences. The conserved amino acids, present in all sequences, are indicated in uppercase.
Chaplins are differentially expressed
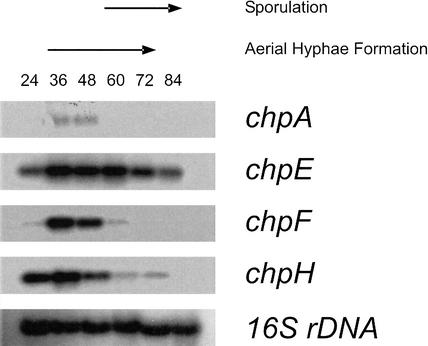
Total RNA from cultures at different stages of morphological differentiation was hybridized with the coding sequences of the chp genes (Fig. 3). Genes chpE and chpH were highly expressed in 24-hour-old solid cultures growing submerged only. Expression remained high during formation of aerial hyphae (between 36 and 72 h) but decreased when aerial growth ceased. In contrast, mRNA of chpD (data not shown), chpF (Fig. 3), and chpG (data not shown) did not accumulate during submerged growth. However, these mRNA accumulated to high levels during formation of aerial hyphae. Expression of chpA (Fig. 3), chpB, and chpC (data not shown) also coincided with formation of aerial hyphae, but mRNA levels were 10- to 25-fold lower compared with those of the small chaplins. Thus, expression of chpA, chpB, chpC, chpD, chpF, and chpG is strictly correlated with formation of aerial hyphae, whereas chpE and chpH are expressed in submerged hyphae as well.
Figure 3.
Temporal expression of the chp genes in S. coelicolor grown on solid NMMP medium as determined by Northern analysis. Genes chpE and chpH were expressed after 24 h of growth, when cultures still grew submerged, and during formation of aerial hyphae. In contrast, chpA and chpF were expressed during formation of aerial hyphae only. Expression of chpA was 10- to 25-fold lower compared with that of chpE, chpF, and chpH. Blots were rehybridized with 16S rDNA to confirm that lanes contained equal amounts of RNA.
To monitor expression of chp genes in colonies, we cloned promoter regions of chpA, chpC, chpG and chpH in vector pIJ8630 in front of the coding sequence of the eGFP gene (Sun et al. 1999). The resulting constructs were introduced in S. coelicolor. Cross-sections of colonies of transformants and the wild-type strain grown on agar plates were analyzed by confocal laser scanning microscopy (Fig. 4). Strong fluorescence was observed in aerial hyphae of strains with eGFP expression regulated by the chpG and chpH promoter, whereas fluorescence was weak in the case of chpA and chpC. Fluorescence in wild-type aerial hyphae was not observed. Fluorescence of eGFP in submerged hyphae of transformants and the wild-type strain could not be detected throughout growth. Similar results were obtained in liquid standing cultures (data not shown).
Figure 4.
Spatial expression of chpC (A) and chpH (B) in S. coelicolor grown on solid medium for 2 d and visualized using eGFP as a reporter. GFP expression was observed in aerial hyphae (AH) of transformants but not in submerged hyphae (SH). Fluorescence due to the chpC promoter was relatively low compared with that of chpH. (C) Wild-type hyphae served as a control. Bar, 10 μm.
Chaplins are involved in formation of aerial hyphae
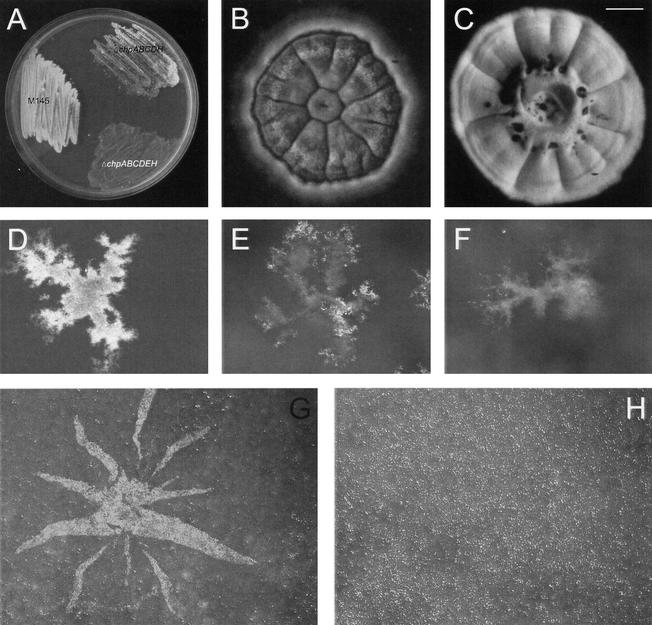
Up to six of the chp genes were deleted in S. coelicolor strain M145 using the Redirect technology (Gust et al. 2002, 2003). Because chpA and chpD as well as chpC and chpH are adjacent to each other on the bacterial chromosome, these genes were deleted pairwise. In contrast to the ΔchpAD and the ΔchpB strains, formation of aerial hyphae was strongly delayed in mutants lacking chpC and chpH (ΔchpCH, ΔchpACDH, and ΔchpABCDH strains) on all solid media tested (Fig. 5A). Besides the delay in onset of development, the number of aerial hyphae formed was also strongly reduced (Fig. 5B,C). However, the aerial hyphae formed were not affected in sporulation. Viability of spores after lysozyme treatment, desiccation, or heat exposure was unaffected (data not shown).
Figure 5.
Chaplins are involved in formation of aerial hyphae. (A) After 3 d of growth on solid minimal medium, the wild-type strain formed a confluent layer of white aerial hyphae, whereas the ΔchpABCDH and the ΔchpABCDEH were strongly affected in aerial growth. (B) The ΔchpABCDH strain formed only few aerial hyphae on solid complete R5 medium even after 8 d of growth, the wild-type strain serving as a control (C). Colonies of the wild-type strain at the water–air interface of liquid standing minimal medium showed a confluent layer of aerial hyphae after 4 d of growth (D), in contrast to the ΔchpABCDH strain (E) and the ΔchpABCDEH strain (F). Formation of aerial hyphae, as indicated by a white fluffy layer, could be restored by applying a mixture of chaplins extracted from a 3-day-old culture of the ΔrdlAB strain at the surface of a colony of the ΔchpABCDEH strain grown on solid medium (G), an extract of the ΔchpABCDEH strain serving as a control (H). Bars: B,C,G,H, 1 mm; D–F, 400 μm.
When chpE was deleted in the ΔchpABCDH strain, aerial hyphae formation was further delayed and even more affected (Fig. 5A). Colonies were bald on all minimal media tested, and only few aerial hyphae were observed on rich media, provided that colonies were allowed to grow for more than 1 wk (data not shown). The few aerial hyphae formed did differentiate into spores and possessed the characteristic rodlet layer (data not shown).
The chp deletion strains were also grown in liquid standing medium. Results were similar to those observed on solid medium. Wild-type colonies at the water–air interface formed a confluent layer of aerial hyphae after 4 d of growth (Fig. 5D). However, formation of aerial hyphae was delayed and reduced in the ΔchpABCDH strain (Fig. 5E), and the ΔchpABCDEH strain remained bald even after prolonged incubation (Fig. 5F). The rigid membrane-fixing colonies at the surface of liquid standing cultures (van Keulen et al. 2003) was observed in all cases.
Extracellular complementation of the chp mutants by purified chaplins
To see whether the chp mutant strains could be complemented extracellularly with the small chaplins, we applied aqueous solutions of TFA extracts of SDS-treated cell walls of 3-day-old cultures of the ΔrdlAB strain forming aerial hyphae at the colony surface after 24 h. This restored aerial hyphae formation in the ΔchpABCDH (data not shown) and the ΔchpABCDEH strains (Fig. 5G) at the spot where the chaplins were applied. Formation of aerial hyphae started only 4 h after application, although the number of aerial hyphae did not reach wild-type levels. Complementation was observed with 0.1–20 μg of chaplin. Mutant colonies were neither complemented with an extract isolated from 3-day-old cultures of the ΔchpABCDEH strain (Fig. 5H) nor when the wild-type strain was grown in close proximity (data not shown).
Development of the wild-type strain was accelerated at spots where chaplins were applied. Also, in this case, aerial hyphae were formed 4 h after applying the protein mixture to a 1-day-old culture. In contrast, none of a variety of bald mutants (bldA, bldD, bldH, and bldJ) tested could be complemented.
Chaplins are highly surface active and form functional amyloid-like fibrils
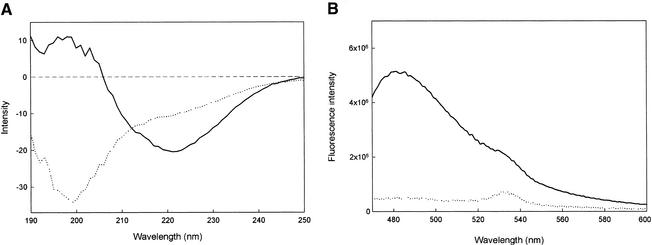
The secondary structure of the mixture of chaplins as obtained from cell walls of 3-day-old cultures of S. coelicolor strain M145ΔrdlAB was studied using circular dichroism. The spectrum of an aqueous solution of the mixture of small chaplins was indicative for random coiled proteins (Fig. 6A). When the aqueous solution was vortexed, thus creating a large surface of air–water interface, the protein mixture adopted a conformation rich in β-sheet. This change in conformation coincided with the formation of a white precipitate. A similar change into a β-sheet-rich state was observed when the aqueous solution of chaplins was allowed to dry in the cuvette (data not shown). In contrast, proteins in the extract of 3-day-old cultures of the ΔchpABCDEH strain did not change their conformation on vortexing (data not shown).
Figure 6.
Conformational changes of a mixture of chaplins (ChpD–H) isolated from cell walls of the M145ΔrdlAB strain. (A) The circular dichroism spectrum of an aqueous solution of the mixture of small chaplins was indicative for random coiled proteins (broken line), and the protein mixture adopted a conformation rich in β-sheet on vortexing (solid line). (B) Chaplins in the β-sheet rich state interact with the fluorescent dye thioflavin T (solid line), increasing its fluorescence 10-fold compared with water that served as a control (broken line).
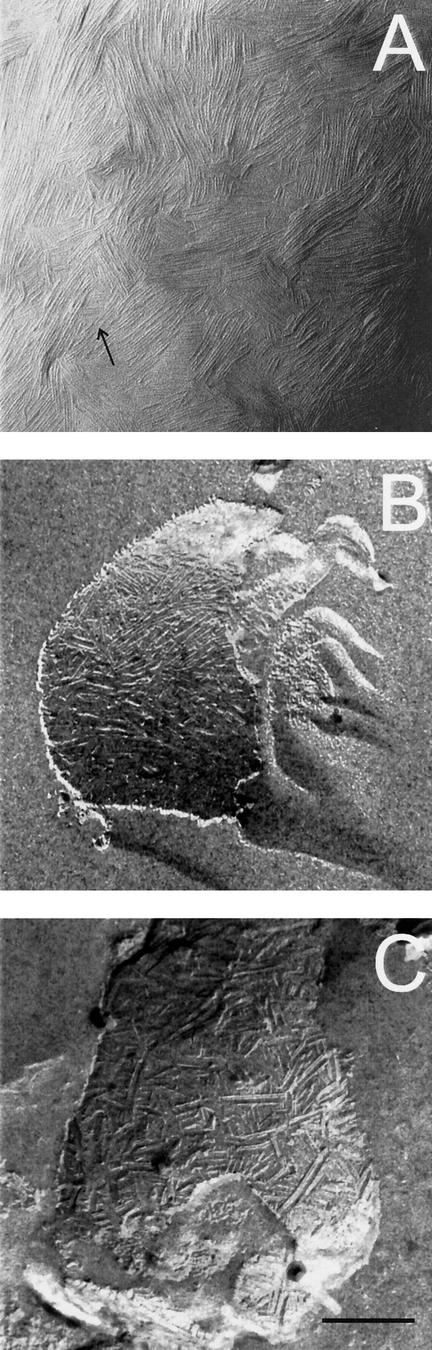
In order to investigate whether the change to the β-sheet-rich state was accompanied with a change in ultrastructure, we dried the protein mixture down on Formvar-coated grids followed by shadowing. A mosaic of 4–6-nm-wide parallel fibrils was observed (Fig. 7A). A similar mosaic was observed when an aqueous solution of a TFA extract of SDS-treated cell walls of the wild-type strain was used (data not shown). Thus, rodlins do not affect formation of chaplin fibrils. The fibrils formed by the chaplins were different from the rodlets observed at wild-type aerial hyphae and spores (Fig. 7C) but resembled those observed at aerial structures of the M145ΔrdlAB strain (Fig. 7B). From this, we conclude that chaplins assemble into fibrils at surfaces of aerial structures.
Figure 7.
(A) Four- to six-nanometer-wide fibrils are formed after drying down an aqueous solution of chaplins. The fibrils resemble those present at the outer surface of the M145ΔrdlAB strain (B) and are different from the rodlets present on aerial hyphae and spores of the wild-type strain (C). Arrow indicates direction of shadowing. Bar, 200 μm.
Formation of β-sheet rich fibrils at the water–air interface suggested that amyloid-like structures are formed (Wösten and de Vocht 2000). Therefore, the interaction of chaplins with the amyloid-specific fluorescent dye thioflavin T (ThT) was studied. Chaplins in the β-sheet state increased fluorescence of ThT significantly (Fig. 6B). At 482 nm, a 10-fold increase of ThT fluorescence was observed. In contrast, no increase was observed with extracts of the ΔchpABCDEH strain (data not shown). This shows that chaplins form amyloid-like structures.
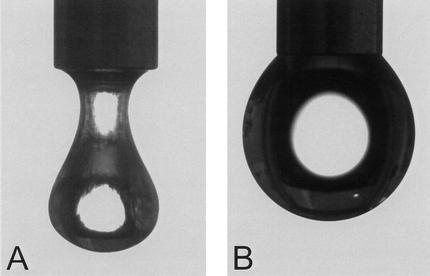
Surface activity of the mixture of small chaplins (200–300 μg/mL) was studied using the pendant droplet technique. At the concentration used, the water surface tension decreased within 15 min from 72 (±1) to 26 (±2) mJ/m2. This was accompanied by the appearance of a light-reflecting film at the water surface and a dramatic change of the shape of the water droplet (Fig. 8). From this we conclude that the conformational change of chaplins at the water–air interface coincides with a tremendous drop in surface tension.
Figure 8.
Surface activity of a mixture of chaplins (A), as indicated by the shape of an 8-μL droplet, water serving as a control (B).
Discussion
Formation of aerial (reproductive) structures in both filamentous bacteria and filamentous fungi was described as a two-step process (Wösten et al. 1999; Wösten and Willey 2000). This model is an oversimplification, given the genetic complexity of the differentiation process (Chater 2001), but provides a first basis to understand the mechanisms involved in aerial growth. In the first step, hyphae breach the water–air interface to escape the aqueous environment to grow into the air. In the second step, aerial hyphae are covered with one or more surface layers, rendering hydrophobicity and possibly providing rigidity and protection. In contrast to filamentous fungi (Wösten 2001), little is known about the molecules that allow aerial growth in streptomycetes. We here report for the first time a class of proteins, called chaplins, that are essential for aerial growth in S. coelicolor on all media tested. These proteins are involved in escape of hyphae from the aqueous environment into the air but also provide aerial hyphae with a hydrophobic coating. Chaplins function as amyloid-like proteins, and are the first example of functional amyloid-like fibrils in Gram-positive bacteria, and only the second example in the bacterial domain (Chapman et al. 2002).
To identify proteins that render aerial hyphae hydrophobic, we isolated proteins from cell walls of differentiating cultures of a S. coelicolor wild-type strain and a ΔrdlAB strain. We adopted a procedure previously used to isolate such proteins from filamentous fungi (de Vries et al. 1993; Wösten et al. 1993). In this way, five proteins were identified. They had masses of 5071–5994 Da and hardly stained with CBB or silver in SDS-PAGE, which explains why they were not reported by Claessen et al. (2002). Yet, considerable amounts of these proteins are produced. They represent about 1% of the mass of the cell wall. Masses and N-terminal sequences of the peptides corresponded to mature forms of deduced hydrophobic secreted proteins encoded by five genes in the S. coelicolor genome (Bentley et al. 2002). These genes were called chpD–H. Three additional homologs, chpA–C, encoding secreted proteins of about 16–20 kD, were identified in the genome. Their mature forms contain two domains highly similar to those of the extractable chaplins. These domains are followed by a hydrophilic region of sufficient length to traverse the cell wall. The C-terminal part of the large nonextractable chaplins ChpA–C contain an LAXTG sequence followed by a membrane-spanning domain and a charged C-terminal tail. Proteins with this motif, such as protein A of Staphylococcus aureus (Ton-That et al. 1997; Mazmanian et al. 1999), are often cell-surface proteins anchored to the peptidoglycan by sortase-like proteins (Mazmanian et al. 1999; Pallen et al. 2001). A covalent linkage of ChpA–C to the cell wall would explain why they have not been physically isolated using the TFA extraction.
The chp genes were deleted one by one or pairwise. The ΔchpCH, the ΔchpACDH, and the ΔchpABCDH strain showed a delay of several days in onset of development on all media tested. Furthermore, the number of aerial hyphae that eventually formed was dramatically reduced. Formation of aerial hyphae on minimal media was almost completely abolished when chpE was also deleted (ΔchpABCDEH strain). Similar results were obtained in liquid standing cultures. Although the ΔchpABCDEH strain formed colonies at the liquid–air interface, it did not form aerial hyphae. All deletion strains formed the rigid film fixing colonies at the water–air interface (van Keulen et al. 2003). This shows that this film of unknown nature is not sufficient to allow aerial growth.
Deletion studies indicated the importance of chpE and chpH. These genes were highly expressed after 24 h of growth on solid media, which is in agreement with array data presented by Elliot et al. (2003). At this time, only a submerged mycelium was formed. These chaplins, but not the other members of this class of proteins, were identified in the medium of liquid shaken cultures (D. Claessen, unpubl.).
Mixtures of ChpD–H isolated from cell walls of aerial hyphae were shown to be highly surface active. Within a few minutes the surface tension was lowered from 72 to 26 mJ/m2. This was accompanied by a conformational change in a state rich in β-sheet, resulting in the formation of a light-reflecting film at the water surface. The ultrastructure of the film was composed of a mosaic of parallel 4–6-nm-wide fibrils. These fibrils increased fluorescence of the amyloid-specific dye ThT, showing that chaplins lower the water surface tension by forming amyloid-like fibrils. From these data, we conclude that ChpE and ChpH lower the water surface tension to enable hyphae to breach the medium–air interface to grow into the air. Indeed, development was fully restored when a chaplin mixture was dried down at surfaces of 1-day-old colonies of the ΔchpABCDH and the ΔchpABCDEH strains. The formation of a nondiffusible film of high molecular weight at the water–air interface explains why aerial hyphae only developed at the spot of application and why wild-type strains grown in close vicinity could not complement the mutants. After prolonged incubation, some aerial hyphae developed in the multiple knockout strains. This may be due to other surface-active molecules that are secreted in the culture medium such as SapB (Willey et al. 1991, 1993). Interestingly, chaplins applied at the surface of the wild-type strain accelerated development for at least 8–16 h. In contrast, bld mutants could not be complemented for reasons unknown.
Once escaped into the air, hyphae of streptomycetes are covered with several surface layers. The most outer layer is the rodlet layer. The rodlins RdlA and RdlB are involved in the formation of this structure (Claessen et al. 2002). In the ΔrdlAB strain, the rodlet layer is absent and thus the underlying layer becomes exposed. It is composed of a mosaic of parallel 4–6-nm-wide fibrils. These fibrils resemble those formed by the chaplins at the water–air interface, indicating that chaplins form a surface layer just beneath the rodlet layer. The fact that all chaplins are moderately (chpA–C) to highly (chpD–H) expressed in aerial hyphae, as was shown by GFP reporter studies and confirmed by Northerns, supports this hypothesis. The chaplin layer underneath the rodlet layer confers surface hydrophobicity in the absence of the latter layer and may interact with it.
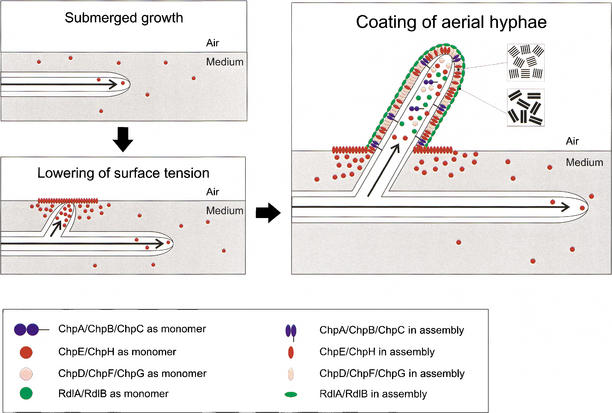
From these and other data, we present a model for the formation of aerial hyphae in S. coelicolor (Fig. 9). This model probably also holds for other streptomycetes because chpD hybridized at stringent conditions with genomic DNA of various members of this group of bacteria. During formation of a feeding submerged mycelium, chpE and chpH are expressed. The proteins diffuse to the water–air interface, where they form a highly insoluble film that lowers the water surface tension. This enables hyphae to escape into the air. In the aerial hyphae, all chp genes are expressed. The proteins diffuse to the outer surface and assemble on contacting air. The resulting layer, composed of a mosaic of parallel 4–6-nm-wide fibrils, confers hydrophobicity to the aerial hypha. It is tempting to speculate that the large chaplins ChpA–C coassemble with the small chaplins ChpD–H with their homologous sequences. The hydrophilic region of the large chaplins may traverse the cell wall, whereas the LAXTG motif is cross-linked to the peptidoglycan. As a result, the small chaplins would be immobilized at the cell surface by the larger chaplins. The chaplin layer provides a hydrophobic surface and this may induce the formation of the rodlet layer formed by RdlA and RdlB.
Figure 9.
The role of chaplins and rodlins in the formation of aerial hyphae in S. coelicolor. Initially, hyphae grow submerged, secreting ChpE and ChpH. These proteins lower the water surface tension dramatically by assembling at the water–air interface. This enables hyphae to grow into the air. Emerging aerial hyphae produce ChpA–H that assemble at the cell wall–air interface into an insoluble protein film characterized by an ultrastructure of 4–6-nm-wide fibrils. The chaplin layer provides a hydrophobic surface and this may induce the formation of the typical rodlet layer formed by RdlA and RdlB.
Chaplins resemble the hydrophobins of filamentous fungi. These proteins, too, enable hyphae to breach the water–air interface by lowering the water surface tension (Wösten et al. 1999) on assembly in a highly insoluble film composed of a mosaic of fibrils (Wösten et al. 1993). Moreover, the assembled hydrophobin is responsible for the hydrophobicity of fungal aerial structures (Wösten et al. 1993, 1994; Wösten 2001). Similar to the chaplins, hydrophobins assemble in an amyloid-like structure (Wösten and de Vocht 2000; Butko et al. 2001; Mackay et al. 2001). Until recently, amyloids were mainly associated with debilitating human diseases like Huntington's disease and Alzheimer's (Chiti et al. 1999; Dobson 1999). Chapman and colleagues (2002) reported that the curli proteins of Escherichia coli are functional amyloids involved in colonization of inert surfaces, biofilm formation, and attachment to a variety of host proteins. Thus, like in fungi, amyloids also serve a wide spectrum of functions in bacterial development.
Materials and methods
Strains and plasmids
Cloning was done in E. coli DH5α, SCS110 or BW25113 (Datsenko and Wanner 2000). E. coli ET12567 containing pUZ8002 was used for conjugation to S. coelicolor. S. coelicolor strains M145 (Kieser et al. 2000), ΔrdlAB6 (Claessen et al. 2002), bldJ261, bldD53 (Willey et al. 1993), bldH109 (Champness 1988), and bldA39 (Piret and Chater 1985) were used in this study as well as Streptomyces lividans TK23 (Kieser et al. 2000), Streptomyces tendae Tü901/8c (Richter et al. 1998), and Streptomyces griseus (DSMZ 40236). Vectors and constructs are summarized in Table 2.
Table 2.
Vectors and constructs used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pGEM-T | E. coli vector for cloning PCR fragments amplified with TAQ DNA polymerase. | Promega |
| pGEM-chpA-H | pGEM-T containing a 244-bp internal fragment of chpA, a 377-bp internal fragment of chpB, a 232-bp internal fragment of chpC, the coding sequence of chpD, a 199-bp internal fragment of chpE, a 178-bp internal fragment of chpF, the coding sequence of chpG, and a 119-bp internal fragment of chpH, respectively. | This work |
| pIJ8630 | E. coli-Streptomyces shuttle vector with pUC18 ori and oriT containing an eGFP gene adapted for codon usage in streptomycetes | Sun et al. 1999 |
| pIJ8630-chpA, C, G, H | pIJ8630 containing the 175-bp promoter region of chpA, the 179-bp promoter region of chpC, the 300-bp promoter region of chpG, and the 132-bp promoter region of chpH, respectively, with NdeI sites at the 3′ end allowing translational fusions. | This work |
Growth conditions and media
Streptomyces strains were grown at 30°C on solid MS or R5 medium (Kieser et al. 2000) or on solid or liquid modified NMMP medium (van Keulen et al. 2003). The solid medium R5 was used for regenerating protoplasts. Yeast extract–malt extract (YEME) medium was used for liquid shaken cultures (Kieser et al. 2000).
Molecular techniques
Standard molecular techniques followed Sambrook et al. (1989). Protoplast preparation and transformation were performed as described by Kieser et al. (2000). Chromosomal DNA of S. coelicolor strains was isolated according to Verhasselt et al. (1989) and modified by Nagy et al. (1995). Total RNA of S. coelicolor was isolated using the SV Total RNA Isolation System (Promega) according to Veenendaal and Wösten (1998) and 5 μg was loaded on gel. DNA and RNA were blotted on Nylon filters (Boehringer) and hybridized at 63°C under conditions described by Church and Gilbert (1984). Under these conditions, cross-hybridization between the different chp genes was weak, if present at all. Probes were radioactively labeled using the Prime-a-Gene Labelling kit (Promega).
Construction of the S. coelicolor M145 chp deletion strains
Gene deletions were made in S. coelicolor M145 using the Redirect technology as described (Gust et al. 2002, 2003). Gene replacements of chpAD, chpB, chpCH, or chpE were made with the apramycin resistance cassette. For the deletion of chpCH in the ΔchpAD strain, the aadA spectinomycin–streptomycin resistance cassette was used, resulting in ΔchpACDH. For the deletion of chpB in the ΔchpACDH strain, the viomycin resistance cassette was used (ΔchpABCDH). The FLP recombinase was used to remove the apramycin cassette in the ΔchpABCDH strain (Gust et al. 2002, 2003), enabling us to reuse this cassette to delete chpE. This resulted in the ΔchpABCDEH strain. Primers used for the Redirect technology are listed in Table 3. Gene deletions were confirmed by Southern analysis.
Table 3.
Primers used for the chp gene deletions using the Redirect technology
| Primer | Sequence (5′-3′) |
|---|---|
| ΔchpAD forward | CGTCATTCAGGCATGACCGCAGAGAAGGGAAAGTTCATGATTCCGGGGATCCGTCGACC |
| ΔchpAD reverse | GGACCGGCCGGGGGCGTCGGGGTGGCGGGGGTGGGGTCATGTAGGCTGGAGCTGCTTC |
| ΔchpB forward | ACTGCCCGCGACCATGTCTGGAAGGCAGGGAGCCTCATGATTCCGGGGATCCGTCGACC |
| ΔchpB reverse | CCGCCCTGACGGCGCGGGTGCCCCCGGTCGACGCGGTCATGTAGGCTGGAGCTGCTTC |
| ΔchpCH forward | CAATTTTGGGATCGCGTTTGCCAACTAGGAGGAACAATGATTCCGGGGATCCGTCGACC |
| ΔchpCH reverse | GGTGCGGAGCCCGCTGCGTGTACCGGTGCGCCCGCCTCATGTAGGCTGGAGCTGCTTC |
| ΔchpE forward | GGCGACTGCCGCGGCGATCGCAAGGAGGGGTTGTAAGTGATTCCGGGGATCCGTCGACC |
| ΔchpE reverse | GCCCGCTGCTGAGGCGGCATTGGGGGGCGGCCTGCGTCATGTAGGCTGGAGCTGCTTC |
Preparation of cell walls and protein extracts
Filaments of S. coelicolor were fragmented at 14,000 psi using an SLM French Pressure Cell Press. The homogenate was treated with 2% SDS for 10 min at 100°C, after which the cell walls were fractionated from the cytoplasmic content by centrifugation at 10,000g for 10 min. The cell wall fraction was extracted with hot 2% SDS once more, extensively washed with water, and freeze dried. SDS-treated cell walls were then extracted with TFA (de Vries et al. 1993; Wösten et al. 1993; Claessen et al. 2002). After evaporating the solvent by a stream of air, extracts were taken up in water for extracellular complementation studies, 0.1% TFA for MALDI-TOF analysis, or SDS sample buffer (2% SDS, 20% glycerol, 0.02% bromophenol blue, 0.1 M Tris-HCl at pH 6.8, 5% β-mercaptoethanol) for SDS-PAGE. If necessary, adjustments of pH were done by addition of 25% ammonia.
Gel electrophoresis and Western blotting
SDS-PAGE was done in 16% gels according to Laemmli (1970). Prestained broad-range molecular weight markers of Bio-Rad were used. After separation, proteins were stained with 0.25% Coomassie Brilliant Blue G-250 (CBB) or silver, or blotted onto a polyvinylidenedifluoride (PVDF) membrane using semidry blotting. For N-terminal sequencing, a PVDF membrane was stained with CBB and slices of the membrane containing the protein were excised. After destaining with 30% methanol, N-terminal sequences were determined on an Applied Biosystems 476A automated sequencer (NAPS Unit, University of British Columbia).
Mass analysis
Matrix assisted laser desorption-ionization mass spectrometry (MALDI-TOF) was used to determine masses of proteins in TFA extracts of SDS-treated cell walls of cultures forming aerial hyphae. Proteins were taken up in 0.1% TFA and mixed with an equal volume of matrix solution [20 mg/mL sinapinic acid in 40/60 (v/v) acetonitril/0.1% TFA]. Two microliters of the mixture was dried down on a target. Spectra were recorded on a TofSpec E&SE Micromass spectrometer.
Electron microscopic techniques
Two-microliter aliquots of aqueous solutions of TFA extracts of SDS-treated cell walls (50 μg protein/mL) were dried down on Formvar-coated nickel grids. Surface shadowing with Pt/C was done at an angle of 45°. For Cryo Scanning Electron Microscopy, cultures grown on solid R5 or MS medium were frozen in a mixture of liquid and solid nitrogen and sputter-coated with gold/palladium. Examination was done at 5.0 kV in a JEOL field emission scanning electron microscope type 6301F. For freeze-fracture observations, spores were frozen in a mixture of solid and liquid nitrogen. Freeze fracturing was done in a Polaron freeze-etch apparatus equipped with a Balzers EVM 052 unit. Replicas of Pt/C were cleaned for 16 h in 40% chromic acid at room temperature.
Circular dichroism and fluorescence spectroscopy
CD spectra of mixtures of ChpD–H were recorded over the wavelength region 190–250 nm on an Aviv 62A DS Circular Dichroism spectrometer, using a 5-mm quartz cuvette. The temperature was kept constant at 25°C and the sample compartment was flushed with N2 continuously. Spectra are the average of three scans using a bandwidth of 1 nm, a step width of 1 nm, and a 5-sec averaging per point. The spectra were corrected by using a reference solution without the protein. Typically, a TFA extract of 5 mg of SDS-treated cell wall material was used. After evaporating the solvent by a stream of air, the extract was taken up in 250 μL of water. The secondary structure at the air–water interface was determined by vortexing aqueous solutions for 1 min. To determine whether ChpD–H form amyloid-like fibrils, we incubated aqueous solutions of the protein mixture with 3 μM ThT. Emission spectra between 470 and 600 nm (excitation = 450 nm) were determined on a Jobin Yvon Horiba Fluorolog.
Surface activity
To measure surface activity, we used the pendant droplet technique. Typically, drops of 5–10 μL were measured with the Krüss Drop Shape Analysis System DSA10 Mk2 according to the instructions of the manufacturer.
Acknowledgments
We thank Bertolt Gust for advice and Plant Bioscience Limited (PBL) for providing materials for the Redirect PCR disruption method. Furthermore, we are indebted to Ietse Stokroos and Jan Zagers for performing freeze-fracturing, scanning electron microscopy, and confocal laser scanning microscopy. D.C. is supported by a grant of the Dutch Programme EET (Economy, Ecology and Technology), a joint initiative of the Ministries of Economic Affairs, Education, Culture, and Sciences, and of Housing, Spatial Planning, and the Environment (EETK01031).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.264303.
References
- Bentley S.D., Chater, K.F., Cerdeño-Tárraga, A.-M., Challis, G.L., Thomson, N.R., James, K.D., Harris, D.E., Quail, M.A., Kieser, H., Harper, D., et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417: 141–147. [DOI] [PubMed] [Google Scholar]
- Butko P., Buford, J.P., Goodwin, J.S., Stroud, P.A., McCormick, C.L., and Cannon, G.C. 2001. Spectroscopic evidence for amyloid-like interfacial self-assembly of hydrophobin Sc3. Biochem. Biophys. Res. Commun. 280: 212–215. [DOI] [PubMed] [Google Scholar]
- Champness W.C. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170: 1168–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M.R., Robinson, L.S., Pinkner, J.S., Roth, R., Heuser, J., Hammar, M., Normark, S., and Hultgren, S.J. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295: 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K.F. 1998. Taking a genetic scalpel to the Streptomyces colony. Microbiology 144: 1465–1478. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): A checkpoint multiplex? Curr. Opin. Microbiol. 4: 667–673. [DOI] [PubMed] [Google Scholar]
- Chiti F., Webster, P., Taddei, N., Clark, A., Stefani, M., Ramponi, G., and Dobson, C.M. 1999. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl. Acad. Sci. 96: 3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G.M. and Gilbert, W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. 81: 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D., Wösten, H.A.B., van Keulen, G., Faber, O.G., Alves, A.M.C.R., Meijer, W.G., and Dijkhuizen, L. 2002. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol. Microbiol. 44: 1483–1492. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A. and Wanner, B.L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries O.M.H., Fekkes, M.P., Wösten, H.A.B., and Wessels, J.G.H. 1993. Insoluble hydrophobin complexes in the walls of Schizophyllum commune and other filamentous fungi. Arch. Microbiol. 159: 330–335. [Google Scholar]
- Dobson C.M. 1999. Protein misfolding, evolution and disease. Trends Biochem. Sci. 24: 329–332. [DOI] [PubMed] [Google Scholar]
- Elliot M.A., Karoonuthaisiri, N., Huang, J., Bibb, M.J., Cohen, S.N., Kao, C.M., and Buttner, M.J. 2003. The chaplins: A family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Gust B., Kieser, T., and Chater, K.F. 2002. REDIRECT technology: PCR-targeting system in Streptomyces coelicolor. The John Innes Centre, Norwich, United Kingdom.
- Gust B., Challis, G.L., Fowler, K., Kieser, T., and Chater, K.F. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. 100: 1541–1546. (Published online ahead of print on January 31, 2003, as 10.1073/pnas.0337542100.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen G.H. and Buttner, M.J. 1998. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1: 656–662. [DOI] [PubMed] [Google Scholar]
- Kieser T., Bibb, M.J., Buttner, M.J., Chater, K.F., and Hopwood, D.A. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- Laemmli U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Mackay J.P., Matthews, J.M., Winefield, R.D., Mackay, L.G., Haverkamp, R.G., and Templeton, M.D. 2001. Structure 9: 83–91. [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Liu, G., Ton-That, H., and Schneewind, O. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285: 760–763. [DOI] [PubMed] [Google Scholar]
- Nagy I., Schoofs, G., Compernolle, F., Proost, P., Vanderleyden, J., and de Mot, R. 1995. Degradation of the thiocarbonate herbicide EPTC (S-ethyl dipropylcarbamotioate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J. Bacteriol. 177: 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen M.J., Lam, A.C., Antonio, M., and Dunbar, K. 2001. An embarrassment of sortases—A richness of substrates? Trends Microbiol. 9: 97–102. [DOI] [PubMed] [Google Scholar]
- Piret J.M. and Chater, K.F. 1985. Phage-mediated cloning of BldA, a region involved in Streptomyces coelicolor morphological development, and its analysis by genetic complementation. J. Bacteriol. 163: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Willey, J.M., Süßmuth, R., Jung, G., and Fiedler, H.-P. 1998. Streptofactin, a novel biosurfactant with aerial mycelium inducing activity from Streptomyces tendae Tü 901/8c. FEMS Microbiol. Lett. 163: 165–171. [Google Scholar]
- Sambrook J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Smucker R.A. and Pfister, R.M. 1978. Characteristics of Streptomyces coelicolor A3(2) aerial spore rodlet mosaic. Can. J. Microbiol. 24: 397–408. [DOI] [PubMed] [Google Scholar]
- Sun J., Kelemen, G.H., Fernández-Abalos, J.M., and Bibb, M.J. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145: 2221–2227. [DOI] [PubMed] [Google Scholar]
- Tillotson R.D., Wösten, H.A.B., Richter, M., and Willey, J.M. 1998. A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophyllum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol. Microbiol. 30: 595–602. [DOI] [PubMed] [Google Scholar]
- Ton-That H., Faull, K.F., and Schneewind, O. 1997. Anchor structure of staphylococcal surface proteins. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J. Biol. Chem. 272: 22285–22292. [DOI] [PubMed] [Google Scholar]
- van Keulen G., Jonkers, H.M., Claessen, D., Dijkhuizen, L., and Wösten, H.A.B. 2003. Differentiation and anaerobiosis in standing liquid cultures of Streptomyces coelicolor. J. Bacteriol. 185: 1455–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenendaal A.K.J. and Wösten, H.A.B. 1998. Total RNA from the Gram-positive bacterium Streptomyces lividans. Promega Benelux News 17: 7. [Google Scholar]
- Verhasselt P., Poncelet, F., Vits, K., and Vanderleyden, J. 1989. Cloning and expression of a Clostridium acetobutylicum alpha-amylase gene in Escherichia coli. FEMS Microbiol. Lett. 59: 135–140. [DOI] [PubMed] [Google Scholar]
- Wildermuth H., Wehrli, E., and Horne, R.W. 1971. The surface structure of spores and aerial mycelium in Streptomyces coelicolor. J. Ultrastruct. Res. 35: 168–180. [DOI] [PubMed] [Google Scholar]
- Willey J.M., Santamaría, R.I., Guijarro, J., Geistlich, M., and Losick, R. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65: 641–650. [DOI] [PubMed] [Google Scholar]
- Willey J.M., Schwedock, J., and Losick, R. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes & Dev. 7: 895–903. [DOI] [PubMed] [Google Scholar]
- Wösten H.A.B. 2001. Hydrophobins: Multipurpose proteins. Annu. Rev. Microbiol. 55: 625–646. [DOI] [PubMed] [Google Scholar]
- Wösten H.A.B. and de Vocht, M.L. 2000. Hydrophobins, the fungal coat unravelled. Biochim. Biophys. Acta 1469: 79–86. [DOI] [PubMed] [Google Scholar]
- Wösten H.A.B. and Willey, J.M. 2000. Surface-active proteins enable microbial aerial hyphae to grow into the air. Microbiology 146: 767–773. [DOI] [PubMed] [Google Scholar]
- Wösten H.A.B., de Vries, O.M.H., and Wessels, J.G.H. 1993. Interfacial self-assembly of a fungal hydrophobin into a hydrophobin rodlet layer. Plant Cell 5: 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten H.A.B., Ásgeirsdóttir, S.A., Krook, J.H., Drenth, J.H.H., and Wessels, J.G.H. 1994. The fungal hydrophobin Sc3p self-assembles at the surface of aerial hyphae as a protein membrane constituting the hydrophobic rodlet layer. Eur. J. Cell Biol. 63: 122–129. [PubMed] [Google Scholar]
- Wösten H.A.B., van Wetter, M.-A., Lugones, L.G., van der Mei, H.C., Busscher, H.J., and Wessels, J.G.H. 1999. How a fungus escapes the water to grow into the air. Curr. Biol. 9: 85–88. [DOI] [PubMed] [Google Scholar]